| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:EML4-ALK |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: EML4-ALK | FusionPDB ID: 26454 | FusionGDB2.0 ID: 26454 | Hgene | Tgene | Gene symbol | EML4 | ALK | Gene ID | 27436 | 238 |
| Gene name | EMAP like 4 | ALK receptor tyrosine kinase | |
| Synonyms | C2orf2|ELP120|EMAP-4|EMAPL4|ROPP120 | CD246|NBLST3 | |
| Cytomap | 2p21 | 2p23.2-p23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | echinoderm microtubule-associated protein-like 4echinoderm microtubule associated protein like 4restrictedly overexpressed proliferation-associated proteinropp 120 | ALK tyrosine kinase receptorCD246 antigenanaplastic lymphoma receptor tyrosine kinasemutant anaplastic lymphoma kinase | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | Q9HC35 | Q96BT7 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000318522, ENST00000401738, ENST00000402711, ENST00000453191, ENST00000482660, | ENST00000389048, ENST00000431873, ENST00000498037, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 22 X 32 X 18=12672 | 56 X 74 X 20=82880 |
| # samples | 43 | 57 | |
| ** MAII score | log2(43/12672*10)=-4.88116377049015 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(57/82880*10)=-7.18391827352181 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: EML4 [Title/Abstract] AND ALK [Title/Abstract] AND fusion [Title/Abstract] Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer (pmid: 17625570) | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | EML4(42552694)-ALK(29446394), # samples:6 | ||
| Anticipated loss of major functional domain due to fusion event. | EML4-ALK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. EML4-ALK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. EML4-ALK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. EML4-ALK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. EML4-ALK seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. EML4-ALK seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. EML4-ALK seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. EML4-ALK seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ALK | GO:0016310 | phosphorylation | 9174053 |
| Tgene | ALK | GO:0046777 | protein autophosphorylation | 9174053 |
 Fusion gene breakpoints across EML4 (5'-gene) Fusion gene breakpoints across EML4 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
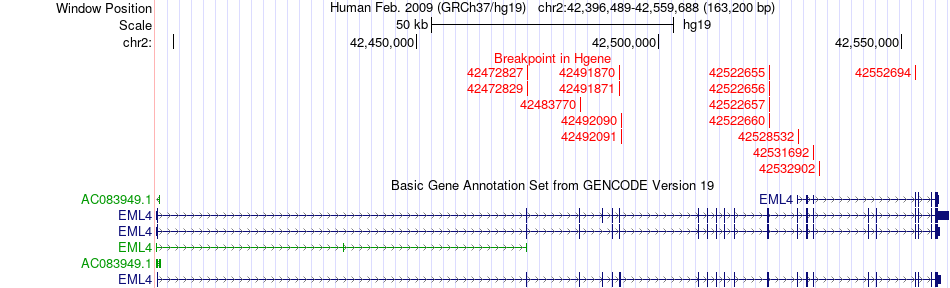 |
 Fusion gene breakpoints across ALK (3'-gene) Fusion gene breakpoints across ALK (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
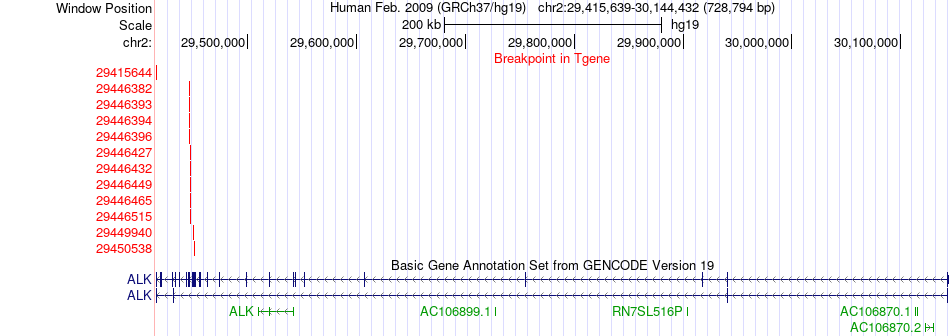 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | acute lymphoblastic leukemia;chronic lymphocytic leukemia;Follicular lymphoma;diffuse large B cell lymphoma;Peripheral T cell lymphoma;Hodgkin disease;adenocarcinoma;adenosquamous carcinoma;squamous cell carcinoma;Nonneoplastic hematologic disorder/lesion | AB274722 | EML4 | chr2 | 42522660 | ALK | chr2 | 29415644 | ||
