| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ACSL6-CDS2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ACSL6-CDS2 | FusionPDB ID: 1520 | FusionGDB2.0 ID: 1520 | Hgene | Tgene | Gene symbol | ACSL6 | CDS2 | Gene ID | 23305 | 8760 |
| Gene name | acyl-CoA synthetase long chain family member 6 | CDP-diacylglycerol synthase 2 | |
| Synonyms | ACS2|FACL6|LACS 6|LACS2|LACS5 | - | |
| Cytomap | 5q31.1 | 20p12.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | long-chain-fatty-acid--CoA ligase 6arachidonate--CoA ligasefatty-acid-Coenzyme A ligase, long-chain 6long fatty acyl-CoA synthetase 2 | phosphatidate cytidylyltransferase 2CDP-DAG synthase 2CDP-DG synthase 2CDP-DG synthetase 2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2CDP-diglyceride diphosphorylase 2CDP-diglyceride pyrophosphorylase 2CDP-diglyceride synthase | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q9UKU0 | O95674 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000296869, ENST00000357096, ENST00000379240, ENST00000379244, ENST00000379246, ENST00000379249, ENST00000379255, ENST00000379264, ENST00000379272, ENST00000431707, ENST00000543479, ENST00000544770, ENST00000477640, | ENST00000379062, ENST00000379070, ENST00000535100, ENST00000460006, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 5 X 4 X 3=60 | 10 X 11 X 7=770 |
| # samples | 4 | 13 | |
| ** MAII score | log2(4/60*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/770*10)=-2.56634682255381 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ACSL6 [Title/Abstract] AND CDS2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ACSL6(131307243)-CDS2(5154169), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ACSL6-CDS2 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. ACSL6-CDS2 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. ACSL6-CDS2 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ACSL6 | GO:0001676 | long-chain fatty acid metabolic process | 24269233 |
 Fusion gene breakpoints across ACSL6 (5'-gene) Fusion gene breakpoints across ACSL6 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
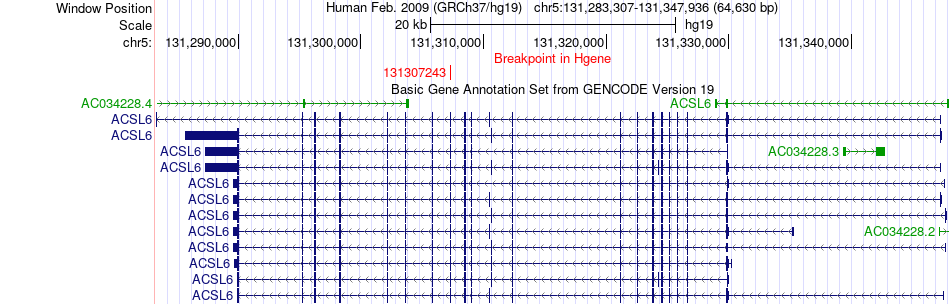 |
 Fusion gene breakpoints across CDS2 (3'-gene) Fusion gene breakpoints across CDS2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
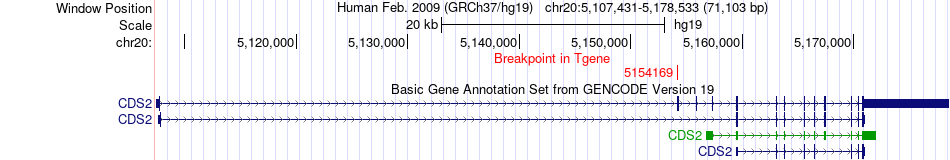 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-ZG-A9LZ-01A | ACSL6 | chr5 | 131307243 | - | CDS2 | chr20 | 5154169 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000379249 | ACSL6 | chr5 | 131307243 | - | ENST00000460006 | CDS2 | chr20 | 5154169 | + | 10437 | 1503 | 3 | 2783 | 926 |
| ENST00000379264 | ACSL6 | chr5 | 131307243 | - | ENST00000460006 | CDS2 | chr20 | 5154169 | + | 10477 | 1543 | 43 | 2823 | 926 |
