| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:CLTC-ROS1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: CLTC-ROS1 | FusionPDB ID: 17408 | FusionGDB2.0 ID: 17408 | Hgene | Tgene | Gene symbol | CLTC | ROS1 | Gene ID | 1213 | 6098 |
| Gene name | clathrin heavy chain | ROS proto-oncogene 1, receptor tyrosine kinase | |
| Synonyms | CHC|CHC17|CLH-17|CLTCL2|Hc|MRD56 | MCF3|ROS|c-ros-1 | |
| Cytomap | 17q23.1 | 6q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | clathrin heavy chain 1clathrin heavy chain on chromosome 17clathrin, heavy polypeptide (Hc)clathrin, heavy polypeptide-like 2 | proto-oncogene tyrosine-protein kinase ROSROS proto-oncogene 1 , receptor tyrosine kinasec-ros oncogene 1 , receptor tyrosine kinaseproto-oncogene c-Ros-1transmembrane tyrosine-specific protein kinasev-ros avian UR2 sarcoma virus oncogene homolog 1 | |
| Modification date | 20200313 | 20200314 | |
| UniProtAcc | P53675 | P08922 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000393043, ENST00000269122, ENST00000579456, ENST00000579815, | ENST00000368507, ENST00000368508, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 27 X 40 X 17=18360 | 18 X 20 X 5=1800 |
| # samples | 84 | 20 | |
| ** MAII score | log2(84/18360*10)=-4.45003292063505 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(20/1800*10)=-3.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: CLTC [Title/Abstract] AND ROS1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | CLTC(57768072)-ROS1(117642557), # samples:4 ROS1(117609849)-CLTC(57771089), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | CLTC-ROS1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CLTC-ROS1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. CLTC-ROS1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CLTC-ROS1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. CLTC-ROS1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. CLTC-ROS1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. CLTC-ROS1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. CLTC-ROS1 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. CLTC-ROS1 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | CLTC | GO:1900126 | negative regulation of hyaluronan biosynthetic process | 24251095 |
| Tgene | ROS1 | GO:0001558 | regulation of cell growth | 16885344 |
| Tgene | ROS1 | GO:0006468 | protein phosphorylation | 16885344 |
| Tgene | ROS1 | GO:0032006 | regulation of TOR signaling | 16885344 |
 Fusion gene breakpoints across CLTC (5'-gene) Fusion gene breakpoints across CLTC (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
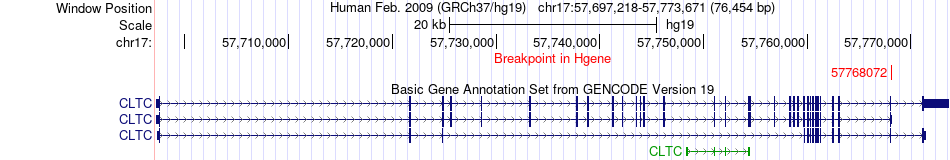 |
 Fusion gene breakpoints across ROS1 (3'-gene) Fusion gene breakpoints across ROS1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
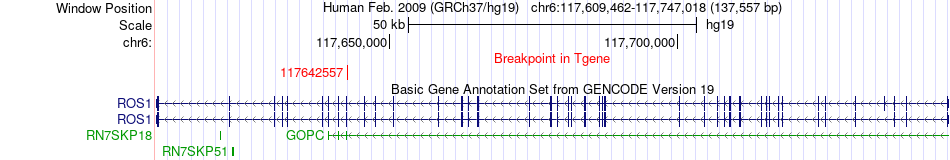 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-44-2665-01A | CLTC | chr17 | 57768072 | - | ROS1 | chr6 | 117642557 | - |
