| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ACY1-BAP1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ACY1-BAP1 | FusionPDB ID: 1916 | FusionGDB2.0 ID: 1916 | Hgene | Tgene | Gene symbol | ACY1 | BAP1 | Gene ID | 95 | 9223 |
| Gene name | aminoacylase 1 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | |
| Synonyms | ACY-1|ACY1D|HEL-S-5 | AIP-3|AIP3|BAIAP1|BAP-1|BAP1|MAGI-1|MAGI-1b|Magi1d|TNRC19|WWP3 | |
| Cytomap | 3p21.2 | 3p14.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | aminoacylase-1N-acyl-L-amino-acid amidohydrolaseacylaseepididymis secretory protein Li 5 | membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1BAI1-associated protein 1WW domain-containing protein 3atrophin-1-interacting protein 3membrane-associated guanylate kinase inverted 1trinucleotide repeat-containing gene 19 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q03154 | Q8IXM2 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000404366, ENST00000458031, ENST00000476351, ENST00000476854, ENST00000494103, ENST00000468068, | ENST00000296288, ENST00000460680, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 1 X 1 X 1=1 | 5 X 5 X 2=50 |
| # samples | 1 | 5 | |
| ** MAII score | log2(1/1*10)=3.32192809488736 | log2(5/50*10)=0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ACY1 [Title/Abstract] AND BAP1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ACY1(52021640)-BAP1(52441476), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | ACY1-BAP1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ACY1-BAP1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | BAP1 | GO:0022409 | positive regulation of cell-cell adhesion | 20298433 |
 Fusion gene breakpoints across ACY1 (5'-gene) Fusion gene breakpoints across ACY1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
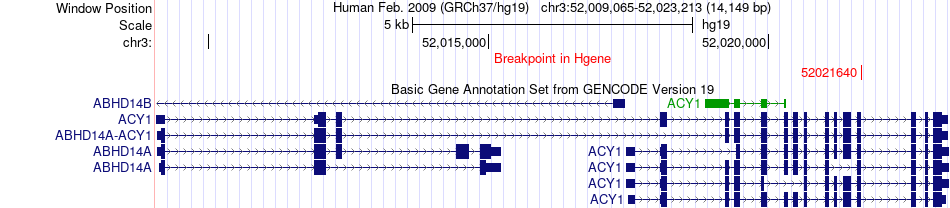 |
 Fusion gene breakpoints across BAP1 (3'-gene) Fusion gene breakpoints across BAP1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
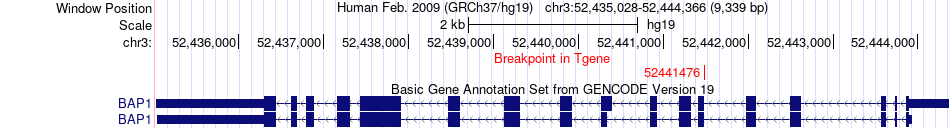 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | MESO | TCGA-LK-A4O5-01A | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000458031 | ACY1 | chr3 | 52021640 | + | ENST00000460680 | BAP1 | chr3 | 52441476 | - | 4512 | 1422 | 231 | 3236 | 1001 |
| ENST00000458031 | ACY1 | chr3 | 52021640 | + | ENST00000296288 | BAP1 | chr3 | 52441476 | - | 4445 | 1422 | 231 | 3182 | 983 |
| ENST00000494103 | ACY1 | chr3 | 52021640 | + | ENST00000460680 | BAP1 | chr3 | 52441476 | - | 3960 | 870 | 165 | 2684 | 839 |
| ENST00000494103 | ACY1 | chr3 | 52021640 | + | ENST00000296288 | BAP1 | chr3 | 52441476 | - | 3893 | 870 | 165 | 2630 | 821 |
| ENST00000476854 | ACY1 | chr3 | 52021640 | + | ENST00000460680 | BAP1 | chr3 | 52441476 | - | 3981 | 891 | 165 | 2705 | 846 |
| ENST00000476854 | ACY1 | chr3 | 52021640 | + | ENST00000296288 | BAP1 | chr3 | 52441476 | - | 3914 | 891 | 165 | 2651 | 828 |
| ENST00000476351 | ACY1 | chr3 | 52021640 | + | ENST00000460680 | BAP1 | chr3 | 52441476 | - | 4071 | 981 | 165 | 2795 | 876 |
| ENST00000476351 | ACY1 | chr3 | 52021640 | + | ENST00000296288 | BAP1 | chr3 | 52441476 | - | 4004 | 981 | 165 | 2741 | 858 |
| ENST00000404366 | ACY1 | chr3 | 52021640 | + | ENST00000460680 | BAP1 | chr3 | 52441476 | - | 4157 | 1067 | 146 | 2881 | 911 |
| ENST00000404366 | ACY1 | chr3 | 52021640 | + | ENST00000296288 | BAP1 | chr3 | 52441476 | - | 4090 | 1067 | 146 | 2827 | 893 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000458031 | ENST00000460680 | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - | 0.004395626 | 0.9956044 |
| ENST00000458031 | ENST00000296288 | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - | 0.004037257 | 0.9959628 |