| ChimerDB4 | COAD | TCGA-AA-3710-01A | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - |
| ChimerDB4 | KIRP | TCGA-2Z-A9JJ-01A | EML4 | chr2 | 42472827 | - | ALK | chr2 | 29446394 | - |
| ChimerDB4 | KIRP | TCGA-2Z-A9JJ-01A | EML4 | chr2 | 42472827 | + | ALK | chr2 | 29446394 | - |
| ChimerDB4 | KIRP | TCGA-2Z-A9JJ | EML4 | chr2 | 42472827 | + | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-50-8460-01A | EML4 | chr2 | 42492090 | + | ALK | chr2 | 29446393 | - |
| ChimerDB4 | LUAD | TCGA-50-8460-01A | EML4 | chr2 | 42492091 | - | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-50-8460-01A | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-67-6215-01A | EML4 | chr2 | 42491870 | + | ALK | chr2 | 29446393 | - |
| ChimerDB4 | LUAD | TCGA-67-6215-01A | EML4 | chr2 | 42491871 | - | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-67-6215-01A | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-67-6215 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-67-6216-01A | EML4 | chr2 | 42491870 | + | ALK | chr2 | 29446393 | - |
| ChimerDB4 | LUAD | TCGA-67-6216-01A | EML4 | chr2 | 42492090 | + | ALK | chr2 | 29446393 | - |
| ChimerDB4 | LUAD | TCGA-67-6216-01A | EML4 | chr2 | 42492091 | - | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-67-6216-01A | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-78-7163-01A | EML4 | chr2 | 42522655 | + | ALK | chr2 | 29446393 | - |
| ChimerDB4 | LUAD | TCGA-78-7163-01A | EML4 | chr2 | 42522656 | - | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-78-7163-01A | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-78-7163 | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-86-A4P8-01A | EML4 | chr2 | 42552694 | - | ALK | chr2 | 29446394 | - |
| ChimerDB4 | LUAD | TCGA-86-A4P8-01A | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - |
| ChimerDB4 | THCA | TCGA-E8-A432-01A | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29449940 | - |
| ChimerDB4 | THCA | TCGA-E8-A432-01A | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29450538 | - |
| ChimerDB4 | THCA | TCGA-E8-A432-01A | EML4 | chr2 | 42492091 | - | ALK | chr2 | 29449940 | - |
| ChimerDB4 | THCA | TCGA-E8-A432-01A | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29449940 | - |
| ChimerDB4 | THCA | TCGA-E8-A432-01A | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29450538 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42345595 | + | ALK | chr2 | 29269593 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42376164 | + | ALK | chr2 | 29269148 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42406198 | + | ALK | chr2 | 29269148 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42472644 | + | ALK | chr2 | 29446394 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42483770 | + | ALK | chr2 | 29415644 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42483770 | + | ALK | chr2 | 29446319 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42491870 | + | ALK | chr2 | 29446393 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446394 