| ChimerDB4 | LUAD | TCGA-44-2665-01A | CLTC | chr17 | 57768072 | + | ROS1 | chr6 | 117642557 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000269122 | CLTC | chr17 | 57768072 | + | ENST00000368508 | ROS1 | chr6 | 117642557 | - | 6772 | 5177 | 274 | 6579 | 2101 |
| ENST00000269122 | CLTC | chr17 | 57768072 | + | ENST00000368507 | ROS1 | chr6 | 117642557 | - | 6772 | 5177 | 274 | 6579 | 2101 |
| ENST00000579456 | CLTC | chr17 | 57768072 | + | ENST00000368508 | ROS1 | chr6 | 117642557 | - | 3509 | 1914 | 200 | 3316 | 1038 |
| ENST00000579456 | CLTC | chr17 | 57768072 | + | ENST00000368507 | ROS1 | chr6 | 117642557 | - | 3509 | 1914 | 200 | 3316 | 1038 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000269122 | ENST00000368508 | CLTC | chr17 | 57768072 | + | ROS1 | chr6 | 117642557 | - | 0.000596105 | 0.99940395 |
| ENST00000269122 | ENST00000368507 | CLTC | chr17 | 57768072 | + | ROS1 | chr6 | 117642557 | - | 0.000596105 | 0.99940395 |
| ENST00000579456 | ENST00000368508 | CLTC | chr17 | 57768072 | + | ROS1 | chr6 | 117642557 | - | 0.001745022 | 0.99825495 |
| ENST00000579456 | ENST00000368507 | CLTC | chr17 | 57768072 | + | ROS1 | chr6 | 117642557 | - | 0.001745022 | 0.99825495 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >17408_17408_1_CLTC-ROS1_CLTC_chr17_57768072_ENST00000269122_ROS1_chr6_117642557_ENST00000368507_length(amino acids)=2101AA_BP=1634 MAQILPIRFQEHLQLQNLGINPANIGFSTLTMESDKFICIREKVGEQAQVVIIDMNDPSNPIRRPISADSAIMNPASKVIALKAGKTLQI FNIEMKSKMKAHTMTDDVTFWKWISLNTVALVTDNAVYHWSMEGESQPVKMFDRHSSLAGCQIINYRTDAKQKWLLLTGISAQQNRVVGA MQLYSVDRKVSQPIEGHAASFAQFKMEGNAEESTLFCFAVRGQAGGKLHIIEVGTPPTGNQPFPKKAVDVFFPPEAQNDFPVAMQISEKH DVVFLITKYGYIHLYDLETGTCIYMNRISGETIFVTAPHEATAGIIGVNRKGQVLSVCVEEENIIPYITNVLQNPDLALRMAVRNNLAGA EELFARKFNALFAQGNYSEAAKVAANAPKGILRTPDTIRRFQSVPAQPGQTSPLLQYFGILLDQGQLNKYESLELCRPVLQQGRKQLLEK WLKEDKLECSEELGDLVKSVDPTLALSVYLRANVPNKVIQCFAETGQVQKIVLYAKKVGYTPDWIFLLRNVMRISPDQGQQFAQMLVQDE EPLADITQIVDVFMEYNLIQQCTAFLLDALKNNRPSEGPLQTRLLEMNLMHAPQVADAILGNQMFTHYDRAHIAQLCEKAGLLQRALEHF TDLYDIKRAVVHTHLLNPEWLVNYFGSLSVEDSLECLRAMLSANIRQNLQICVQVASKYHEQLSTQSLIELFESFKSFEGLFYFLGSIVN FSQDPDVHFKYIQAACKTGQIKEVERICRESNCYDPERVKNFLKEAKLTDQLPLIIVCDRFDFVHDLVLYLYRNNLQKYIEIYVQKVNPS RLPVVIGGLLDVDCSEDVIKNLILVVRGQFSTDELVAEVEKRNRLKLLLPWLEARIHEGCEEPATHNALAKIYIDSNNNPERFLRENPYY DSRVVGKYCEKRDPHLACVAYERGQCDLELINVCNENSLFKSLSRYLVRRKDPELWGSVLLESNPYRRPLIDQVVQTALSETQDPEEVSV TVKAFMTADLPNELIELLEKIVLDNSVFSEHRNLQNLLILTAIKADRTRVMEYINRLDNYDAPDIANIAISNELFEEAFAIFRKFDVNTS AVQVLIEHIGNLDRAYEFAERCNEPAVWSQLAKAQLQKGMVKEAIDSYIKADDPSSYMEVVQAANTSGNWEELVKYLQMARKKARESYVE TELIFALAKTNRLAELEEFINGPNNAHIQQVGDRCYDEKMYDAAKLLYNNVSNFGRLASTLVHLGEYQAAVDGARKANSTRTWKEVCFAC VDGKEFRLAQMCGLHIVVHADELEELINYYQDRGYFEELITMLEAALGLERAHMGMFTELAILYSKFKPQKMREHLELFWSRVNIPKVLR AAEQAHLWAELVFLYDKYEEYDNAIITMMNHPTDAWKEGQFKDIITKVANVELYYRAIQFYLEFKPLLLNDLLMVLSPRLDHTRAVNYFS KVKQLPLVKPYLRSVQNHNNKSVNESLNNLFITEEDYQALRTSIDAYDNFDNISLAQRLEKHELIEFRRIAAYLFKGNNRWKQSVELCKK