| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:CDK6-KMT2A |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: CDK6-KMT2A | FusionPDB ID: 15293 | FusionGDB2.0 ID: 15295 | Hgene | Tgene | Gene symbol | CDK6 | KMT2A | Gene ID | 1021 | 4297 |
| Gene name | cyclin dependent kinase 6 | lysine methyltransferase 2A | |
| Synonyms | MCPH12|PLSTIRE | ALL-1|CXXC7|HRX|HTRX1|MLL|MLL1|MLL1A|TRX1|WDSTS | |
| Cytomap | 7q21.2 | 11q23.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cyclin-dependent kinase 6cell division protein kinase 6serine/threonine-protein kinase PLSTIRE | histone-lysine N-methyltransferase 2ACXXC-type zinc finger protein 7lysine (K)-specific methyltransferase 2Alysine N-methyltransferase 2Amixed lineage leukemia 1myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)trithorax-like | |
| Modification date | 20200329 | 20200319 | |
| UniProtAcc | Q00534 | Q03164 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000265734, ENST00000424848, ENST00000491250, | ENST00000354520, ENST00000420751, ENST00000389506, ENST00000534358, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 12 X 9 X 7=756 | 31 X 72 X 3=6696 |
| # samples | 17 | 79 | |
| ** MAII score | log2(17/756*10)=-2.15285148808337 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(79/6696*10)=-3.08337496948588 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: CDK6 [Title/Abstract] AND KMT2A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | CDK6(92404010)-KMT2A(118360505), # samples:1 CDK6(92364333)-KMT2A(118199863), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | CDK6-KMT2A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CDK6-KMT2A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CDK6-KMT2A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CDK6-KMT2A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CDK6-KMT2A seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. CDK6-KMT2A seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. CDK6-KMT2A seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. CDK6-KMT2A seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. CDK6-KMT2A seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. CDK6-KMT2A seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. CDK6-KMT2A seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. CDK6-KMT2A seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. CDK6-KMT2A seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CDK6 | GO:0001954 | positive regulation of cell-matrix adhesion | 10205165 |
| Hgene | CDK6 | GO:0003323 | type B pancreatic cell development | 20668294 |
| Hgene | CDK6 | GO:0006468 | protein phosphorylation | 8114739 |
| Hgene | CDK6 | GO:0010468 | regulation of gene expression | 15254224 |
| Hgene | CDK6 | GO:0045638 | negative regulation of myeloid cell differentiation | 17431401 |
| Hgene | CDK6 | GO:0045656 | negative regulation of monocyte differentiation | 26542173 |
| Hgene | CDK6 | GO:0045668 | negative regulation of osteoblast differentiation | 15254224 |
| Hgene | CDK6 | GO:0045786 | negative regulation of cell cycle | 14985467 |
| Hgene | CDK6 | GO:2000773 | negative regulation of cellular senescence | 17420273 |
| Tgene | KMT2A | GO:0044648 | histone H3-K4 dimethylation | 25561738 |
| Tgene | KMT2A | GO:0045944 | positive regulation of transcription by RNA polymerase II | 20861184 |
| Tgene | KMT2A | GO:0051568 | histone H3-K4 methylation | 19556245 |
| Tgene | KMT2A | GO:0065003 | protein-containing complex assembly | 15199122 |
| Tgene | KMT2A | GO:0080182 | histone H3-K4 trimethylation | 20861184 |
| Tgene | KMT2A | GO:0097692 | histone H3-K4 monomethylation | 25561738|26324722 |
 Fusion gene breakpoints across CDK6 (5'-gene) Fusion gene breakpoints across CDK6 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
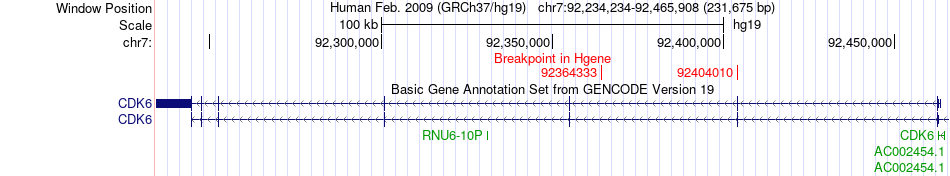 |
 Fusion gene breakpoints across KMT2A (3'-gene) Fusion gene breakpoints across KMT2A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
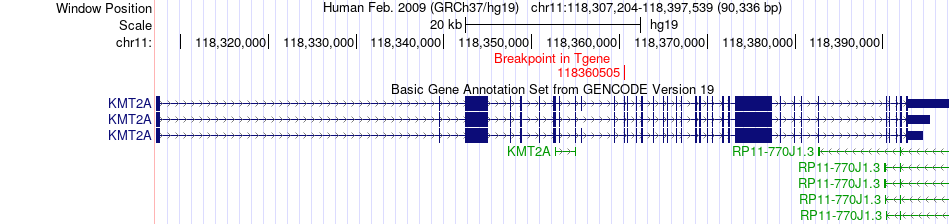 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | CDK6 | chr7 | 92462404 | - | KMT2A | chr11 | 118355576 | + |
| ChiTaRS5.0 | N/A | AF492830 | CDK6 | chr7 | 92404010 | - | KMT2A | chr11 | 118360505 | + |
| ChiTaRS5.0 | N/A | AF492834 | CDK6 | chr7 | 92364333 | - | KMT2A | chr11 | 118199863 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000265734 | CDK6 | chr7 | 92462404 | - | ENST00000534358 | KMT2A | chr11 | 118355576 | + | 12998 | 645 | 453 | 8345 | 2630 |
| ENST00000265734 | CDK6 | chr7 | 92462404 | - | ENST00000389506 | KMT2A | chr11 | 118355576 | + | 10082 | 645 | 453 | 8336 | 2627 |
| ENST00000424848 | CDK6 | chr7 | 92404010 | - | ENST00000534358 | KMT2A | chr11 | 118360505 | + | 12945 | 853 | 484 | 8292 | 2602 |
| ENST00000424848 | CDK6 | chr7 | 92404010 | - | ENST00000389506 | KMT2A | chr11 | 118360505 | + | 10029 | 853 | 484 | 8283 | 2599 |
| ENST00000424848 | CDK6 | chr7 | 92404010 | - | ENST00000354520 | KMT2A | chr11 | 118360505 | + | 10808 | 853 | 484 | 8283 | 2599 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000424848 | ENST00000534358 | CDK6 | chr7 | 92404010 | - | KMT2A | chr11 | 118360505 | + | 0.000419523 | 0.99958044 |
