| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:CHST11-HSP90B1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: CHST11-HSP90B1 | FusionPDB ID: 16683 | FusionGDB2.0 ID: 16683 | Hgene | Tgene | Gene symbol | CHST11 | HSP90B1 | Gene ID | 50515 | 7184 |
| Gene name | carbohydrate sulfotransferase 11 | heat shock protein 90 beta family member 1 | |
| Synonyms | C4ST|C4ST-1|C4ST1|HSA269537|OCBMD | ECGP|GP96|GRP94|HEL-S-125m|HEL35|TRA1 | |
| Cytomap | 12q23.3 | 12q23.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | carbohydrate sulfotransferase 11C4S-1IgH/CHST11 fusioncarbohydrate (chondroitin 4) sulfotransferase 11chondroitin 4-O-sulfotransferase 1 | endoplasmin94 kDa glucose-regulated proteinendothelial cell (HBMEC) glycoproteinepididymis luminal protein 35epididymis secretory sperm binding protein Li 125mheat shock protein 90 kDa beta member 1heat shock protein 90kDa beta (Grp94), member 1hea | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q9NPF2 | P14625 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000303694, ENST00000546689, ENST00000547956, ENST00000549260, ENST00000550711, | ENST00000299767, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 13 X 8 X 6=624 | 21 X 21 X 8=3528 |
| # samples | 15 | 25 | |
| ** MAII score | log2(15/624*10)=-2.05658352836637 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(25/3528*10)=-3.81885056089543 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: CHST11 [Title/Abstract] AND HSP90B1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | CHST11(104995769)-HSP90B1(104333287), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | CHST11-HSP90B1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CHST11-HSP90B1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CHST11 | GO:0030206 | chondroitin sulfate biosynthetic process | 11056388 |
| Tgene | HSP90B1 | GO:0001666 | response to hypoxia | 15620698 |
| Tgene | HSP90B1 | GO:0031247 | actin rod assembly | 19000834 |
| Tgene | HSP90B1 | GO:0043666 | regulation of phosphoprotein phosphatase activity | 19000834 |
| Tgene | HSP90B1 | GO:0071318 | cellular response to ATP | 19000834 |
 Fusion gene breakpoints across CHST11 (5'-gene) Fusion gene breakpoints across CHST11 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
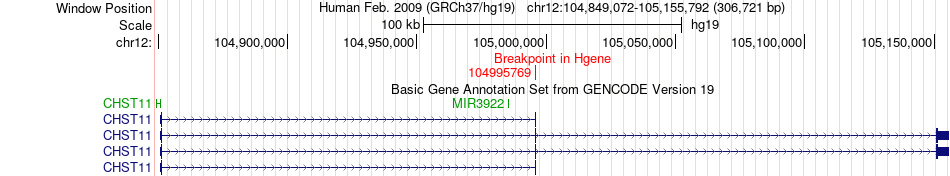 |
 Fusion gene breakpoints across HSP90B1 (3'-gene) Fusion gene breakpoints across HSP90B1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
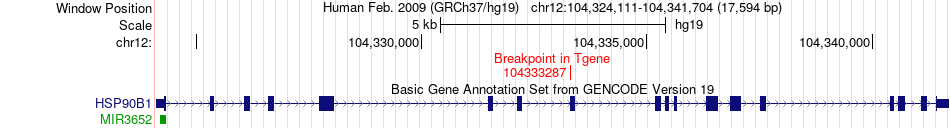 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-23-1028-01A | CHST11 | chr12 | 104995769 | + | HSP90B1 | chr12 | 104333287 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000547956 | CHST11 | chr12 | 104995769 | + | ENST00000299767 | HSP90B1 | chr12 | 104333287 | + | 2551 | 850 | 814 | 2286 | 490 |
| ENST00000549260 | CHST11 | chr12 | 104995769 | + | ENST00000299767 | HSP90B1 | chr12 | 104333287 | + | 2331 | 630 | 12 | 2066 | 684 |
| ENST00000303694 | CHST11 | chr12 | 104995769 | + | ENST00000299767 | HSP90B1 | chr12 | 104333287 | + | 2344 | 643 | 10 | 2079 | 689 |
| ENST00000546689 | CHST11 | chr12 | 104995769 | + | ENST00000299767 | HSP90B1 | chr12 | 104333287 | + | 2371 | 670 | 700 | 2106 | 468 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000547956 | ENST00000299767 | CHST11 | chr12 | 104995769 | + | HSP90B1 | chr12 | 104333287 | + | 0.000506149 | 0.99949384 |