DSLYKDAMQYASESKDTELAEELLQWFLQEEKRECFGACLFTCYDLLRPDVVLETAWRHNIMDFAMPYFIQVMKEYLTKVDKLDASESLR KEEEQATETQPIVYVWHRRLKNQKSAKEGVTVLINEDKELAELRGLAAGVGLANACYAIHTLPTQEEIENLPAFPREKLTLRLLLGSGAF GEVYEGTAVDILGVGSGEIKVAVKTLKKGSTDQEKIEFLKEAHLMSKFNHPNILKQLGVCLLNEPQYIILELMEGGDLLTYLRKARMATF YGPLLTLVDLVDLCVDISKGCVYLERMHFIHRDLAARNCLVSVKDYTSPRIVKIGDFGLARDIYKNDYYRKRGEGLLPVRWMAPESLMDG IFTTQSDVWSFGILIWEILTLGHQPYPAHSNLDVLNYVQTGGRLEPPRNCPDDLWNLMTQCWAQEPDQRPTFHRIQDQLQLFRNFFLNSI YKSRDEANNSGVINESFEGEDGDVICLNSDDIMPVALMETKNREGLNYMVLATECGQGEEKSEGPLGSQESESCGLRKEEKEPHADKDFC -------------------------------------------------------------- >17408_17408_2_CLTC-ROS1_CLTC_chr17_57768072_ENST00000269122_ROS1_chr6_117642557_ENST00000368508_length(amino acids)=2101AA_BP=1634 MAQILPIRFQEHLQLQNLGINPANIGFSTLTMESDKFICIREKVGEQAQVVIIDMNDPSNPIRRPISADSAIMNPASKVIALKAGKTLQI FNIEMKSKMKAHTMTDDVTFWKWISLNTVALVTDNAVYHWSMEGESQPVKMFDRHSSLAGCQIINYRTDAKQKWLLLTGISAQQNRVVGA MQLYSVDRKVSQPIEGHAASFAQFKMEGNAEESTLFCFAVRGQAGGKLHIIEVGTPPTGNQPFPKKAVDVFFPPEAQNDFPVAMQISEKH DVVFLITKYGYIHLYDLETGTCIYMNRISGETIFVTAPHEATAGIIGVNRKGQVLSVCVEEENIIPYITNVLQNPDLALRMAVRNNLAGA EELFARKFNALFAQGNYSEAAKVAANAPKGILRTPDTIRRFQSVPAQPGQTSPLLQYFGILLDQGQLNKYESLELCRPVLQQGRKQLLEK WLKEDKLECSEELGDLVKSVDPTLALSVYLRANVPNKVIQCFAETGQVQKIVLYAKKVGYTPDWIFLLRNVMRISPDQGQQFAQMLVQDE EPLADITQIVDVFMEYNLIQQCTAFLLDALKNNRPSEGPLQTRLLEMNLMHAPQVADAILGNQMFTHYDRAHIAQLCEKAGLLQRALEHF TDLYDIKRAVVHTHLLNPEWLVNYFGSLSVEDSLECLRAMLSANIRQNLQICVQVASKYHEQLSTQSLIELFESFKSFEGLFYFLGSIVN FSQDPDVHFKYIQAACKTGQIKEVERICRESNCYDPERVKNFLKEAKLTDQLPLIIVCDRFDFVHDLVLYLYRNNLQKYIEIYVQKVNPS RLPVVIGGLLDVDCSEDVIKNLILVVRGQFSTDELVAEVEKRNRLKLLLPWLEARIHEGCEEPATHNALAKIYIDSNNNPERFLRENPYY DSRVVGKYCEKRDPHLACVAYERGQCDLELINVCNENSLFKSLSRYLVRRKDPELWGSVLLESNPYRRPLIDQVVQTALSETQDPEEVSV TVKAFMTADLPNELIELLEKIVLDNSVFSEHRNLQNLLILTAIKADRTRVMEYINRLDNYDAPDIANIAISNELFEEAFAIFRKFDVNTS AVQVLIEHIGNLDRAYEFAERCNEPAVWSQLAKAQLQKGMVKEAIDSYIKADDPSSYMEVVQAANTSGNWEELVKYLQMARKKARESYVE TELIFALAKTNRLAELEEFINGPNNAHIQQVGDRCYDEKMYDAAKLLYNNVSNFGRLASTLVHLGEYQAAVDGARKANSTRTWKEVCFAC VDGKEFRLAQMCGLHIVVHADELEELINYYQDRGYFEELITMLEAALGLERAHMGMFTELAILYSKFKPQKMREHLELFWSRVNIPKVLR AAEQAHLWAELVFLYDKYEEYDNAIITMMNHPTDAWKEGQFKDIITKVANVELYYRAIQFYLEFKPLLLNDLLMVLSPRLDHTRAVNYFS KVKQLPLVKPYLRSVQNHNNKSVNESLNNLFITEEDYQALRTSIDAYDNFDNISLAQRLEKHELIEFRRIAAYLFKGNNRWKQSVELCKK DSLYKDAMQYASESKDTELAEELLQWFLQEEKRECFGACLFTCYDLLRPDVVLETAWRHNIMDFAMPYFIQVMKEYLTKVDKLDASESLR KEEEQATETQPIVYVWHRRLKNQKSAKEGVTVLINEDKELAELRGLAAGVGLANACYAIHTLPTQEEIENLPAFPREKLTLRLLLGSGAF GEVYEGTAVDILGVGSGEIKVAVKTLKKGSTDQEKIEFLKEAHLMSKFNHPNILKQLGVCLLNEPQYIILELMEGGDLLTYLRKARMATF YGPLLTLVDLVDLCVDISKGCVYLERMHFIHRDLAARNCLVSVKDYTSPRIVKIGDFGLARDIYKNDYYRKRGEGLLPVRWMAPESLMDG IFTTQSDVWSFGILIWEILTLGHQPYPAHSNLDVLNYVQTGGRLEPPRNCPDDLWNLMTQCWAQEPDQRPTFHRIQDQLQLFRNFFLNSI YKSRDEANNSGVINESFEGEDGDVICLNSDDIMPVALMETKNREGLNYMVLATECGQGEEKSEGPLGSQESESCGLRKEEKEPHADKDFC -------------------------------------------------------------- >17408_17408_3_CLTC-ROS1_CLTC_chr17_57768072_ENST00000579456_ROS1_chr6_117642557_ENST00000368507_length(amino