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42492090 | + | ALK | chr2 | 29446393 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29415644 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29416042 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29445266 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29446394 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42522655 | + | ALK | chr2 | 29416090 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29443701 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29445473 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29446393 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29446394 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42522657 | + | ALK | chr2 | 29416041 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42522657 | + | ALK | chr2 | 29445266 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42522660 | + | ALK | chr2 | 29446319 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42526287 | + | ALK | chr2 | 29446394 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42528532 | + | ALK | chr2 | 29445266 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42528532 | + | ALK | chr2 | 29446319 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42531692 | + | ALK | chr2 | 29446319 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42531692 | + | ALK | chr2 | 29446393 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42532902 | + | ALK | chr2 | 29446394 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29416090 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29445266 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29449940 | - |
| ChimerKB3 | . | . | EML4 | chr2 | 42553392 | + | ALK | chr2 | 29446394 | - |
| ChiTaRS5.0 | N/A | AB274722 | EML4 | chr2 | 42522660 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | AB275889 | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - |
| ChiTaRS5.0 | N/A | AB374361 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | AB374362 | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | AB374364 | EML4 | chr2 | 42472827 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | AB374365 | EML4 | chr2 | 42472829 | + | ALK | chr2 | 29446515 | - |
| ChiTaRS5.0 | N/A | AB462411 | EML4 | chr2 | 42522657 | + | ALK | chr2 | 29446465 | - |
| ChiTaRS5.0 | N/A | AB462412 | EML4 | chr2 | 42528532 | + | ALK | chr2 | 29446382 | - |
| ChiTaRS5.0 | N/A | DI348820 | EML4 | chr2 | 42472829 | + | ALK | chr2 | 29446515 | - |
| ChiTaRS5.0 | N/A | EU236948 | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | GU797894 | EML4 | chr2 | 42531692 | + | ALK | chr2 | 29446427 | - |
| ChiTaRS5.0 | N/A | GU797895 | EML4 | chr2 | 42532902 | + | ALK | chr2 | 29446432 | - |
| ChiTaRS5.0 | N/A | HC056544 | EML4 | chr2 | 42522660 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | HC056549 | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - |
| ChiTaRS5.0 | N/A | HC056650 | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | HC056652 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | HC056672 | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29446396 | - |