| ENST00000549260 | ENST00000299767 | CHST11 | chr12 | 104995769 | + | HSP90B1 | chr12 | 104333287 | + | 0.001145799 | 0.99885416 |
| ENST00000303694 | ENST00000299767 | CHST11 | chr12 | 104995769 | + | HSP90B1 | chr12 | 104333287 | + | 0.001691926 | 0.99830806 |
| ENST00000546689 | ENST00000299767 | CHST11 | chr12 | 104995769 | + | HSP90B1 | chr12 | 104333287 | + | 0.000510725 | 0.9994893 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >16683_16683_1_CHST11-HSP90B1_CHST11_chr12_104995769_ENST00000303694_HSP90B1_chr12_104333287_ENST00000299767_length(amino acids)=689AA_BP=210 MERNEVAFGEKGEGPSDSHSGQRSGSGGGTTITARTPAARPATRQRRPPAGSEEAAERRGGGAGARSQRVHASQHFQTNSGTFHTPARAG GSESGREATPILPLSLAQLCPAPPGLRSARRGPCSCAPGALPGHPGPRSQDKAMKPALLEVMRMNRICRMVLATCLGSFILVIFYFQSML HPVMRRNPFGVDICCRKGSRSPLQELYNPIQVEKTVWDWELMNDIKPIWQRPSKEVEEDEYKAFYKSFSKESDDPMAYIHFTAEGEVTFK SILFVPTSAPRGLFDEYGSKKSDYIKLYVRRVFITDDFHDMMPKYLNFVKGVVDSDDLPLNVSRETLQQHKLLKVIRKKLVRKTLDMIKK IADDKYNDTFWKEFGTNIKLGVIEDHSNRTRLAKLLRFQSSHHPTDITSLDQYVERMKEKQDKIYFMAGSSRKEAESSPFVERLLKKGYE VIYLTEPVDEYCIQALPEFDGKRFQNVAKEGVKFDESEKTKESREAVEKEFEPLLNWMKDKALKDKIEKAVVSQRLTESPCALVASQYGW SGNMERIMKAQAYQTGKDISTNYYASQKKTFEINPRHPLIRDMLRRIKEDEDDKTVLDLAVVLFETATLRSGYLLPDTKAYGDRIERMLR -------------------------------------------------------------- >16683_16683_2_CHST11-HSP90B1_CHST11_chr12_104995769_ENST00000546689_HSP90B1_chr12_104333287_ENST00000299767_length(amino acids)=468AA_BP= MNDIKPIWQRPSKEVEEDEYKAFYKSFSKESDDPMAYIHFTAEGEVTFKSILFVPTSAPRGLFDEYGSKKSDYIKLYVRRVFITDDFHDM MPKYLNFVKGVVDSDDLPLNVSRETLQQHKLLKVIRKKLVRKTLDMIKKIADDKYNDTFWKEFGTNIKLGVIEDHSNRTRLAKLLRFQSS HHPTDITSLDQYVERMKEKQDKIYFMAGSSRKEAESSPFVERLLKKGYEVIYLTEPVDEYCIQALPEFDGKRFQNVAKEGVKFDESEKTK ESREAVEKEFEPLLNWMKDKALKDKIEKAVVSQRLTESPCALVASQYGWSGNMERIMKAQAYQTGKDISTNYYASQKKTFEINPRHPLIR DMLRRIKEDEDDKTVLDLAVVLFETATLRSGYLLPDTKAYGDRIERMLRLSLNIDPDAKVEEEPEEEPEETAEDTTEDTEQDEDEEMDVG -------------------------------------------------------------- >16683_16683_3_CHST11-HSP90B1_CHST11_chr12_104995769_ENST00000547956_HSP90B1_chr12_104333287_ENST00000299767_length(amino acids)=490AA_BP=12 MRLTVSSSSNGTVEKTVWDWELMNDIKPIWQRPSKEVEEDEYKAFYKSFSKESDDPMAYIHFTAEGEVTFKSILFVPTSAPRGLFDEYGS KKSDYIKLYVRRVFITDDFHDMMPKYLNFVKGVVDSDDLPLNVSRETLQQHKLLKVIRKKLVRKTLDMIKKIADDKYNDTFWKEFGTNIK LGVIEDHSNRTRLAKLLRFQSSHHPTDITSLDQYVERMKEKQDKIYFMAGSSRKEAESSPFVERLLKKGYEVIYLTEPVDEYCIQALPEF DGKRFQNVAKEGVKFDESEKTKESREAVEKEFEPLLNWMKDKALKDKIEKAVVSQRLTESPCALVASQYGWSGNMERIMKAQAYQTGKDI STNYYASQKKTFEINPRHPLIRDMLRRIKEDEDDKTVLDLAVVLFETATLRSGYLLPDTKAYGDRIERMLRLSLNIDPDAKVEEEPEEEP -------------------------------------------------------------- >16683_16683_4_CHST11-HSP90B1_CHST11_chr12_104995769_ENST00000549260_HSP90B1_chr12_104333287_ENST00000299767_length(amino acids)=684AA_BP=205 MERNEVAFGEKGEGPSDSHSGQRSGSGGGTTITARTPAARPATRQRRPPAGSEEAAERRGGGAGARSQRVHASQHFQTNSGTFHTPARAG GSESGREATPILPLSLAQLCPAPPGLRSARRGPCSCAPGALPGHPGPRSQDKAMKPALLEVMRMNRICRMVLATCLGSFILVIFYFQIMR RNPFGVDICCRKGSRSPLQELYNPIQVEKTVWDWELMNDIKPIWQRPSKEVEEDEYKAFYKSFSKESDDPMAYIHFTAEGEVTFKSILFV PTSAPRGLFDEYGSKKSDYIKLYVRRVFITDDFHDMMPKYLNFVKGVVDSDDLPLNVSRETLQQHKLLKVIRKKLVRKTLDMIKKIADDK YNDTFWKEFGTNIKLGVIEDHSNRTRLAKLLRFQSSHHPTDITSLDQYVERMKEKQDKIYFMAGSSRKEAESSPFVERLLKKGYEVIYLT EPVDEYCIQALPEFDGKRFQNVAKEGVKFDESEKTKESREAVEKEFEPLLNWMKDKALKDKIEKAVVSQRLTESPCALVASQYGWSGNME RIMKAQAYQTGKDISTNYYASQKKTFEINPRHPLIRDMLRRIKEDEDDKTVLDLAVVLFETATLRSGYLLPDTKAYGDRIERMLRLSLNI -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:104995769/chr12:104333287) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CHST11 | HSP90B1 |