acids)=1038AA_BP=571 MAQILPIRFQEHLQLQNLGINPANIGFSTLTMESDKFICIREKVGEQAQVVIIDMNDPSNPIRRPISADSAIMNPASKVIALKAGKTLQI FNIEMKSKMKARESYVETELIFALAKTNRLAELEEFINGPNNAHIQQVGDRCYDEKMYDAAKLLYNNVSNFGRLASTLVHLGEYQAAVDG ARKANSTRTWKEVCFACVDGKEFRLAQMCGLHIVVHADELEELINYYQDRGYFEELITMLEAALGLERAHMGMFTELAILYSKFKPQKMR EHLELFWSRVNIPKVLRAAEQAHLWAELVFLYDKYEEYDNAIITMMNHPTDAWKEGQFKDIITKVANVELYYRAIQFYLEFKPLLLNDLL MVLSPRLDHTRAVNYFSKVKQLPLVKPYLRSVQNHNNKSVNESLNNLFITEEDYQALRTSIDAYDNFDNISLAQRLEKHELIEFRRIAAY LFKGNNRWKQSVELCKKDSLYKDAMQYASESKDTELAEELLQWFLQEEKRECFGACLFTCYDLLRPDVVLETAWRHNIMDFAMPYFIQVM KEYLTKVDKLDASESLRKEEEQATETQPIVYVWHRRLKNQKSAKEGVTVLINEDKELAELRGLAAGVGLANACYAIHTLPTQEEIENLPA FPREKLTLRLLLGSGAFGEVYEGTAVDILGVGSGEIKVAVKTLKKGSTDQEKIEFLKEAHLMSKFNHPNILKQLGVCLLNEPQYIILELM EGGDLLTYLRKARMATFYGPLLTLVDLVDLCVDISKGCVYLERMHFIHRDLAARNCLVSVKDYTSPRIVKIGDFGLARDIYKNDYYRKRG EGLLPVRWMAPESLMDGIFTTQSDVWSFGILIWEILTLGHQPYPAHSNLDVLNYVQTGGRLEPPRNCPDDLWNLMTQCWAQEPDQRPTFH RIQDQLQLFRNFFLNSIYKSRDEANNSGVINESFEGEDGDVICLNSDDIMPVALMETKNREGLNYMVLATECGQGEEKSEGPLGSQESES -------------------------------------------------------------- >17408_17408_4_CLTC-ROS1_CLTC_chr17_57768072_ENST00000579456_ROS1_chr6_117642557_ENST00000368508_length(amino acids)=1038AA_BP=571 MAQILPIRFQEHLQLQNLGINPANIGFSTLTMESDKFICIREKVGEQAQVVIIDMNDPSNPIRRPISADSAIMNPASKVIALKAGKTLQI FNIEMKSKMKARESYVETELIFALAKTNRLAELEEFINGPNNAHIQQVGDRCYDEKMYDAAKLLYNNVSNFGRLASTLVHLGEYQAAVDG ARKANSTRTWKEVCFACVDGKEFRLAQMCGLHIVVHADELEELINYYQDRGYFEELITMLEAALGLERAHMGMFTELAILYSKFKPQKMR EHLELFWSRVNIPKVLRAAEQAHLWAELVFLYDKYEEYDNAIITMMNHPTDAWKEGQFKDIITKVANVELYYRAIQFYLEFKPLLLNDLL MVLSPRLDHTRAVNYFSKVKQLPLVKPYLRSVQNHNNKSVNESLNNLFITEEDYQALRTSIDAYDNFDNISLAQRLEKHELIEFRRIAAY LFKGNNRWKQSVELCKKDSLYKDAMQYASESKDTELAEELLQWFLQEEKRECFGACLFTCYDLLRPDVVLETAWRHNIMDFAMPYFIQVM KEYLTKVDKLDASESLRKEEEQATETQPIVYVWHRRLKNQKSAKEGVTVLINEDKELAELRGLAAGVGLANACYAIHTLPTQEEIENLPA FPREKLTLRLLLGSGAFGEVYEGTAVDILGVGSGEIKVAVKTLKKGSTDQEKIEFLKEAHLMSKFNHPNILKQLGVCLLNEPQYIILELM EGGDLLTYLRKARMATFYGPLLTLVDLVDLCVDISKGCVYLERMHFIHRDLAARNCLVSVKDYTSPRIVKIGDFGLARDIYKNDYYRKRG EGLLPVRWMAPESLMDGIFTTQSDVWSFGILIWEILTLGHQPYPAHSNLDVLNYVQTGGRLEPPRNCPDDLWNLMTQCWAQEPDQRPTFH RIQDQLQLFRNFFLNSIYKSRDEANNSGVINESFEGEDGDVICLNSDDIMPVALMETKNREGLNYMVLATECGQGEEKSEGPLGSQESES -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:57768072/chr6:117642557) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| CLTC | ROS1 |
| FUNCTION: Clathrin is the major protein of the polyhedral coat of coated pits and vesicles. Two different adapter protein complexes link the clathrin lattice either to the plasma membrane or to the trans-Golgi network (By similarity). {ECO:0000250}. | FUNCTION: Orphan receptor tyrosine kinase (RTK) that plays a role in epithelial cell differentiation and regionalization of the proximal epididymal epithelium. May activate several downstream signaling pathways related to cell differentiation, proliferation, growth and survival including the PI3 kinase-mTOR signaling pathway. Mediates the phosphorylation