| ENST00000424848 | ENST00000389506 | CDK6 | chr7 | 92404010 | - | KMT2A | chr11 | 118360505 | + | 0.000722561 | 0.9992774 |
| ENST00000424848 | ENST00000354520 | CDK6 | chr7 | 92404010 | - | KMT2A | chr11 | 118360505 | + | 0.000794911 | 0.99920505 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >15293_15293_1_CDK6-KMT2A_CDK6_chr7_92404010_ENST00000424848_KMT2A_chr11_118360505_ENST00000354520_length(amino acids)=2599AA_BP=123 MEKDGLCRADQQYECVAEIGEGAYGKVFKARDLKNGGRFVALKRVRVQTGEEGMPLSTIREVAVLRHLETFEHPNVVRLFDVCTVSRTDR ETKLTLVFEHVDQDLTTYLDKVPEPGVPTETIKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWS HDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKEL QISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDI VKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLM KKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEV FEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVD FEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHD ENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGD PLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTV VTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFS SKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPG QVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALK ESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFG LTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGV DDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRV HTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQ LPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMT PDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIV VNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQ SSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPS QLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTT TPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAA SPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSV PGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVT QNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLS FAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEE VQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDS -------------------------------------------------------------- >15293_15293_2_CDK6-KMT2A_CDK6_chr7_92404010_ENST00000424848_KMT2A_chr11_118360505_ENST00000389506_length(amino acids)=2599AA_BP=123 MEKDGLCRADQQYECVAEIGEGAYGKVFKARDLKNGGRFVALKRVRVQTGEEGMPLSTIREVAVLRHLETFEHPNVVRLFDVCTVSRTDR ETKLTLVFEHVDQDLTTYLDKVPEPGVPTETIKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWS HDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKEL QISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDI VKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLM KKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEV FEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVD FEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHD ENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGD PLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTV VTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFS SKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPG QVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALK ESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFG LTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGV DDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRV HTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQ LPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMT PDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIV VNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQ SSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPS QLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTT TPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAA SPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSV PGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVT QNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLS FAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEE VQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDS -------------------------------------------------------------- >15293_15293_3_CDK6-KMT2A_CDK6_chr7_92404010_ENST00000424848_KMT2A_chr11_118360505_ENST00000534358_length(amino acids)=2602AA_BP=123 MEKDGLCRADQQYECVAEIGEGAYGKVFKARDLKNGGRFVALKRVRVQTGEEGMPLSTIREVAVLRHLETFEHPNVVRLFDVCTVSRTDR ETKLTLVFEHVDQDLTTYLDKVPEPGVPTETIKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWS HDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALE KELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFS DDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQP PLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWS AEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRV FVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTV EHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVT VGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQ RTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSV SFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFL EPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKN ALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNM FFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQL DGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESS RRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVF SQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEG HMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEK LIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPA ATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINF TPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQ TTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGI TAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSAS PSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEV QVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLK QLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEE EEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRI -------------------------------------------------------------- >15293_15293_4_CDK6-KMT2A_CDK6_chr7_92462404_ENST00000265734_KMT2A_chr11_118355576_ENST00000389506_length(amino acids)=2627AA_BP=63 MRGGDRGGRLWEGVQGPRLEERRPFRGVEARAGADRRGGHAALHHPRGGGAEAPGDLRAPQRGQEDCEAENVWEMGGLGILTSVPITPRV VCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKK KVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSN LPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDD QQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNA VLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDD SANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKV YCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDAR KRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSP TQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKV VDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLA PQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSS RDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKEL QAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPD GPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSG GEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDN CHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSS ELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTI TEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTD SPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTS QSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNK KLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQD TSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTS QTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALS SAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQD TASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQIC AESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRK SAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNV IRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDAS -------------------------------------------------------------- >15293_15293_5_CDK6-KMT2A_CDK6_chr7_92462404_ENST00000265734_KMT2A_chr11_118355576_ENST00000534358_length(amino acids)=2630AA_BP=63 MRGGDRGGRLWEGVQGPRLEERRPFRGVEARAGADRRGGHAALHHPRGGGAEAPGDLRAPQRGQEDCEAENVWEMGGLGILTSVPITPRV VCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKK KVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEI LSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSK QDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGML PNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTY GDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDD KKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTT DARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPS YSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESS AKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIP KLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMG SSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSA KELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLM LPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVI SSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPK MDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPES SSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKR VTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPN STDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSL PTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTR KNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTI SQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQL GTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNK ALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAE QQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGF QICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVH LRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYA GNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIE -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:92404010/chr11:118360505) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CDK6 | KMT2A |