| ENST00000494103 | ENST00000460680 | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - | 0.012677126 | 0.98732287 |
| ENST00000494103 | ENST00000296288 | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - | 0.013093281 | 0.98690665 |
| ENST00000476854 | ENST00000460680 | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - | 0.013475177 | 0.9865248 |
| ENST00000476854 | ENST00000296288 | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - | 0.014417702 | 0.9855823 |
| ENST00000476351 | ENST00000460680 | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - | 0.008514651 | 0.9914854 |
| ENST00000476351 | ENST00000296288 | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - | 0.008284329 | 0.9917157 |
| ENST00000404366 | ENST00000460680 | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - | 0.006538075 | 0.99346197 |
| ENST00000404366 | ENST00000296288 | ACY1 | chr3 | 52021640 | + | BAP1 | chr3 | 52441476 | - | 0.007768508 | 0.9922315 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >1916_1916_1_ACY1-BAP1_ACY1_chr3_52021640_ENST00000404366_BAP1_chr3_52441476_ENST00000296288_length(amino acids)=893AA_BP=307 MTSKGPEEEHPSVTLFRQYLRIRTVQPKPDYGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEH WSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANP TDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMS ASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQSKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITG RLFELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESK SASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEED LAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRYKGKGTGKPGALSGSADGQLSVLQPNTINVLAEKLKESQKDLSIPLSIK TSSGAGSPAVAVPTHSQPSPTPSNESTDTASEIGSAFNSPLRSPIRSANPTRPSSPVTSHISKVLFGEDDSLLRVDCIRYNRAVRDLGPV ISTGLLHLAEDGVLSPLALTEGGKGSSPSIRPIQGSQGSSSPVEKEVVEATDSREKTGMVRPGEPLSGEKYSPKELLALLKCVEAEIANY -------------------------------------------------------------- >1916_1916_2_ACY1-BAP1_ACY1_chr3_52021640_ENST00000404366_BAP1_chr3_52441476_ENST00000460680_length(amino acids)=911AA_BP=307 MTSKGPEEEHPSVTLFRQYLRIRTVQPKPDYGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEH WSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANP TDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMS ASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQSKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITG RLFELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQP ELIQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLP AFLDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRYKGKGTGKPGALSGSADGQLSVLQPNTINVL AEKLKESQKDLSIPLSIKTSSGAGSPAVAVPTHSQPSPTPSNESTDTASEIGSAFNSPLRSPIRSANPTRPSSPVTSHISKVLFGEDDSL LRVDCIRYNRAVRDLGPVISTGLLHLAEDGVLSPLALTEGGKGSSPSIRPIQGSQGSSSPVEKEVVEATDSREKTGMVRPGEPLSGEKYS PKELLALLKCVEAEIANYEACLKEEVEKRKKFKIDDQRRTHNYDEFICTFISMLAQEGMLANLVEQNISVRRRQGVSIGRLHKQRKPDRR -------------------------------------------------------------- >1916_1916_3_ACY1-BAP1_ACY1_chr3_52021640_ENST00000458031_BAP1_chr3_52441476_ENST00000296288_length(amino acids)=983AA_BP=397 MWGCQAPLSRLPASGETPMSQSWLVSPLATRPSFTARCSHSTRHTGWRWCCFMERPLTLTRGSSWAHCSYCHRGATGPWPLTFQLTTRSA MTSKGPEEEHPSVTLFRQYLRIRTVQPKPDYGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEH WSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANP TDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMS ASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQSKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITG RLFELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESK SASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEED LAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRYKGKGTGKPGALSGSADGQLSVLQPNTINVLAEKLKESQKDLSIPLSIK TSSGAGSPAVAVPTHSQPSPTPSNESTDTASEIGSAFNSPLRSPIRSANPTRPSSPVTSHISKVLFGEDDSLLRVDCIRYNRAVRDLGPV