of PTPN11, an activator of this pathway. May also phosphorylate and activate the transcription factor STAT3 to control anchorage-independent cell growth. Mediates the phosphorylation and the activation of VAV3, a guanine nucleotide exchange factor regulating cell morphology. May activate other downstream signaling proteins including AKT1, MAPK1, MAPK3, IRS1 and PLCG2. {ECO:0000269|PubMed:11094073, ECO:0000269|PubMed:16885344}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 108_149 | 1634.3333333333333 | 1676.0 | Region | Note=WD40-like repeat 3 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 1213_1522 | 1634.3333333333333 | 1676.0 | Region | Involved in binding clathrin light chain |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 150_195 | 1634.3333333333333 | 1676.0 | Region | Note=WD40-like repeat 4 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 196_257 | 1634.3333333333333 | 1676.0 | Region | Note=WD40-like repeat 5 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 24_67 | 1634.3333333333333 | 1676.0 | Region | Note=WD40-like repeat 1 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 258_301 | 1634.3333333333333 | 1676.0 | Region | Note=WD40-like repeat 6 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 2_479 | 1634.3333333333333 | 1676.0 | Region | Note=Globular terminal domain |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 302_330 | 1634.3333333333333 | 1676.0 | Region | Note=WD40-like repeat 7 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 449_465 | 1634.3333333333333 | 1676.0 | Region | Binding site for the uncoating ATPase%2C involved in lattice disassembly |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 480_523 | 1634.3333333333333 | 1676.0 | Region | Note=Flexible linker |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 524_634 | 1634.3333333333333 | 1676.0 | Region | Note=Distal segment |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 68_107 | 1634.3333333333333 | 1676.0 | Region | Note=WD40-like repeat 2 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 1128_1269 | 1634.3333333333333 | 1676.0 | Repeat | CHCR 5 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 1274_1420 | 1634.3333333333333 | 1676.0 | Repeat | CHCR 6 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 1423_1566 | 1634.3333333333333 | 1676.0 | Repeat | CHCR 7 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 537_683 | 1634.3333333333333 | 1676.0 | Repeat | CHCR 1 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 686_828 | 1634.3333333333333 | 1676.0 | Repeat | CHCR 2 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 833_972 | 1634.3333333333333 | 1676.0 | Repeat | CHCR 3 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 979_1124 | 1634.3333333333333 | 1676.0 | Repeat | CHCR 4 |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 1945_2222 | 1880.3333333333333 | 2348.0 | Domain | Protein kinase | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 