| ENST00000379272 | ACSL6 | chr5 | 131307243 | - | ENST00000460006 | CDS2 | chr20 | 5154169 | + | 10482 | 1548 | 3 | 2828 | 941 |
| ENST00000296869 | ACSL6 | chr5 | 131307243 | - | ENST00000460006 | CDS2 | chr20 | 5154169 | + | 10428 | 1494 | 51 | 2774 | 907 |
| ENST00000379246 | ACSL6 | chr5 | 131307243 | - | ENST00000460006 | CDS2 | chr20 | 5154169 | + | 10492 | 1558 | 166 | 2838 | 890 |
| ENST00000357096 | ACSL6 | chr5 | 131307243 | - | ENST00000460006 | CDS2 | chr20 | 5154169 | + | 10068 | 1134 | 0 | 2414 | 804 |
| ENST00000431707 | ACSL6 | chr5 | 131307243 | - | ENST00000460006 | CDS2 | chr20 | 5154169 | + | 10259 | 1325 | 17 | 2605 | 862 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000379249 | ENST00000460006 | ACSL6 | chr5 | 131307243 | - | CDS2 | chr20 | 5154169 | + | 0.000194974 | 0.99980503 |
| ENST00000379264 | ENST00000460006 | ACSL6 | chr5 | 131307243 | - | CDS2 | chr20 | 5154169 | + | 0.000195785 | 0.9998042 |
| ENST00000379272 | ENST00000460006 | ACSL6 | chr5 | 131307243 | - | CDS2 | chr20 | 5154169 | + | 0.000191068 | 0.99980897 |
| ENST00000296869 | ENST00000460006 | ACSL6 | chr5 | 131307243 | - | CDS2 | chr20 | 5154169 | + | 0.000193955 | 0.9998061 |
| ENST00000379246 | ENST00000460006 | ACSL6 | chr5 | 131307243 | - | CDS2 | chr20 | 5154169 | + | 0.000478111 | 0.99952185 |
| ENST00000357096 | ENST00000460006 | ACSL6 | chr5 | 131307243 | - | CDS2 | chr20 | 5154169 | + | 0.000297173 | 0.99970275 |
| ENST00000431707 | ENST00000460006 | ACSL6 | chr5 | 131307243 | - | CDS2 | chr20 | 5154169 | + | 0.000219066 | 0.99978095 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >1520_1520_1_ACSL6-CDS2_ACSL6_chr5_131307243_ENST00000296869_CDS2_chr20_5154169_ENST00000460006_length(amino acids)=907AA_BP=481 MTAMLTFFLVSGGSLWLFVEFVLSLLEKMQTQEILRILRLPELGDLGQFFRSLSATTLVSMGALAAILAYWFTHRPKALQPPCNLLMQSE EVEDSGGARRSVIGSGPQLLTHYYDDARTMYQVFRRGLSISGNGPCLGFRKPKQPYQWLSYQEVADRAEFLGSGLLQHNCKACTDQFIGV FAQNRPEWIIVELACYTYSMVVVPLYDTLGPGAIRYIINTADISTVIVDKPQKAVLLLEHVERKETPGLKLIILMDPFEEALKERGQKCG VVIKSMQAVEDCGQENHQAPVPPQPDDLSIVCFTSGTTGNPKGAMLTHGNVVADFSGFLKVTESQWAPTCADVHISYLPLAHMFERMVQS VVYCHGGRVGFFQGDIRLLSDDMKALCPTIFPVVPRLLNRMYDKIFSQANTPLKRWLLEFAAKRKQAEVRSGIIRNDSIWDELFFNKIQA SLGGCVRMIVTGAAPASPTVLGFLRAALGCQESESEAKVDGETASDSESRAESAPLPVSADDTPEVLNRALSNLSSRWKNWWVRGILTLA MIAFFFIIIYLGPMVLMIIVMCVQIKCFHEIITIGYNVYHSYDLPWFRTLSWYFLLCVNYFFYGETVTDYFFTLVQREEPLRILSKYHRF ISFTLYLIGFCMFVLSLVKKHYRLQFYMFGWTHVTLLIVVTQSHLVIHNLFEGMIWFIVPISCVICNDIMAYMFGFFFGRTPLIKLSPKK TWEGFIGGFFATVVFGLLLSYVMSGYRCFVCPVEYNNDTNSFTVDCEPSDLFRLQEYNIPGVIQSVIGWKTVRMYPFQIHSIALSTFASL IGPFGGFFASGFKRAFKIKDFANTIPGHGGIMDRFDCQYLMATFVNVYIASFIRGPNPSKLIQQFLTLRPDQQLHIFNTLRSHLIDKGML -------------------------------------------------------------- >1520_1520_2_ACSL6-CDS2_ACSL6_chr5_131307243_ENST00000357096_CDS2_chr20_5154169_ENST00000460006_length(amino acids)=804AA_BP=378 MQTQEILRILRLPELGDLGQFFRSLSATTLDSGGARRSVIGSGPQLLTHYYDDARTMYQVFRRGLSISGNGPCLGFRKPKQPYQWLSYQE VADRAEFLGSGLLQHNCKACTDQFIGVFAQNRPEWIIVELACYTYSMVVVPLYDTLGPGAIRYIINTADISTVIVDKPQKAVLLLEHVER KETPGLKLIILMDPFEEALKERGQKCGVVIKSMQAVEDCGQENHQAPVPPQPDDLSIVCFTSGTTGNPKGAMLTHGNVVADFSGFLKVTE GDIRLLSDDMKALCPTIFPVVPRLLNRMYDKIFSQANTPLKRWLLEFAAKRKQAEVRSGIIRNDSIWDELFFNKIQASLGGCVRMIVTGA APASPTVLGFLRAALGCQESESEAKVDGETASDSESRAESAPLPVSADDTPEVLNRALSNLSSRWKNWWVRGILTLAMIAFFFIIIYLGP MVLMIIVMCVQIKCFHEIITIGYNVYHSYDLPWFRTLSWYFLLCVNYFFYGETVTDYFFTLVQREEPLRILSKYHRFISFTLYLIGFCMF VLSLVKKHYRLQFYMFGWTHVTLLIVVTQSHLVIHNLFEGMIWFIVPISCVICNDIMAYMFGFFFGRTPLIKLSPKKTWEGFIGGFFATV VFGLLLSYVMSGYRCFVCPVEYNNDTNSFTVDCEPSDLFRLQEYNIPGVIQSVIGWKTVRMYPFQIHSIALSTFASLIGPFGGFFASGFK -------------------------------------------------------------- >1520_1520_3_ACSL6-CDS2_ACSL6_chr5_131307243_ENST00000379246_CDS2_chr20_5154169_ENST00000460006_length(amino acids)=890AA_BP=464 MPEFVLSLLEKMQTQEILRILRLPELGDLGQFFRSLSATTLVSMGALAAILAYWFTHRPKALQPPCNLLMQSEEVEDSGGARRSVIGSGP QLLTHYYDDARTMYQVFRRGLSISGNGPCLGFRKPKQPYQWLSYQEVADRAEFLGSGLLQHNCKACTDQFIGVFAQNRPEWIIVELACYT YSMVVVPLYDTLGPGAIRYIINTADISTVIVDKPQKAVLLLEHVERKETPGLKLIILMDPFEEALKERGQKCGVVIKSMQAVEDCGQENH QAPVPPQPDDLSIVCFTSGTTGNPKGAMLTHGNVVADFSGFLKVTEKVIFPRQDDVLISFLPLAHMFERVIQSVVYCHGGRVGFFQGDIR LLSDDMKALCPTIFPVVPRLLNRMYDKIFSQANTPLKRWLLEFAAKRKQAEVRSGIIRNDSIWDELFFNKIQASLGGCVRMIVTGAAPAS PTVLGFLRAALGCQESESEAKVDGETASDSESRAESAPLPVSADDTPEVLNRALSNLSSRWKNWWVRGILTLAMIAFFFIIIYLGPMVLM IIVMCVQIKCFHEIITIGYNVYHSYDLPWFRTLSWYFLLCVNYFFYGETVTDYFFTLVQREEPLRILSKYHRFISFTLYLIGFCMFVLSL VKKHYRLQFYMFGWTHVTLLIVVTQSHLVIHNLFEGMIWFIVPISCVICNDIMAYMFGFFFGRTPLIKLSPKKTWEGFIGGFFATVVFGL LLSYVMSGYRCFVCPVEYNNDTNSFTVDCEPSDLFRLQEYNIPGVIQSVIGWKTVRMYPFQIHSIALSTFASLIGPFGGFFASGFKRAFK -------------------------------------------------------------- >1520_1520_4_ACSL6-CDS2_ACSL6_chr5_131307243_ENST00000379249_CDS2_chr20_5154169_ENST00000460006_length(amino acids)=926AA_BP=500 MRGCGAAGRGGRAGPGRAPLTAMLTFFLVSGGSLWLFVEFVLSLLEKMQTQEILRILRLPELGDLGQFFRSLSATTLVSMGALAAILAYW FTHRPKALQPPCNLLMQSEEVEDSGGARRSVIGSGPQLLTHYYDDARTMYQVFRRGLSISGNGPCLGFRKPKQPYQWLSYQEVADRAEFL GSGLLQHNCKACTDQFIGVFAQNRPEWIIVELACYTYSMVVVPLYDTLGPGAIRYIINTADISTVIVDKPQKAVLLLEHVERKETPGLKL IILMDPFEEALKERGQKCGVVIKSMQAVEDCGQENHQAPVPPQPDDLSIVCFTSGTTGNPKGAMLTHGNVVADFSGFLKVTESQWAPTCA DVHISYLPLAHMFERMVQSVVYCHGGRVGFFQGDIRLLSDDMKALCPTIFPVVPRLLNRMYDKIFSQANTPLKRWLLEFAAKRKQAEVRS GIIRNDSIWDELFFNKIQASLGGCVRMIVTGAAPASPTVLGFLRAALGCQESESEAKVDGETASDSESRAESAPLPVSADDTPEVLNRAL SNLSSRWKNWWVRGILTLAMIAFFFIIIYLGPMVLMIIVMCVQIKCFHEIITIGYNVYHSYDLPWFRTLSWYFLLCVNYFFYGETVTDYF