| FUNCTION: Serine/threonine-protein kinase involved in the control of the cell cycle and differentiation; promotes G1/S transition. Phosphorylates pRB/RB1 and NPM1. Interacts with D-type G1 cyclins during interphase at G1 to form a pRB/RB1 kinase and controls the entrance into the cell cycle. Involved in initiation and maintenance of cell cycle exit during cell differentiation; prevents cell proliferation and regulates negatively cell differentiation, but is required for the proliferation of specific cell types (e.g. erythroid and hematopoietic cells). Essential for cell proliferation within the dentate gyrus of the hippocampus and the subventricular zone of the lateral ventricles. Required during thymocyte development. Promotes the production of newborn neurons, probably by modulating G1 length. Promotes, at least in astrocytes, changes in patterns of gene expression, changes in the actin cytoskeleton including loss of stress fibers, and enhanced motility during cell differentiation. Prevents myeloid differentiation by interfering with RUNX1 and reducing its transcription transactivation activity, but promotes proliferation of normal myeloid progenitors. Delays senescence. Promotes the proliferation of beta-cells in pancreatic islets of Langerhans. May play a role in the centrosome organization during the cell cycle phases (PubMed:23918663). {ECO:0000269|PubMed:12833137, ECO:0000269|PubMed:14985467, ECO:0000269|PubMed:15254224, ECO:0000269|PubMed:15809340, ECO:0000269|PubMed:17420273, ECO:0000269|PubMed:17431401, ECO:0000269|PubMed:20333249, ECO:0000269|PubMed:20668294, ECO:0000269|PubMed:23918663, ECO:0000269|PubMed:8114739}. | FUNCTION: Histone methyltransferase that plays an essential role in early development and hematopoiesis (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalytic subunit of the MLL1/MLL complex, a multiprotein complex that mediates both methylation of 'Lys-4' of histone H3 (H3K4me) complex and acetylation of 'Lys-16' of histone H4 (H4K16ac) (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:24235145, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalyzes methyl group transfer from S-adenosyl-L-methionine to the epsilon-amino group of 'Lys-4' of histone H3 (H3K4) via a non-processive mechanism. Part of chromatin remodeling machinery predominantly forms H3K4me1 and H3K4me2 methylation marks at active chromatin sites where transcription and DNA repair take place (PubMed:25561738, PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Has weak methyltransferase activity by itself, and requires other component of the MLL1/MLL complex to obtain full methyltransferase activity (PubMed:19187761, PubMed:26886794). Has no activity toward histone H3 phosphorylated on 'Thr-3', less activity toward H3 dimethylated on 'Arg-8' or 'Lys-9', while it has higher activity toward H3 acetylated on 'Lys-9' (PubMed:19187761). Binds to unmethylated CpG elements in the promoter of target genes and helps maintain them in the nonmethylated state (PubMed:20010842). Required for transcriptional activation of HOXA9 (PubMed:12453419, PubMed:20677832, PubMed:20010842). Promotes PPP1R15A-induced apoptosis (PubMed:10490642). Plays a critical role in the control of circadian gene expression and is essential for the transcriptional activation mediated by the CLOCK-ARNTL/BMAL1 heterodimer (By similarity). Establishes a permissive chromatin state for circadian transcription by mediating a rhythmic methylation of 'Lys-4' of histone H3 (H3K4me) and this histone modification directs the circadian acetylation at H3K9 and H3K14 allowing the recruitment of CLOCK-ARNTL/BMAL1 to chromatin (By similarity). Also has auto-methylation activity on Cys-3882 in absence of histone H3 substrate (PubMed:24235145). {ECO:0000250|UniProtKB:P55200, ECO:0000269|PubMed:10490642, ECO:0000269|PubMed:12453419, ECO:0000269|PubMed:15960975, ECO:0000269|PubMed:19187761, ECO:0000269|PubMed:19556245, ECO:0000269|PubMed:20010842, ECO:0000269|PubMed:21220120, ECO:0000269|PubMed:24235145, ECO:0000269|PubMed:26886794, ECO:0000305|PubMed:20677832}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDK6 | chr7:92404010 | chr11:118360505 | ENST00000265734 | - | 3 | 8 | 19_27 | 123.0 | 327.0 | Nucleotide binding | ATP |
| Hgene | CDK6 | chr7:92404010 | chr11:118360505 | ENST00000424848 | - | 3 | 8 | 19_27 | 123.0 | 327.0 | Nucleotide binding | ATP |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 1703_1748 | 1455.0 | 3932.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 2018_2074 | 1455.0 | 3932.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 3666_3747 | 1455.0 | 3932.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 3829_3945 | 1455.0 | 3932.0 | Domain | SET | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 3953_3969 | 1455.0 | 3932.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 1703_1748 | 1493.0 | 3970.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 2018_2074 | 1493.0 | 3970.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 3666_3747 | 1493.0 | 3970.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 3829_3945 | 1493.0 | 3970.0 | Domain | SET | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 3953_3969 | 1493.0 | 3970.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 1703_1748 | 1493.0 | 3973.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 2018_2074 | 1493.0 | 3973.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 3666_3747 | 1493.0 | 3973.