1951_1959 | 1880.3333333333333 | 2348.0 | Nucleotide binding | ATP | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 1883_2347 | 1880.3333333333333 | 2348.0 | Topological domain | Cytoplasmic |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 1550_1675 | 1634.3333333333333 | 1676.0 | Region | Trimerization |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 524_1675 | 1634.3333333333333 | 1676.0 | Region | Note=Heavy chain arm |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000269122 | + | 31 | 32 | 639_1675 | 1634.3333333333333 | 1676.0 | Region | Note=Proximal segment |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 108_149 | 0 | 1640.0 | Region | Note=WD40-like repeat 3 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 1213_1522 | 0 | 1640.0 | Region | Involved in binding clathrin light chain |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 150_195 | 0 | 1640.0 | Region | Note=WD40-like repeat 4 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 1550_1675 | 0 | 1640.0 | Region | Trimerization |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 196_257 | 0 | 1640.0 | Region | Note=WD40-like repeat 5 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 24_67 | 0 | 1640.0 | Region | Note=WD40-like repeat 1 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 258_301 | 0 | 1640.0 | Region | Note=WD40-like repeat 6 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 2_479 | 0 | 1640.0 | Region | Note=Globular terminal domain |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 302_330 | 0 | 1640.0 | Region | Note=WD40-like repeat 7 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 449_465 | 0 | 1640.0 | Region | Binding site for the uncoating ATPase%2C involved in lattice disassembly |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 480_523 | 0 | 1640.0 | Region | Note=Flexible linker |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 524_1675 | 0 | 1640.0 | Region | Note=Heavy chain arm |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 524_634 | 0 | 1640.0 | Region | Note=Distal segment |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 639_1675 | 0 | 1640.0 | Region | Note=Proximal segment |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 68_107 | 0 | 1640.0 | Region | Note=WD40-like repeat 2 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 1128_1269 | 0 | 1640.0 | Repeat | CHCR 5 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 1274_1420 | 0 | 1640.0 | Repeat | CHCR 6 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 1423_1566 | 0 | 1640.0 | Repeat | CHCR 7 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 537_683 | 0 | 1640.0 | Repeat | CHCR 1 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 686_828 | 0 | 1640.0 | Repeat | CHCR 2 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 833_972 | 0 | 1640.0 | Repeat | CHCR 3 |