FTLVQREEPLRILSKYHRFISFTLYLIGFCMFVLSLVKKHYRLQFYMFGWTHVTLLIVVTQSHLVIHNLFEGMIWFIVPISCVICNDIMA YMFGFFFGRTPLIKLSPKKTWEGFIGGFFATVVFGLLLSYVMSGYRCFVCPVEYNNDTNSFTVDCEPSDLFRLQEYNIPGVIQSVIGWKT VRMYPFQIHSIALSTFASLIGPFGGFFASGFKRAFKIKDFANTIPGHGGIMDRFDCQYLMATFVNVYIASFIRGPNPSKLIQQFLTLRPD -------------------------------------------------------------- >1520_1520_5_ACSL6-CDS2_ACSL6_chr5_131307243_ENST00000379264_CDS2_chr20_5154169_ENST00000460006_length(amino acids)=926AA_BP=500 MRGCGAAGRGGRAGPGRAPLTAMLTFFLVSGGSLWLFVEFVLSLLEKMQTQEILRILRLPELGDLGQFFRSLSATTLVSMGALAAILAYW FTHRPKALQPPCNLLMQSEEVEDSGGARRSVIGSGPQLLTHYYDDARTMYQVFRRGLSISGNGPCLGFRKPKQPYQWLSYQEVADRAEFL GSGLLQHNCKACTDQFIGVFAQNRPEWIIVELACYTYSMVVVPLYDTLGPGAIRYIINTADISTVIVDKPQKAVLLLEHVERKETPGLKL IILMDPFEEALKERGQKCGVVIKSMQAVEDCGQENHQAPVPPQPDDLSIVCFTSGTTGNPKGAMLTHGNVVADFSGFLKVTEKVIFPRQD DVLISFLPLAHMFERVIQSVVYCHGGRVGFFQGDIRLLSDDMKALCPTIFPVVPRLLNRMYDKIFSQANTPLKRWLLEFAAKRKQAEVRS GIIRNDSIWDELFFNKIQASLGGCVRMIVTGAAPASPTVLGFLRAALGCQESESEAKVDGETASDSESRAESAPLPVSADDTPEVLNRAL SNLSSRWKNWWVRGILTLAMIAFFFIIIYLGPMVLMIIVMCVQIKCFHEIITIGYNVYHSYDLPWFRTLSWYFLLCVNYFFYGETVTDYF FTLVQREEPLRILSKYHRFISFTLYLIGFCMFVLSLVKKHYRLQFYMFGWTHVTLLIVVTQSHLVIHNLFEGMIWFIVPISCVICNDIMA YMFGFFFGRTPLIKLSPKKTWEGFIGGFFATVVFGLLLSYVMSGYRCFVCPVEYNNDTNSFTVDCEPSDLFRLQEYNIPGVIQSVIGWKT VRMYPFQIHSIALSTFASLIGPFGGFFASGFKRAFKIKDFANTIPGHGGIMDRFDCQYLMATFVNVYIASFIRGPNPSKLIQQFLTLRPD -------------------------------------------------------------- >1520_1520_6_ACSL6-CDS2_ACSL6_chr5_131307243_ENST00000379272_CDS2_chr20_5154169_ENST00000460006_length(amino acids)=941AA_BP=515 MRGCGAAGRGGRAGPGRAPLTAMLTFFLVSGGSLWLFVEFVLSLLEKMQTQEILRILRLPELGDLGQFFRSLSATTLVSMGALAAILAYW FTHRPKALQPPCNLLMQSEEVEDSGGARRSVIGSGPQLLTHYYDDARTMYQVFRRGLSISGNGPCLGFRKPKQPYQWLSYQEVADRAEFL GSGLLQHNCKACTDQFIGVFAQNRPEWIIVELACYTYSMVVVPLYDTLGPGAIRYIINTGLSCQEGASATASTQADISTVIVDKPQKAVL LLEHVERKETPGLKLIILMDPFEEALKERGQKCGVVIKSMQAVEDCGQENHQAPVPPQPDDLSIVCFTSGTTGNPKGAMLTHGNVVADFS GFLKVTEKVIFPRQDDVLISFLPLAHMFERVIQSVVYCHGGRVGFFQGDIRLLSDDMKALCPTIFPVVPRLLNRMYDKIFSQANTPLKRW LLEFAAKRKQAEVRSGIIRNDSIWDELFFNKIQASLGGCVRMIVTGAAPASPTVLGFLRAALGCQESESEAKVDGETASDSESRAESAPL PVSADDTPEVLNRALSNLSSRWKNWWVRGILTLAMIAFFFIIIYLGPMVLMIIVMCVQIKCFHEIITIGYNVYHSYDLPWFRTLSWYFLL CVNYFFYGETVTDYFFTLVQREEPLRILSKYHRFISFTLYLIGFCMFVLSLVKKHYRLQFYMFGWTHVTLLIVVTQSHLVIHNLFEGMIW FIVPISCVICNDIMAYMFGFFFGRTPLIKLSPKKTWEGFIGGFFATVVFGLLLSYVMSGYRCFVCPVEYNNDTNSFTVDCEPSDLFRLQE YNIPGVIQSVIGWKTVRMYPFQIHSIALSTFASLIGPFGGFFASGFKRAFKIKDFANTIPGHGGIMDRFDCQYLMATFVNVYIASFIRGP -------------------------------------------------------------- >1520_1520_7_ACSL6-CDS2_ACSL6_chr5_131307243_ENST00000431707_CDS2_chr20_5154169_ENST00000460006_length(amino acids)=862AA_BP=436 MEKMQTQEILRILRLPELGDLGQFFRSLSATTLDSGGARRSVIGSGPQLLTHYYDDARTMYQVFRRGLSISGNGPCLGFRKPKQPYQWLS