ISTGLLHLAEDGVLSPLALTEGGKGSSPSIRPIQGSQGSSSPVEKEVVEATDSREKTGMVRPGEPLSGEKYSPKELLALLKCVEAEIANY -------------------------------------------------------------- >1916_1916_4_ACY1-BAP1_ACY1_chr3_52021640_ENST00000458031_BAP1_chr3_52441476_ENST00000460680_length(amino acids)=1001AA_BP=397 MWGCQAPLSRLPASGETPMSQSWLVSPLATRPSFTARCSHSTRHTGWRWCCFMERPLTLTRGSSWAHCSYCHRGATGPWPLTFQLTTRSA MTSKGPEEEHPSVTLFRQYLRIRTVQPKPDYGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEH WSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANP TDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMS ASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQSKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITG RLFELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQP ELIQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLP AFLDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRYKGKGTGKPGALSGSADGQLSVLQPNTINVL AEKLKESQKDLSIPLSIKTSSGAGSPAVAVPTHSQPSPTPSNESTDTASEIGSAFNSPLRSPIRSANPTRPSSPVTSHISKVLFGEDDSL LRVDCIRYNRAVRDLGPVISTGLLHLAEDGVLSPLALTEGGKGSSPSIRPIQGSQGSSSPVEKEVVEATDSREKTGMVRPGEPLSGEKYS PKELLALLKCVEAEIANYEACLKEEVEKRKKFKIDDQRRTHNYDEFICTFISMLAQEGMLANLVEQNISVRRRQGVSIGRLHKQRKPDRR -------------------------------------------------------------- >1916_1916_5_ACY1-BAP1_ACY1_chr3_52021640_ENST00000476351_BAP1_chr3_52441476_ENST00000296288_length(amino acids)=858AA_BP=272 MTSKGPEEEHPSVTLFRQYLRIRTVQPKPDYGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAV RRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTA AEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEF AQSKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRLFELDGLKVYPIDHGPWGEDEEWTDKARRVIMER IGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCA QAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQN TNSALRYKGKGTGKPGALSGSADGQLSVLQPNTINVLAEKLKESQKDLSIPLSIKTSSGAGSPAVAVPTHSQPSPTPSNESTDTASEIGS AFNSPLRSPIRSANPTRPSSPVTSHISKVLFGEDDSLLRVDCIRYNRAVRDLGPVISTGLLHLAEDGVLSPLALTEGGKGSSPSIRPIQG SQGSSSPVEKEVVEATDSREKTGMVRPGEPLSGEKYSPKELLALLKCVEAEIANYEACLKEEVEKRKKFKIDDQRRTHNYDEFICTFISM -------------------------------------------------------------- >1916_1916_6_ACY1-BAP1_ACY1_chr3_52021640_ENST00000476351_BAP1_chr3_52441476_ENST00000460680_length(amino acids)=876AA_BP=272 MTSKGPEEEHPSVTLFRQYLRIRTVQPKPDYGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAV RRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTA AEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEF AQSKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRLFELDGLKVYPIDHGPWGEDEEWTDKARRVIMER IGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSASNKSPLVLEANRAPA ASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQ YSDDEDDYEDDEEDDVQNTNSALRYKGKGTGKPGALSGSADGQLSVLQPNTINVLAEKLKESQKDLSIPLSIKTSSGAGSPAVAVPTHSQ PSPTPSNESTDTASEIGSAFNSPLRSPIRSANPTRPSSPVTSHISKVLFGEDDSLLRVDCIRYNRAVRDLGPVISTGLLHLAEDGVLSPL ALTEGGKGSSPSIRPIQGSQGSSSPVEKEVVEATDSREKTGMVRPGEPLSGEKYSPKELLALLKCVEAEIANYEACLKEEVEKRKKFKID -------------------------------------------------------------- >1916_1916_7_ACY1-BAP1_ACY1_chr3_52021640_ENST00000476854_BAP1_chr3_52441476_ENST00000296288_length(amino acids)=828AA_BP=242 MTSKGPEEEHPSVTLFRQYLRIRTVQPKPDYGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEH WSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANP TDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQSKGYAIGNAPELAKAHNSHARPEPRHLP EKQNGLSAVRTMEAFHFVSYVPITGRLFELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQ LIRVTQPELIQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPT PIVQRLPAFLDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRYKGKGTGKPGALSGSADGQLSVLQ PNTINVLAEKLKESQKDLSIPLSIKTSSGAGSPAVAVPTHSQPSPTPSNESTDTASEIGSAFNSPLRSPIRSANPTRPSSPVTSHISKVL