| FUNCTION: Catalyzes the transfer of sulfate to position 4 of the N-acetylgalactosamine (GalNAc) residue of chondroitin. Chondroitin sulfate constitutes the predominant proteoglycan present in cartilage and is distributed on the surfaces of many cells and extracellular matrices. Can also sulfate Gal residues in desulfated dermatan sulfate. Preferentially sulfates in GlcA->GalNAc unit than in IdoA->GalNAc unit. Does not form 4, 6-di-O-sulfated GalNAc when chondroitin sulfate C is used as an acceptor. | FUNCTION: Molecular chaperone that functions in the processing and transport of secreted proteins (By similarity). When associated with CNPY3, required for proper folding of Toll-like receptors (By similarity). Functions in endoplasmic reticulum associated degradation (ERAD) (PubMed:18264092). Has ATPase activity (By similarity). May participate in the unfolding of cytosolic leaderless cargos (lacking the secretion signal sequence) such as the interleukin 1/IL-1 to facilitate their translocation into the ERGIC (endoplasmic reticulum-Golgi intermediate compartment) and secretion; the translocation process is mediated by the cargo receptor TMED10 (PubMed:32272059). {ECO:0000250|UniProtKB:P08113, ECO:0000269|PubMed:18264092, ECO:0000269|PubMed:32272059}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHST11 | chr12:104995769 | chr12:104333287 | ENST00000303694 | + | 2 | 3 | 1_16 | 68.0 | 353.0 | Topological domain | Cytoplasmic |
| Hgene | CHST11 | chr12:104995769 | chr12:104333287 | ENST00000549260 | + | 2 | 3 | 1_16 | 63.0 | 348.0 | Topological domain | Cytoplasmic |
| Hgene | CHST11 | chr12:104995769 | chr12:104333287 | ENST00000303694 | + | 2 | 3 | 17_37 | 68.0 | 353.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | CHST11 | chr12:104995769 | chr12:104333287 | ENST00000549260 | + | 2 | 3 | 17_37 | 63.0 | 348.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Tgene | HSP90B1 | chr12:104995769 | chr12:104333287 | ENST00000299767 | 6 | 18 | 800_803 | 325.0 | 804.0 | Motif | Note=Prevents secretion from ER |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CHST11 | chr12:104995769 | chr12:104333287 | ENST00000303694 | + | 2 | 3 | 124_130 | 68.0 | 353.0 | Nucleotide binding | PAPS |
| Hgene | CHST11 | chr12:104995769 | chr12:104333287 | ENST00000303694 | + | 2 | 3 | 186_194 | 68.0 | 353.0 | Nucleotide binding | PAPS |
| Hgene | CHST11 | chr12:104995769 | chr12:104333287 | ENST00000549260 | + | 2 | 3 | 124_130 | 63.0 | 348.0 | Nucleotide binding | PAPS |
| Hgene | CHST11 | chr12:104995769 | chr12:104333287 | ENST00000549260 | + | 2 | 3 | 186_194 | 63.0 | 348.0 | Nucleotide binding | PAPS |
| Hgene | CHST11 | chr12:104995769 | chr12:104333287 | ENST00000303694 | + | 2 | 3 | 38_352 | 68.0 | 353.0 | Topological domain | Lumenal |
| Hgene | CHST11 | chr12:104995769 | chr12:104333287 | ENST00000549260 | + | 2 | 3 | 38_352 | 63.0 | 348.0 | Topological domain | Lumenal |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| CHST11 | |
| HSP90B1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to CHST11-HSP90B1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to CHST11-HSP90B1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