YQEVADRAEFLGSGLLQHNCKACTDQFIGVFAQNRPEWIIVELACYTYSMVVVPLYDTLGPGAIRYIINTGLSCQEGASATASTQADIST VIVDKPQKAVLLLEHVERKETPGLKLIILMDPFEEALKERGQKCGVVIKSMQAVEDCGQENHQAPVPPQPDDLSIVCFTSGTTGNPKGAM LTHGNVVADFSGFLKVTEKVIFPRQDDVLISFLPLAHMFERVIQSVVYCHGGRVGFFQGDIRLLSDDMKALCPTIFPVVPRLLNRMYDKI FSQANTPLKRWLLEFAAKRKQAEVRSGIIRNDSIWDELFFNKIQASLGGCVRMIVTGAAPASPTVLGFLRAALGCQESESEAKVDGETAS DSESRAESAPLPVSADDTPEVLNRALSNLSSRWKNWWVRGILTLAMIAFFFIIIYLGPMVLMIIVMCVQIKCFHEIITIGYNVYHSYDLP WFRTLSWYFLLCVNYFFYGETVTDYFFTLVQREEPLRILSKYHRFISFTLYLIGFCMFVLSLVKKHYRLQFYMFGWTHVTLLIVVTQSHL VIHNLFEGMIWFIVPISCVICNDIMAYMFGFFFGRTPLIKLSPKKTWEGFIGGFFATVVFGLLLSYVMSGYRCFVCPVEYNNDTNSFTVD CEPSDLFRLQEYNIPGVIQSVIGWKTVRMYPFQIHSIALSTFASLIGPFGGFFASGFKRAFKIKDFANTIPGHGGIMDRFDCQYLMATFV -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:131307243/chr20:5154169) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ACSL6 | CDS2 |
| FUNCTION: Catalyzes the conversion of long-chain fatty acids to their active form acyl-CoA for both synthesis of cellular lipids, and degradation via beta-oxidation (PubMed:22633490, PubMed:24269233). Plays an important role in fatty acid metabolism in brain and the acyl-CoAs produced may be utilized exclusively for the synthesis of the brain lipid. {ECO:0000269|PubMed:22633490, ECO:0000269|PubMed:24269233}. | FUNCTION: Catalyzes the conversion of phosphatidic acid (PA) to CDP-diacylglycerol (CDP-DAG), an essential intermediate in the synthesis of phosphatidylglycerol, cardiolipin and phosphatidylinositol (PubMed:25375833). Exhibits specificity for the nature of the acyl chains at the sn-1 and sn-2 positions in the substrate, PA and the preferred acyl chain composition is 1-stearoyl-2-arachidonoyl-sn-phosphatidic acid (PubMed:25375833). Plays an important role in regulating the growth and maturation of lipid droplets which are storage organelles at the center of lipid and energy homeostasis (PubMed:26946540, PubMed:31548309). {ECO:0000269|PubMed:25375833, ECO:0000269|PubMed:26946540, ECO:0000269|PubMed:31548309}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000296869 | - | 14 | 21 | 25_45 | 478.0 | 723.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000357096 | - | 12 | 19 | 25_45 | 378.0 | 623.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379240 | - | 14 | 21 | 25_45 | 453.0 | 698.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379244 | - | 14 | 21 | 25_45 | 453.0 | 698.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379246 | - | 14 | 21 | 25_45 | 464.0 | 709.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379249 | - | 14 | 21 | 25_45 | 453.0 | 668.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379255 | - | 13 | 20 | 25_45 | 378.0 | 623.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379264 | - | 14 | 21 | 25_45 | 478.0 | 723.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379272 | - | 15 | 22 | 25_45 | 468.0 | 713.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000543479 | - | 14 | 21 | 25_45 | 453.0 | 698.0 | Transmembrane | Helical%3B Signal-anchor for type III membrane protein |