| Hgene | CLTC | chr17:57768072 | chr6:117642557 | ENST00000393043 | + | 1 | 31 | 979_1124 | 0 | 1640.0 | Repeat | CHCR 4 |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 101_196 | 1880.3333333333333 | 2348.0 | Domain | Fibronectin type-III 1 | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 1043_1150 | 1880.3333333333333 | 2348.0 | Domain | Fibronectin type-III 5 | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 1450_1556 | 1880.3333333333333 | 2348.0 | Domain | Fibronectin type-III 6 | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 1557_1656 | 1880.3333333333333 | 2348.0 | Domain | Fibronectin type-III 7 | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 1658_1751 | 1880.3333333333333 | 2348.0 | Domain | Fibronectin type-III 8 | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 1752_1854 | 1880.3333333333333 | 2348.0 | Domain | Fibronectin type-III 9 | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 197_285 | 1880.3333333333333 | 2348.0 | Domain | Fibronectin type-III 2 | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 557_671 | 1880.3333333333333 | 2348.0 | Domain | Fibronectin type-III 3 | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 947_1042 | 1880.3333333333333 | 2348.0 | Domain | Fibronectin type-III 4 | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 28_1859 | 1880.3333333333333 | 2348.0 | Topological domain | Extracellular | |
| Tgene | ROS1 | chr17:57768072 | chr6:117642557 | ENST00000368508 | 33 | 43 | 1860_1882 | 1880.3333333333333 | 2348.0 | Transmembrane | Helical |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| CLTC | HGS, MCC, IKBKE, TFE3, SNX5, TOM1, SNX9, DNAJC6, ARMCX3, C10orf88, EPN1, EHD1, GGA1, GGA2, GGA3, KIT, AMPH, CLINT1, TNK2, LDLRAP1, HIP1, AP1B1, AP2A2, AP3B2, MAP3K10, PICALM, EPN2, Epn1, VCL, Arrb2, STAMBP, STAMBPL1, STAM, MAP1LC3A, SNCA, PRKACA, WNK1, Mapk13, HDAC5, SPG20, RICTOR, Erh, Cd2ap, Pttg1, EBNA-LP, PCM1, NDRG1, PPP1CA, ITSN1, ARRB1, SIRT7, SH3KBP1, DYRK1A, SNAP91, AP2B1, BIN1, AP1G1, RAC1, TOM1L1, TOM1L2, ISG15, CUL3, CUL5, CUL2, CDK2, CUL1, COPS5, DCUN1D1, CAND1, NEDD8, XRCC6, HERC1, ATG16L1, SYNJ1, LRRK2, Nfe2l2, AKT1, GRB2, SHC1, AP2M1, MYO6, SRC, FYN, Htt, DNAJA3, RPSA, ESR1, PAXIP1, PPP1CC, SMURF1, ARRB2, ERBB2, FN1, VCAM1, Fcho2, DAB2, UBL4A, ITGA4, MYBL2, SVIL, PAN2, NPM1, PTAFR, ATP4A, EP300, QARS, YWHAQ, FBXO6, RPA1, RPA2, RPA3, ERG, EPHA2, LGR4, Fbxl16, STAU1, TP53, HUWE1, FBXW11, GJB5, NXF1, ADRB2, AVPR2, CUL7, OBSL1, CCDC8, EED, ZFYVE9, OCRL, ATR, SCYL2, HTR6, HNRNPA1, UNK, ATP1A1, ATP1A2, ATP1A3, ATP5C1, CCDC47, CCT3, CCT6A, BLOC1S1, CAD, CANX, CCT6B, CISD1, CLTA, CYC1, DHX38, EHD2, FASTKD2, HSP90B1, IPO5, MTCH2, NDUFB9, NDUFS1, NLRP1, RAB5B, RANBP2, RANBP6, SLC25A3, SSR3, TECR, TOMM22, UQCRC2, VDAC1, VDAC2, VDAC3, ZMPSTE24, DDOST, EIF6, COX2, PHB, PHB2, PPP2R1A, PPP2R1B, RPN1, TMCO1, TUFM, UQCRQ, NTRK1, OFD1, CEP97, CNTRL, FBF1, NPHP1, DCTN1, POC5, POC1B, PPME1, ABCA2, PARP4, AP2A1, ATP6V1B2, ATP6V0A1, CAV1, CLTB, CAPZA2, DBN1, FLNA, GAK, MYH9, DNAJC7, IQGAP1, PDLIM7, SEC16A, SYNPO, MAPRE1, LIMA1, GTSE1, ANLN, MYO19, ARX, MYO18A, CSNK1A1, CSNK1E, STOM, EPS15, HSPA1A, HSPA4, NDUFA9, CHMP1A, PIK3C2A, PRKAR2A, SEC13, SOAT1, CLTCL1, SORBS2, PKP4, USO1, NUMB, TNK1, RAB11A, HIP1R, ATP6V0D1, XPR1, SEC24C, KIAA0430, IST1, HELZ, MVP, TROAP, SEC24B, RBM14, SEC23A, VTI1B, TRAFD1, PKP3, AAK1, BTBD3, TNRC6B, EDC4, DSTYK, PTPN23, NECAP1, TNRC6A, EHD4, EGFL7, ATP6V1D, PTRH2, PLEKHA5, CPVL, BMP2K, DCP1A, ERBB2IP, CHMP1B, KIDINS220, MTUS1, DENND1A, CRTC3, CPSF7, EDC3, CCDC115, FAM83A, REPS1, STON2, FCHO2, CHMP4B, MITD1, CRTC2, FAM83B, FAM83H, SLC35B2, Actb, Myh9, Myo1c, Ppp1cb, Tpm1, Coro1c, Cul3, Spast, Tmod3, Dctn3, Uso1, Lima1, Samm50, Calml3, Myh10, Strn3, Smap1, Ptpn23, Nhsl1, Sec24c, Flnb, IKBKB, MAP3K7, MCM2, AP3D1, U2AF2, CDC73, ZNF746, ARF6, CYLD, LASP1, BRCA1, TES, MTF1, BRD1, CFTR, ZNF598, PDGFRB, FBXO7, TMPO, PTCH1, HSPA8, MAP2K1, PIK3CA, APOE, BMPR1A, JUP, UQCRB, UBE2M, RAD18, PRPF8, EFTUD2, AAR2, PIH1D1, TNIP2, LARP7, RNF4, TNF, SPDL1, RIOK1, PRKCZ, HEXIM1, MEPCE, RUNX1, RNF123, PPT1, HERC2, UBE3A, AGR2, RECQL4, SMAP1, MYC, CDK9, Prkaa1, Prkab1, CDK5RAP2, GRWD1, KRT17, KIAA1429, PLEKHA2, RC3H2, ACTC1, TET2, NR2C2, UCHL3, PRDM16, MECOM, AGRN, BMH1, BMH2, ATXN3, TCF7L2, STX2, GSK3B, ABCC6, BIRC3, LMBR1L, WWP2, Hsp22, TRIM28, GJA1, TEX101, ARIH2, PLEKHA4, PINK1, YAP1, TFCP2, FANCD2, Clta, PXN, RALBP1, PTCHD3, C18orf54, ANKRD55, E, M, nsp13, nsp14, nsp4, nsp5, nsp6, ORF14, ORF3a, ORF6, ORF7a, ORF7b, ABCC2, CDCA5, ACO1, OPTN, NEK4, LRRC31, DUX4, CIT, AURKB, ECT2, KIF14, KIF20A, MAD2L2, KIF23, PRC1, PRNP, CHCHD1, HAX1, MFN2, SLC25A46, LDLR, SUMO2, PRKCB, Rnf183, AIMP2, BRD4, NINL, AFTPH, NUPR1, EPHA3, CIC, Apc2, RBM39, LGALS9, RIN3, EIF3H, DNAJA1, HSPA1L, HSPA2, HSPA6, BAG5, AR, UFL1, DDRGK1, CD3EAP, LAMTOR1, POLR2C, TRIM37, HTRA4, DRD2, ADRBK1, WDR5, PAGE4, NUDCD2, BGLT3, EDEM1, BAG2, DTX3, BTF3, IGF1R, NEDD4, FBXW7, TFRC, IFITM1, BSG, IFITM3, TMPRSS11B, CLEC4E, nsp15, ORF10, FGD5, MAP1LC3B, NBR1, CALCOCO2, SQSTM1, TOLLIP, Chrna4, SIRT6, PER2, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| CLTC |  |
| ROS1 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to CLTC-ROS1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to CLTC-ROS1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | CLTC | C4518356 | MiT family translocation renal cell carcinoma | 2 | ORPHANET |
| Hgene | CLTC | C4693389 | MENTAL RETARDATION, AUTOSOMAL DOMINANT 56 | 2 | GENOMICS_ENGLAND;UNIPROT |
| Hgene | CLTC | C0079744 | Diffuse Large B-Cell Lymphoma | 1 | CTD_human |
| Hgene | CLTC | C0334121 | Inflammatory Myofibroblastic Tumor | 1 | ORPHANET |
| Tgene | ROS1 | C0007131 | Non-Small Cell Lung Carcinoma | 4 | CTD_human |
| Tgene | ROS1 | C0017638 | Glioma | 1 | CTD_human |
| Tgene | ROS1 | C0025202 | melanoma | 1 | CTD_human |
| Tgene | ROS1 | C0152013 | Adenocarcinoma of lung (disorder) | 1 | CGI;CTD_human |
| Tgene | ROS1 | C0259783 | mixed gliomas | 1 | CTD_human |
| Tgene | ROS1 | C0555198 | Malignant Glioma | 1 | CTD_human |