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 3829_3945 | 1493.0 | 3973.0 | Domain | SET | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 3953_3969 | 1493.0 | 3973.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 3906_3907 | 1455.0 | 3932.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 3906_3907 | 1493.0 | 3970.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 3906_3907 | 1493.0 | 3973.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 1479_1533 | 1455.0 | 3932.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 1566_1627 | 1455.0 | 3932.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 1870_1910 | 1455.0 | 3932.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 1931_1978 | 1455.0 | 3932.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 1566_1627 | 1493.0 | 3970.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 1870_1910 | 1493.0 | 3970.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 1931_1978 | 1493.0 | 3970.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 1566_1627 | 1493.0 | 3973.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 1870_1910 | 1493.0 | 3973.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 1931_1978 | 1493.0 | 3973.0 | Zinc finger | PHD-type 4 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CDK6 | chr7:92404010 | chr11:118360505 | ENST00000265734 | - | 3 | 8 | 13_300 | 123.0 | 327.0 | Domain | Protein kinase |
| Hgene | CDK6 | chr7:92404010 | chr11:118360505 | ENST00000424848 | - | 3 | 8 | 13_300 | 123.0 | 327.0 | Domain | Protein kinase |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 137_143 | 1455.0 | 3932.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 17_102 | 1455.0 | 3932.0 | Compositional bias | Note=Ala/Gly/Ser-rich | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 561_564 | 1455.0 | 3932.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 568_571 | 1455.0 | 3932.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 137_143 | 1493.0 | 3970.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 17_102 | 1493.0 | 3970.0 | Compositional bias | Note=Ala/Gly/Ser-rich | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 561_564 | 1493.0 | 3970.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 568_571 | 1493.0 | 3970.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 137_143 | 1493.0 | 3973.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 17_102 | 1493.0 | 3973.0 | Compositional bias | Note=Ala/Gly/Ser-rich | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 561_564 | 1493.0 | 3973.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 568_571 | 1493.0 | 3973.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 169_180 | 1455.0 | 3932.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 217_227 | 1455.0 | 3932.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 301_309 | 1455.0 | 3932.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 169_180 | 1493.0 | 3970.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 217_227 | 1493.0 | 3970.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 301_309 | 1493.0 | 3970.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 169_180 | 1493.0 | 3973.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 217_227 | 1493.0 | 3973.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 301_309 | 1493.0 | 3973.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 123_134 | 1455.0 | 3932.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 147_152 | 1455.0 | 3932.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 6_25 | 1455.0 | 3932.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 123_134 | 1493.0 | 3970.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 147_152 | 1493.0 | 3970.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 6_25 | 1493.0 | 3970.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 123_134 | 1493.0 | 3973.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 147_152 | 1493.0 | 3973.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 6_25 | 1493.0 | 3973.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 1147_1195 | 1455.0 | 3932.0 | Zinc finger | CXXC-type | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000354520 | 9 | 35 | 1431_1482 | 1455.0 | 3932.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 1147_1195 | 1493.0 | 3970.0 | Zinc finger | CXXC-type | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 1431_1482 | 1493.0 | 3970.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000389506 | 10 | 36 | 1479_1533 | 1493.0 | 3970.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 1147_1195 | 1493.0 | 3973.0 | Zinc finger | CXXC-type | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 1431_1482 | 1493.0 | 3973.