| ChiTaRS5.0 | N/A | HW349733 | EML4 | chr2 | 42472829 | + | ALK | chr2 | 29446515 | - |
| ChiTaRS5.0 | N/A | JQ828841 | EML4 | chr2 | 42483770 | + | ALK | chr2 | 29446449 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000318522 | EML4 | chr2 | 42491870 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 3070 | 929 | 262 | 2619 | 785 |
| ENST00000402711 | EML4 | chr2 | 42491870 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 2893 | 752 | 259 | 2442 | 727 |
| ENST00000401738 | EML4 | chr2 | 42491870 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 2959 | 818 | 151 | 2508 | 785 |
| ENST00000318522 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3070 | 929 | 262 | 2619 | 785 |
| ENST00000402711 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2893 | 752 | 259 | 2442 | 727 |
| ENST00000401738 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2959 | 818 | 151 | 2508 | 785 |
| ENST00000401738 | EML4 | chr2 | 42492090 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 2992 | 851 | 151 | 2541 | 796 |
| ENST00000401738 | EML4 | chr2 | 42492091 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2992 | 851 | 151 | 2541 | 796 |
| ENST00000318522 | EML4 | chr2 | 42522656 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 3892 | 1751 | 262 | 3441 | 1059 |
| ENST00000402711 | EML4 | chr2 | 42522656 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 3715 | 1574 | 259 | 3264 | 1001 |
| ENST00000401738 | EML4 | chr2 | 42522656 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 3814 | 1673 | 151 | 3363 | 1070 |
| ENST00000318522 | EML4 | chr2 | 42522656 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3892 | 1751 | 262 | 3441 | 1059 |
| ENST00000402711 | EML4 | chr2 | 42522656 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3715 | 1574 | 259 | 3264 | 1001 |
| ENST00000401738 | EML4 | chr2 | 42522656 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3814 | 1673 | 151 | 3363 | 1070 |
| ENST00000318522 | EML4 | chr2 | 42522660 | + | ENST00000389048 | ALK | chr2 | 29446319 | - | 3817 | 1751 | 262 | 1842 | 526 |
| ENST00000402711 | EML4 | chr2 | 42522660 | + | ENST00000389048 | ALK | chr2 | 29446319 | - | 3640 | 1574 | 259 | 1665 | 468 |
| ENST00000401738 | EML4 | chr2 | 42522660 | + | ENST00000389048 | ALK | chr2 | 29446319 | - | 3739 | 1673 | 151 | 1764 | 537 |
| ENST00000318522 | EML4 | chr2 | 42528532 | + | ENST00000389048 | ALK | chr2 | 29445266 | - | 3758 | 1903 | 262 | 2601 | 779 |
| ENST00000402711 | EML4 | chr2 | 42528532 | + | ENST00000389048 | ALK | chr2 | 29445266 | - | 3581 | 1726 | 259 | 2424 | 721 |
| ENST00000401738 | EML4 | chr2 | 42528532 | + | ENST00000389048 | ALK | chr2 | 29445266 | - | 3680 | 1825 | 151 | 2523 | 790 |
| ENST00000318522 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 4645 | 2504 | 262 | 4194 | 1310 |
| ENST00000402711 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 4468 | 2327 | 259 | 4017 | 1252 |