| Tgene | CDS2 | chr5:131307243 | chr20:5154169 | ENST00000460006 | 0 | 13 | 132_152 | 19.0 | 446.0 | Transmembrane | Helical | |
| Tgene | CDS2 | chr5:131307243 | chr20:5154169 | ENST00000460006 | 0 | 13 | 166_186 | 19.0 | 446.0 | Transmembrane | Helical | |
| Tgene | CDS2 | chr5:131307243 | chr20:5154169 | ENST00000460006 | 0 | 13 | 213_233 | 19.0 | 446.0 | Transmembrane | Helical | |
| Tgene | CDS2 | chr5:131307243 | chr20:5154169 | ENST00000460006 | 0 | 13 | 262_282 | 19.0 | 446.0 | Transmembrane | Helical | |
| Tgene | CDS2 | chr5:131307243 | chr20:5154169 | ENST00000460006 | 0 | 13 | 340_360 | 19.0 | 446.0 | Transmembrane | Helical | |
| Tgene | CDS2 | chr5:131307243 | chr20:5154169 | ENST00000460006 | 0 | 13 | 79_99 | 19.0 | 446.0 | Transmembrane | Helical |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000296869 | - | 14 | 21 | 46_697 | 478.0 | 723.0 | Topological domain | Cytoplasmic |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000357096 | - | 12 | 19 | 46_697 | 378.0 | 623.0 | Topological domain | Cytoplasmic |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379240 | - | 14 | 21 | 46_697 | 453.0 | 698.0 | Topological domain | Cytoplasmic |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379244 | - | 14 | 21 | 46_697 | 453.0 | 698.0 | Topological domain | Cytoplasmic |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379246 | - | 14 | 21 | 46_697 | 464.0 | 709.0 | Topological domain | Cytoplasmic |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379249 | - | 14 | 21 | 46_697 | 453.0 | 668.0 | Topological domain | Cytoplasmic |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379255 | - | 13 | 20 | 46_697 | 378.0 | 623.0 | Topological domain | Cytoplasmic |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379264 | - | 14 | 21 | 46_697 | 478.0 | 723.0 | Topological domain | Cytoplasmic |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000379272 | - | 15 | 22 | 46_697 | 468.0 | 713.0 | Topological domain | Cytoplasmic |
| Hgene | ACSL6 | chr5:131307243 | chr20:5154169 | ENST00000543479 | - | 14 | 21 | 46_697 | 453.0 | 698.0 | Topological domain | Cytoplasmic |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ACSL6 | |
| CDS2 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to ACSL6-CDS2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ACSL6-CDS2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