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr7:92404010 | chr11:118360505 | ENST00000534358 | 10 | 36 | 1479_1533 | 1493.0 | 3973.0 | Zinc finger | PHD-type 2 |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (1164) >>>1164.pdbFusion protein BP residue: 63 CIF file (1164) >>>1164.cif | CDK6 | chr7 | 92462404 | - | KMT2A | chr11 | 118355576 | + | MRGGDRGGRLWEGVQGPRLEERRPFRGVEARAGADRRGGHAALHHPRGGG AEAPGDLRAPQRGQEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGH VEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQAT KQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPG KGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCD RWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQI SLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPP VLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDG GQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNA VLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLH PPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLY IGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGAT VGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFE VFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCED KLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDEN RTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKV PRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGS RRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKV VDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSS ASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRE LNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSS PDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHS SKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGL SMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKES SPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLP VQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGE EDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSG GEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPK RNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQ GQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCG NILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMG LFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVP SQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPD HFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQ NQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQ TLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTS QSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPP DPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSN IAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSS IMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLN VVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGI QDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASP SGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNS MGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKR FQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQD TASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNF SSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKS LTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRN YKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYN PNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRG LFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEV VDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTY | 2627 |
| 3D view using mol* of 1164 (AA BP:63) | ||||||||||
 | ||||||||||
| PDB file (1166) >>>1166.pdbFusion protein BP residue: 63 CIF file (1166) >>>1166.cif | CDK6 | chr7 | 92462404 | - | KMT2A | chr11 | 118355576 | + | MRGGDRGGRLWEGVQGPRLEERRPFRGVEARAGADRRGGHAALHHPRGGG AEAPGDLRAPQRGQEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGH VEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQAT KQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPG KGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCD RWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKE LQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGP DPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAIN SDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGML PNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPT PLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGR LLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKP GATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPEN GFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSD CEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEH DENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVI SKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRS IGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESS AKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMK QSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTT SRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSA NSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKE KHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGD KGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNAL KESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQ NLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRS YGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVI SSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQ IPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRV KTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSD DCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREK DMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSP TVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHM TPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTV PIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLY VLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSL PTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQ PPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSST PSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRS KSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSS VLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQS LGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITA ASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDA PNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPK TKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAE QQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEE SNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDA WKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKH CRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPP EYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIH GRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDD SEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEE | 2630 |
| 3D view using mol* of 1166 (AA BP:63) | ||||||||||
 | ||||||||||
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
CDK6_pLDDT.png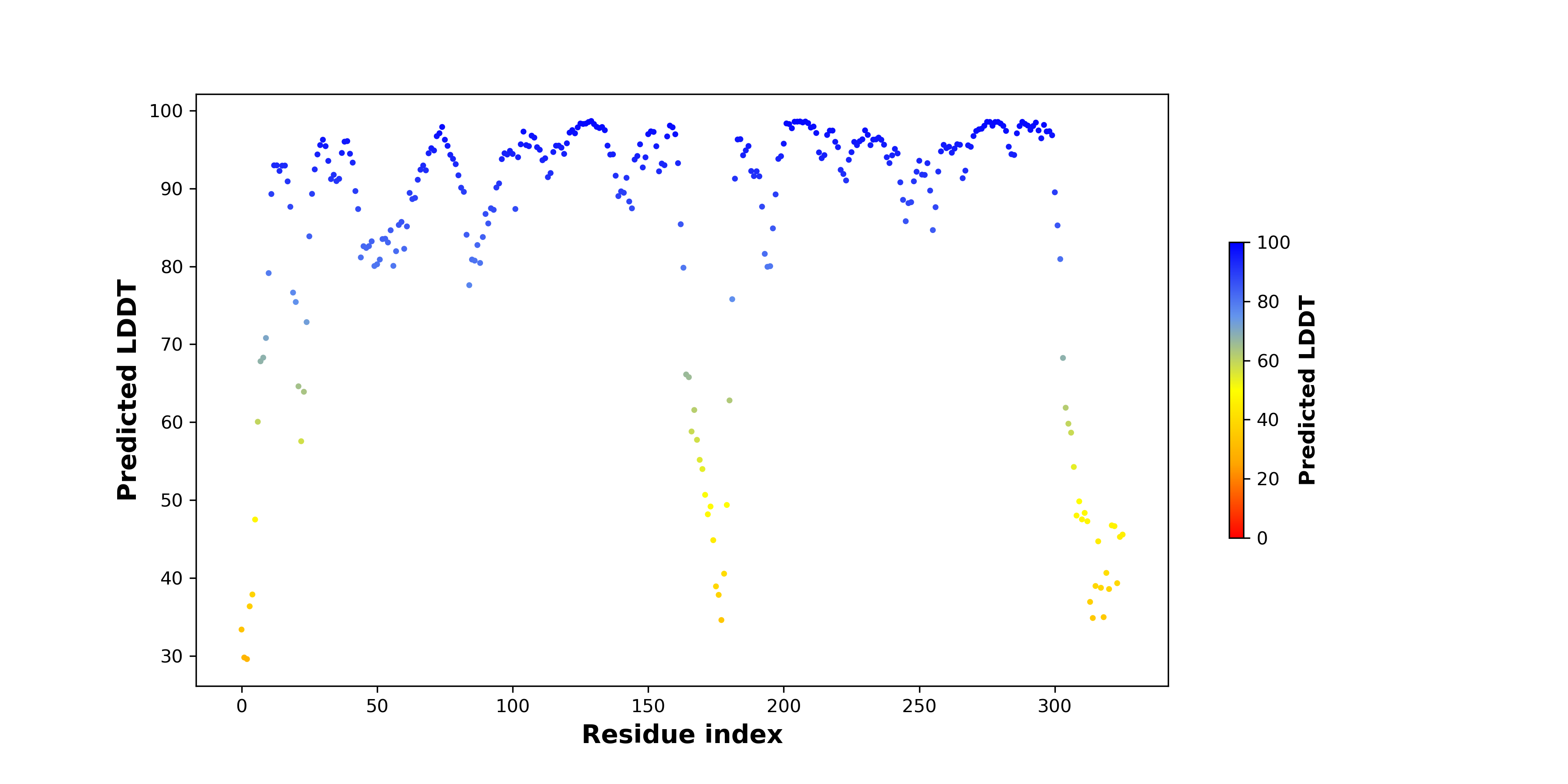 |
KMT2A_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
CDK6_KMT2A_1164_pLDDT.png (AA BP:63)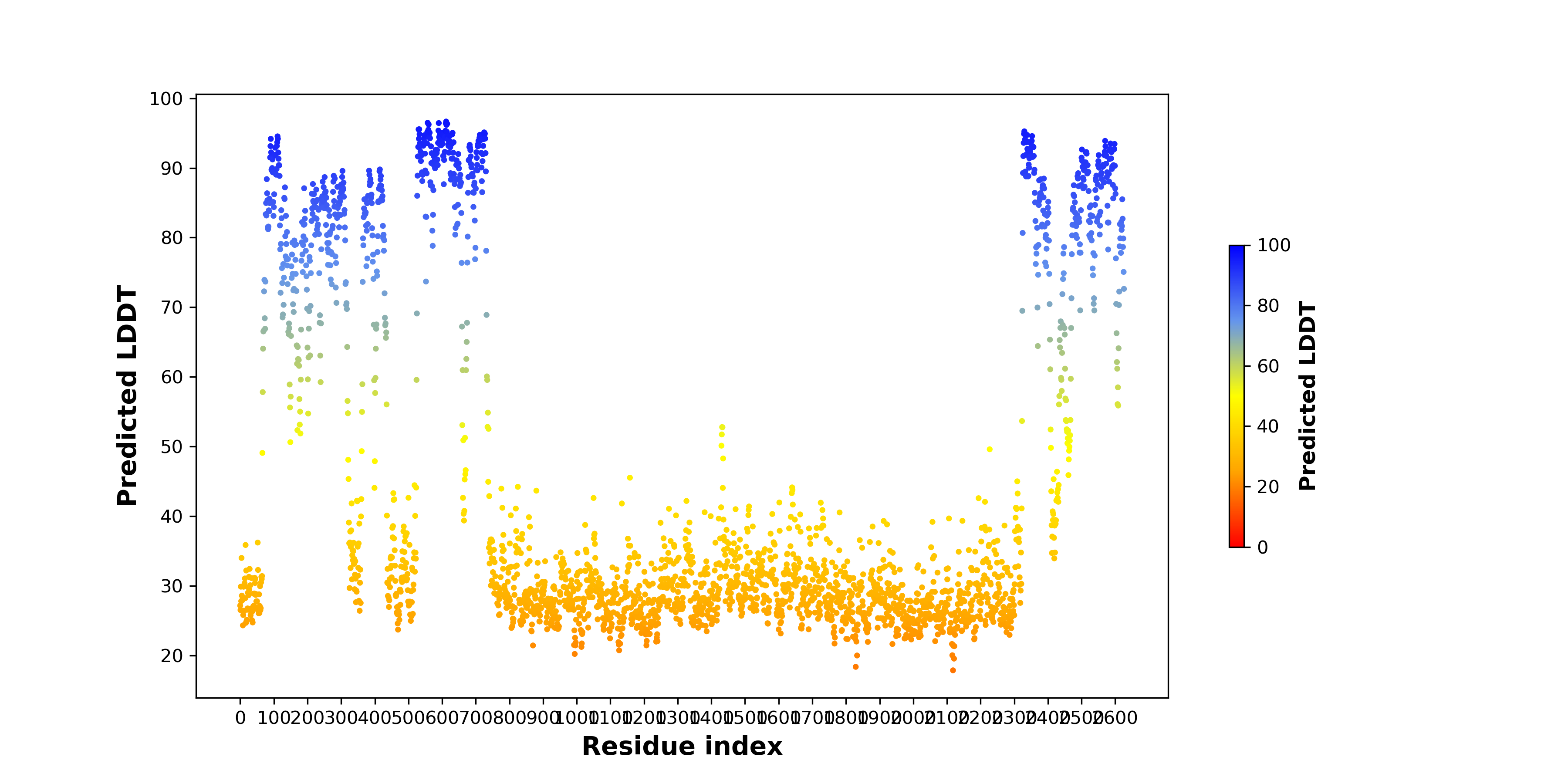 |
CDK6_KMT2A_1166_pLDDT.png (AA BP:63)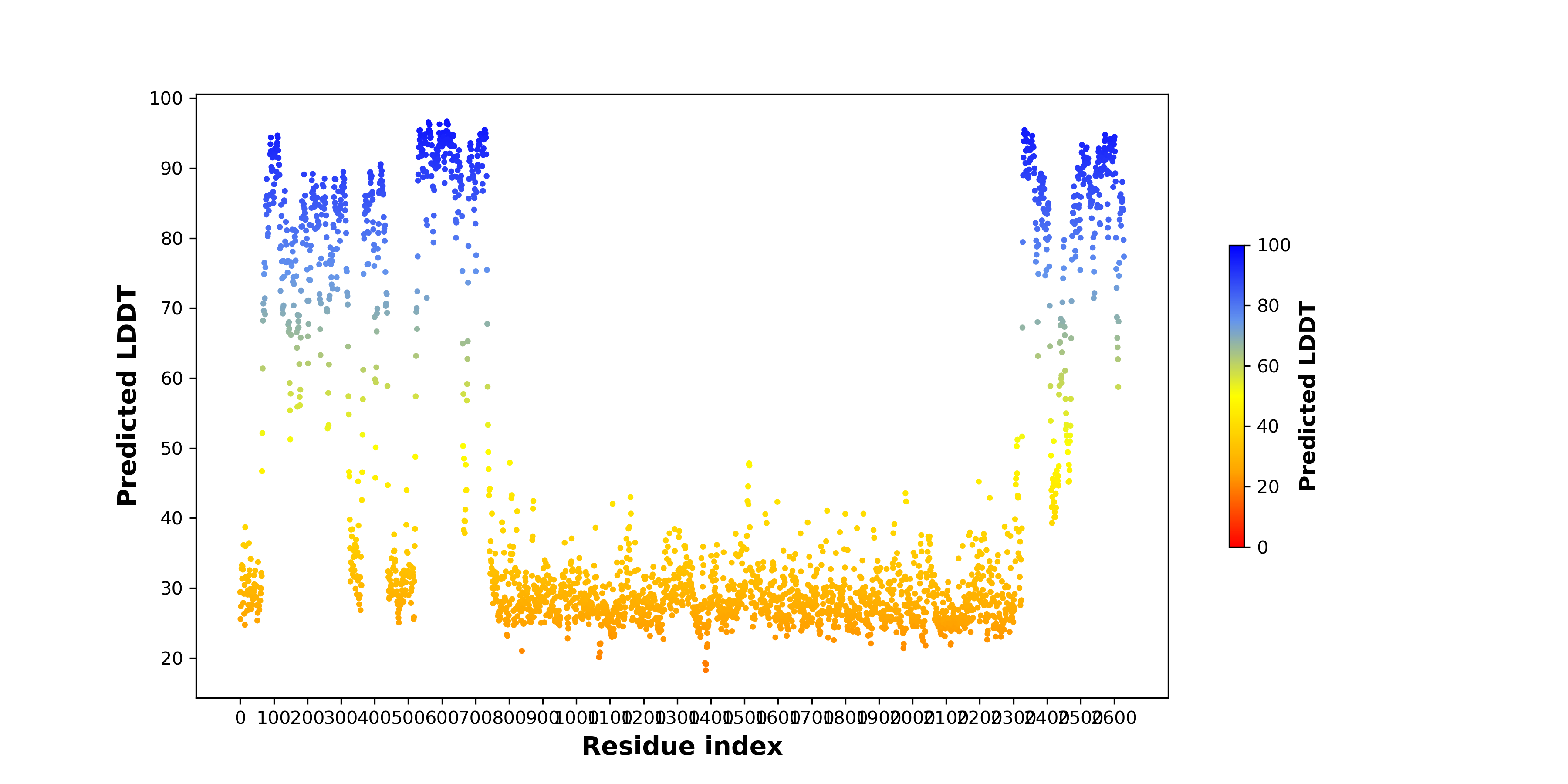 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| CDK6 | CDKN2C, CDKN2A, CCND1, CDKN2D, PPP2CA, PPM1B, CDK2, CCND2, CCND3, CDKN1B, CDC37, FBXO7, CDKN1A, UHRF2, CDK5R1, ELAVL1, CCNE1, CCNA2, KIF26B, RB1, CDK7, ATXN1, HIST1H1A, PML, HSP90AA1, FOXM1, ZSCAN1, SYNPO2, DEDD2, SRSF12, ZNF101, SNIP1, ZNF335, PRX, EBF4, EIF4ENIF1, RBM23, BCL11A, TCEB3B, PPHLN1, TRA2A, SENP3, NIPBL, CBY1, SSBP2, SIRT1, POGZ, LPIN1, ANKRD12, TPX2, TRAK1, CASC3, CLASRP, PPARGC1A, MSL3, SORBS1, ABI2, N4BP1, VGLL4, TJP2, SRSF11, ZMYM3, ZNF174, SLBP, ZFP36, MZF1, KLF10, TFDP1, ZEB1, SOX5, SOX10, SRSF1, SRSF2, SRSF7, TRA2B, RBL1, RBL2, NUMA1, NFATC3, MYC, MEF2D, MLLT3, ISL1, HSF1, EZH2, FOXO3, ELK1, ATF6B, DDIT3, CDC6, MCM2, MCM10, UCHL1, CDKN2B, WDR33, CDKL3, CDK6, DMBT1, AMY2A, PIGR, CDK4, DAB1, PSMA3, RTFDC1, HSP90AA5P, UBE2W, CCNT2, CCNT1, RCHY1, EIF2AK1, CDKN1C, NTRK1, RUNX1, CTNNB1, AR, VHL, FGFR3, USP17L2, MCF2L2, PAK4, FKBP5, FAM26E, TRIM25, HHV8GK18_gp82, EGLN2, RBPMS, HEXIM1, LARP7, KIAA1429, GBF1, SUMO1, UBE2I, ARNT, BECN1, AKT1, CD44, EPHA2, FGFR4, LATS2, STK11, FZR1, GLIS2, MAP2K3, MAP2K5, NF2, RAF1, RASSF1, ARAF, CBLC, KEAP1, MAP3K5, TEAD2, TERT, TSC1, ERBB2, GLIS1, GRM1, HGF, HIF1A, KAT2A, KDELR2, MAPK14, MET, PDGFRA, TNFRSF1A, TP53, TRIM28, APOBEC1, CCNI, CCNH, OTP, TRIM15, UBE2N, FASN, ACACA, NUPR1, USP51, ATG7, BSPRY, KLC3, CA4, NEU2, METTL21B, PPM1M, FAM96A, EPB41L5, HSP90AB1, PCNA, MAP1LC3B, CALCOCO2, |
| KMT2A | PPIE, PPP1R15A, KMT2A, ASH2L, HCFC1, HCFC2, MEN1, RBBP5, WDR5, AVP, INS, OXT, MAP3K5, HDAC1, CTBP1, CBX4, BMI1, CREBBP, SMARCB1, CXXC1, MYB, CTNNB1, SNW1, E2F2, E2F4, E2F6, PSIP1, MLLT4, POLR2A, KAT8, RNF2, TP53, tat, SBF1, MTM1, SET, HIST1H3A, HIST1H4A, KAT6A, ELL, AFF1, AFF4, CDK9, CCNT1, CTR9, LEO1, PAF1, CDC73, WDR61, MLLT3, DOT1L, SKP2, HIST3H3, SVIL, HIST2H3C, SIN3A, MLLT1, RUNX1, CBFB, H3F3A, SIRT7, ASB2, TCEB1, TCEB2, CBX8, TOP1, TAF6, NCL, HECW2, LGR4, CSNK2A2, SENP3, SYMPK, PKN1, PIH1D1, KRAS, TAF1, CHD3, SMARCA2, SMARCC2, SMARCC1, HDAC2, RBBP4, RBBP7, TBP, MBD3, SAP30, RAN, TAF9, TASP1, HIST1H2BG, EWSR1, DYNLT1, KIF11, ING4, Cbx1, Set, ZNF131, ASB7, AR, CSNK2A1, OGT, BRD4, KDM5B, KDM5D, KDM6B, CKS1B, CKS2, CHD4, ESR2, AGR2, EZH2, FBXW7, SOX2, MED17, WDR82, ACTR8, GEMIN5, RUVBL1, MCM2, RTF1, MED15, MED8, MRGBP, MED14, HIST1H2BB, HIST1H2AB, KIAA1429, SP1, MYC, NR2C2, IRF2, PLEKHA4, DPY30, PTEN, NPM1, RPL13, CCT6A, TMCO1, ITGB1, ESR1, MYCN, SUPT5H, CIT, Pik3r2, FASN, LDLR, CDC27, KMT2B, SETD1A, KDM6A, KANSL2, BUB3, SETD1B, KANSL1, RERE, MCRS1, KMT2C, ZZZ3, MBIP, YEATS2, PAXIP1, KANSL3, KMT2D, BOD1L1, CSRP2BP, ZNF608, ATN1, PHF20L1, PHF20, NCOA6, TADA3, APEX1, ASF1A, CBX3, CD3EAP, CENPA, NUP50, POLR1E, TERF2IP, ZNF330, NAA40, PRKRA, SRSF4, KRR1, RPL13A, ABT1, CSNK2B, OASL, SRSF5, SRSF6, RPS9, FGFBP1, RPLP0, RPL36, KAL1, RPL3, PML, ELF1, ELF2, FEV, NHLH1, EN1, KLF12, KLF3, NFIX, NFYC, YY1, BRD3, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| CDK6 |  |
| KMT2A |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to CDK6-KMT2A |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to CDK6-KMT2A |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CDK6 | C0004238 | Atrial Fibrillation | 2 | CTD_human |
| Hgene | CDK6 | C0025149 | Medulloblastoma | 2 | CTD_human |
| Hgene | CDK6 | C0205833 | Medullomyoblastoma | 2 | CTD_human |
| Hgene | CDK6 | C0235480 | Paroxysmal atrial fibrillation | 2 | CTD_human |
| Hgene | CDK6 | C0278510 | Childhood Medulloblastoma | 2 | CTD_human |
| Hgene | CDK6 | C0278876 | Adult Medulloblastoma | 2 | CTD_human |
| Hgene | CDK6 | C0751291 | Desmoplastic Medulloblastoma | 2 | CTD_human |
| Hgene | CDK6 | C1275668 | Melanotic medulloblastoma | 2 | CTD_human |