| ENST00000401738 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 4567 | 2426 | 151 | 4116 | 1321 |
| ENST00000453191 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2695 | 554 | 409 | 2244 | 611 |
| ENST00000318522 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29449940 | - | 4903 | 2504 | 262 | 4452 | 1396 |
| ENST00000402711 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29449940 | - | 4726 | 2327 | 259 | 4275 | 1338 |
| ENST00000401738 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29449940 | - | 4825 | 2426 | 151 | 4374 | 1407 |
| ENST00000453191 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29449940 | - | 2953 | 554 | 409 | 2502 | 697 |
| ENST00000318522 | EML4 | chr2 | 42553392 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 4744 | 2603 | 262 | 4293 | 1343 |
| ENST00000402711 | EML4 | chr2 | 42553392 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 4567 | 2426 | 259 | 4116 | 1285 |
| ENST00000401738 | EML4 | chr2 | 42553392 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 4666 | 2525 | 151 | 4215 | 1354 |
| ENST00000453191 | EML4 | chr2 | 42553392 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2794 | 653 | 409 | 2343 | 644 |
| ENST00000318522 | EML4 | chr2 | 42472827 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2611 | 470 | 262 | 2160 | 632 |
| ENST00000402711 | EML4 | chr2 | 42472827 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2608 | 467 | 259 | 2157 | 632 |
| ENST00000401738 | EML4 | chr2 | 42472827 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2500 | 359 | 151 | 2049 | 632 |
| ENST00000401738 | EML4 | chr2 | 42492091 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2992 | 851 | 151 | 2541 | 796 |
| ENST00000318522 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3070 | 929 | 262 | 2619 | 785 |
| ENST00000402711 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2893 | 752 | 259 | 2442 | 727 |
| ENST00000401738 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2959 | 818 | 151 | 2508 | 785 |
| ENST00000318522 | EML4 | chr2 | 42522656 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3892 | 1751 | 262 | 3441 | 1059 |
| ENST00000402711 | EML4 | chr2 | 42522656 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3715 | 1574 | 259 | 3264 | 1001 |
| ENST00000401738 | EML4 | chr2 | 42522656 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3814 | 1673 | 151 | 3363 | 1070 |
| ENST00000318522 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 4645 | 2504 | 262 | 4194 | 1310 |
| ENST00000402711 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 4468 | 2327 | 259 | 4017 | 1252 |
| ENST00000401738 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 4567 | 2426 | 151 | 4116 | 1321 |
| ENST00000453191 | EML4 | chr2 | 42552694 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2695 | 554 | 409 | 2244 | 611 |
| ENST00000401738 | EML4 | chr2 | 42492091 | + | ENST00000389048 | ALK | chr2 | 29449940 | - | 3250 | 851 | 151 | 2799 | 882 |