FGEDDSLLRVDCIRYNRAVRDLGPVISTGLLHLAEDGVLSPLALTEGGKGSSPSIRPIQGSQGSSSPVEKEVVEATDSREKTGMVRPGEP LSGEKYSPKELLALLKCVEAEIANYEACLKEEVEKRKKFKIDDQRRTHNYDEFICTFISMLAQEGMLANLVEQNISVRRRQGVSIGRLHK -------------------------------------------------------------- >1916_1916_8_ACY1-BAP1_ACY1_chr3_52021640_ENST00000476854_BAP1_chr3_52441476_ENST00000460680_length(amino acids)=846AA_BP=242 MTSKGPEEEHPSVTLFRQYLRIRTVQPKPDYGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEH WSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANP TDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQSKGYAIGNAPELAKAHNSHARPEPRHLP EKQNGLSAVRTMEAFHFVSYVPITGRLFELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEA RLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPK LVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRYKGKGT GKPGALSGSADGQLSVLQPNTINVLAEKLKESQKDLSIPLSIKTSSGAGSPAVAVPTHSQPSPTPSNESTDTASEIGSAFNSPLRSPIRS ANPTRPSSPVTSHISKVLFGEDDSLLRVDCIRYNRAVRDLGPVISTGLLHLAEDGVLSPLALTEGGKGSSPSIRPIQGSQGSSSPVEKEV VEATDSREKTGMVRPGEPLSGEKYSPKELLALLKCVEAEIANYEACLKEEVEKRKKFKIDDQRRTHNYDEFICTFISMLAQEGMLANLVE -------------------------------------------------------------- >1916_1916_9_ACY1-BAP1_ACY1_chr3_52021640_ENST00000494103_BAP1_chr3_52441476_ENST00000296288_length(amino acids)=821AA_BP=235 MTSKGPEEEHPSVTLFRQYLRIRTVQPKPDYGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEH WSHDPFEAFKDSEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVN LTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQSKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLS AVRTMEAFHFVSYVPITGRLFELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQP ELIQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLP AFLDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRYKGKGTGKPGALSGSADGQLSVLQPNTINVL AEKLKESQKDLSIPLSIKTSSGAGSPAVAVPTHSQPSPTPSNESTDTASEIGSAFNSPLRSPIRSANPTRPSSPVTSHISKVLFGEDDSL LRVDCIRYNRAVRDLGPVISTGLLHLAEDGVLSPLALTEGGKGSSPSIRPIQGSQGSSSPVEKEVVEATDSREKTGMVRPGEPLSGEKYS PKELLALLKCVEAEIANYEACLKEEVEKRKKFKIDDQRRTHNYDEFICTFISMLAQEGMLANLVEQNISVRRRQGVSIGRLHKQRKPDRR -------------------------------------------------------------- >1916_1916_10_ACY1-BAP1_ACY1_chr3_52021640_ENST00000494103_BAP1_chr3_52441476_ENST00000460680_length(amino acids)=839AA_BP=235 MTSKGPEEEHPSVTLFRQYLRIRTVQPKPDYGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEH WSHDPFEAFKDSEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVN LTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQSKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLS AVRTMEAFHFVSYVPITGRLFELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKV NRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPG SSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRYKGKGTGKPGALS GSADGQLSVLQPNTINVLAEKLKESQKDLSIPLSIKTSSGAGSPAVAVPTHSQPSPTPSNESTDTASEIGSAFNSPLRSPIRSANPTRPS SPVTSHISKVLFGEDDSLLRVDCIRYNRAVRDLGPVISTGLLHLAEDGVLSPLALTEGGKGSSPSIRPIQGSQGSSSPVEKEVVEATDSR EKTGMVRPGEPLSGEKYSPKELLALLKCVEAEIANYEACLKEEVEKRKKFKIDDQRRTHNYDEFICTFISMLAQEGMLANLVEQNISVRR -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:52021640/chr3:52441476) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ACY1 | BAP1 |
| FUNCTION: Involved in the hydrolysis of N-acylated or N-acetylated amino acids (except L-aspartate). {ECO:0000269|PubMed:12933810}. | FUNCTION: Component of chromatin complexes such as the MLL1/MLL and NURF complexes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | BAP1 | chr3:52021640 | chr3:52441476 | ENST00000460680 | 4 | 17 | 630_661 | 125.0 | 730.0 | Coiled coil | Ontology_term=ECO:0000255 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ACY1 | |
| BAP1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to ACY1-BAP1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ACY1-BAP1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