| Hgene | CDK6 | C2585653 | Persistent atrial fibrillation | 2 | CTD_human |
| Hgene | CDK6 | C3468561 | familial atrial fibrillation | 2 | CTD_human |
| Hgene | CDK6 | C0002871 | Anemia | 1 | CTD_human |
| Hgene | CDK6 | C0003873 | Rheumatoid Arthritis | 1 | CTD_human |
| Hgene | CDK6 | C0004403 | Autosome Abnormalities | 1 | CTD_human |
| Hgene | CDK6 | C0008625 | Chromosome Aberrations | 1 | CTD_human |
| Hgene | CDK6 | C0017636 | Glioblastoma | 1 | CTD_human |
| Hgene | CDK6 | C0023452 | Childhood Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | CDK6 | C0023453 | L2 Acute Lymphoblastic Leukemia | 1 | CTD_human |
| Hgene | CDK6 | C0023467 | Leukemia, Myelocytic, Acute | 1 | CTD_human |
| Hgene | CDK6 | C0024668 | Mammary Neoplasms, Experimental | 1 | CTD_human |
| Hgene | CDK6 | C0026998 | Acute Myeloid Leukemia, M1 | 1 | CTD_human |
| Hgene | CDK6 | C0038454 | Cerebrovascular accident | 1 | CTD_human |
| Hgene | CDK6 | C0263454 | Chloracne | 1 | CTD_human |
| Hgene | CDK6 | C0334588 | Giant Cell Glioblastoma | 1 | CTD_human |
| Hgene | CDK6 | C0345967 | Malignant mesothelioma | 1 | CTD_human |
| Hgene | CDK6 | C0677866 | Brain Stem Neoplasms | 1 | CTD_human |
| Hgene | CDK6 | C0751886 | Brain Stem Neoplasms, Primary | 1 | CTD_human |
| Hgene | CDK6 | C0751887 | Medullary Neoplasms | 1 | CTD_human |
| Hgene | CDK6 | C0751888 | Mesencephalic Neoplasms | 1 | CTD_human |
| Hgene | CDK6 | C0751889 | Pontine Tumors | 1 | CTD_human |
| Hgene | CDK6 | C0751956 | Acute Cerebrovascular Accidents | 1 | CTD_human |
| Hgene | CDK6 | C1168401 | Squamous cell carcinoma of the head and neck | 1 | CTD_human |
| Hgene | CDK6 | C1621958 | Glioblastoma Multiforme | 1 | CTD_human |
| Hgene | CDK6 | C1879321 | Acute Myeloid Leukemia (AML-M2) | 1 | CTD_human |
| Hgene | CDK6 | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 1 | CTD_human |
| Hgene | CDK6 | C3711387 | Autosomal Recessive Primary Microcephaly | 1 | ORPHANET |
| Hgene | CDK6 | C4015156 | MICROCEPHALY 12, PRIMARY, AUTOSOMAL RECESSIVE | 1 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | KMT2A | C2826025 | Mixed phenotype acute leukemia | 3 | ORPHANET |
| Tgene | KMT2A | C0023418 | leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023452 | Childhood Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023453 | L2 Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023466 | Leukemia, Monocytic, Chronic | 2 | CTD_human |
| Tgene | KMT2A | C0023467 | Leukemia, Myelocytic, Acute | 2 | CTD_human |
| Tgene | KMT2A | C0023470 | Myeloid Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0026998 | Acute Myeloid Leukemia, M1 | 2 | CTD_human |
| Tgene | KMT2A | C1854630 | Growth Deficiency and Mental Retardation with Facial Dysmorphism | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | KMT2A | C1879321 | Acute Myeloid Leukemia (AML-M2) | 2 | CTD_human |
| Tgene | KMT2A | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 2 | CTD_human |
| Tgene | KMT2A | C0001418 | Adenocarcinoma | 1 | CTD_human |
| Tgene | KMT2A | C0004403 | Autosome Abnormalities | 1 | CTD_human |
| Tgene | KMT2A | C0005684 | Malignant neoplasm of urinary bladder | 1 | CTD_human |
| Tgene | KMT2A | C0005695 | Bladder Neoplasm | 1 | CTD_human |
| Tgene | KMT2A | C0007138 | Carcinoma, Transitional Cell | 1 | CTD_human |
| Tgene | KMT2A | C0008625 | Chromosome Aberrations | 1 | CTD_human |
| Tgene | KMT2A | C0023448 | Lymphoid leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0023465 | Acute monocytic leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0023479 | Acute myelomonocytic leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human |
| Tgene | KMT2A | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Tgene | KMT2A | C0036341 | Schizophrenia | 1 | PSYGENET |
| Tgene | KMT2A | C0038356 | Stomach Neoplasms | 1 | CTD_human |
| Tgene | KMT2A | C0149925 | Small cell carcinoma of lung | 1 | CTD_human |
| Tgene | KMT2A | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| Tgene | KMT2A | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| Tgene | KMT2A | C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| Tgene | KMT2A | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| Tgene | KMT2A | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| Tgene | KMT2A | C0270972 | Cornelia De Lange Syndrome | 1 | ORPHANET |
| Tgene | KMT2A | C0280141 | Acute Undifferentiated Leukemia | 1 | ORPHANET |
| Tgene | KMT2A | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| Tgene | KMT2A | C0856823 | Undifferentiated type acute leukemia | 1 | ORPHANET |
| Tgene | KMT2A | C1535926 | Neurodevelopmental Disorders | 1 | CTD_human |
| Tgene | KMT2A | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human |
| Tgene | KMT2A | C2239176 | Liver carcinoma | 1 | CTD_human |
| Tgene | KMT2A | C2930974 | Acute erythroleukemia | 1 | CTD_human |
| Tgene | KMT2A | C2930975 | Acute erythroleukemia - M6a subtype | 1 | CTD_human |
| Tgene | KMT2A | C2930976 | Acute myeloid leukemia FAB-M6 | 1 | CTD_human |
| Tgene | KMT2A | C2930977 | Acute erythroleukemia - M6b subtype | 1 | CTD_human |