| ENST00000318522 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29449940 | - | 3328 | 929 | 262 | 2877 | 871 |
| ENST00000402711 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29449940 | - | 3151 | 752 | 259 | 2700 | 813 |
| ENST00000401738 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29449940 | - | 3217 | 818 | 151 | 2766 | 871 |
| ENST00000318522 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29450538 | - | 3427 | 929 | 262 | 2976 | 904 |
| ENST00000402711 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29450538 | - | 3250 | 752 | 259 | 2799 | 846 |
| ENST00000401738 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29450538 | - | 3316 | 818 | 151 | 2865 | 904 |
| ENST00000401738 | EML4 | chr2 | 42492091 | + | ENST00000389048 | ALK | chr2 | 29450538 | - | 3349 | 851 | 151 | 2898 | 915 |
| ENST00000401738 | EML4 | chr2 | 42492090 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 2992 | 851 | 151 | 2541 | 796 |
| ENST00000318522 | EML4 | chr2 | 42491870 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 3070 | 929 | 262 | 2619 | 785 |
| ENST00000402711 | EML4 | chr2 | 42491870 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 2893 | 752 | 259 | 2442 | 727 |
| ENST00000401738 | EML4 | chr2 | 42491870 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 2959 | 818 | 151 | 2508 | 785 |
| ENST00000318522 | EML4 | chr2 | 42522655 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 3892 | 1751 | 262 | 3441 | 1059 |
| ENST00000402711 | EML4 | chr2 | 42522655 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 3715 | 1574 | 259 | 3264 | 1001 |
| ENST00000401738 | EML4 | chr2 | 42522655 | + | ENST00000389048 | ALK | chr2 | 29446393 | - | 3814 | 1673 | 151 | 3363 | 1070 |
| ENST00000318522 | EML4 | chr2 | 42522660 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 3892 | 1751 | 262 | 3441 | 1059 |
| ENST00000402711 | EML4 | chr2 | 42522660 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 3715 | 1574 | 259 | 3264 | 1001 |
| ENST00000401738 | EML4 | chr2 | 42522660 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 3814 | 1673 | 151 | 3363 | 1070 |
| ENST00000318522 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 3070 | 929 | 262 | 2619 | 785 |
| ENST00000402711 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 2893 | 752 | 259 | 2442 | 727 |
| ENST00000401738 | EML4 | chr2 | 42491871 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 2959 | 818 | 151 | 2508 | 785 |
| ENST00000401738 | EML4 | chr2 | 42492091 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 2992 | 851 | 151 | 2541 | 796 |
| ENST00000318522 | EML4 | chr2 | 42472827 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 2611 | 470 | 262 | 2160 | 632 |
| ENST00000402711 | EML4 | chr2 | 42472827 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 2608 | 467 | 259 | 2157 | 632 |
| ENST00000401738 | EML4 | chr2 | 42472827 | + | ENST00000389048 | ALK | chr2 | 29446396 | - | 2500 | 359 | 151 | 2049 | 632 |
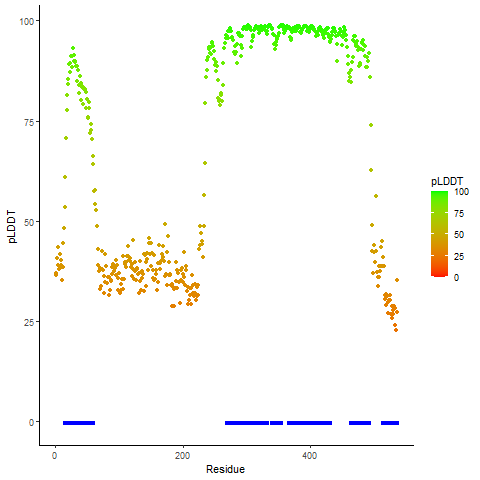
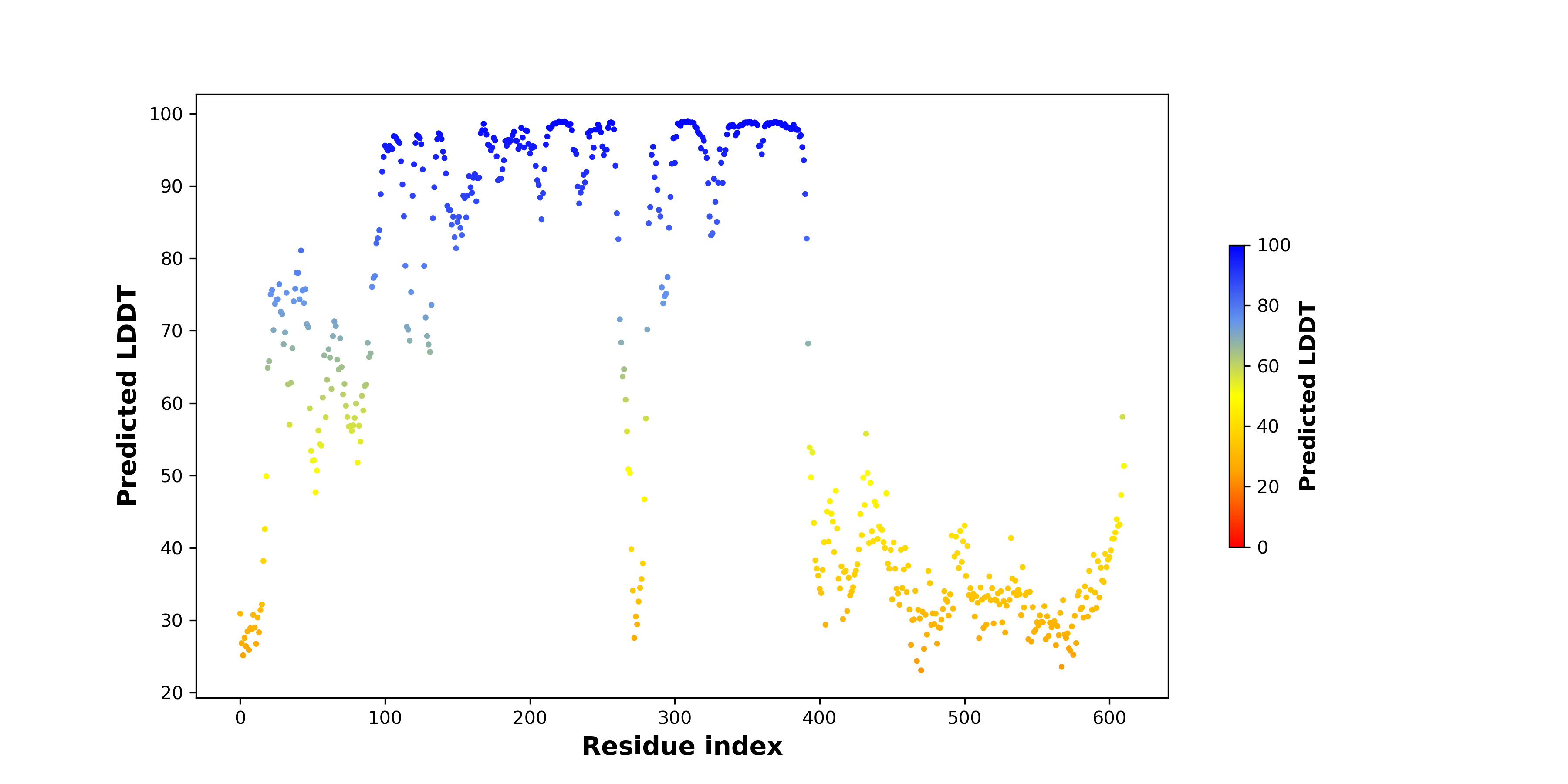
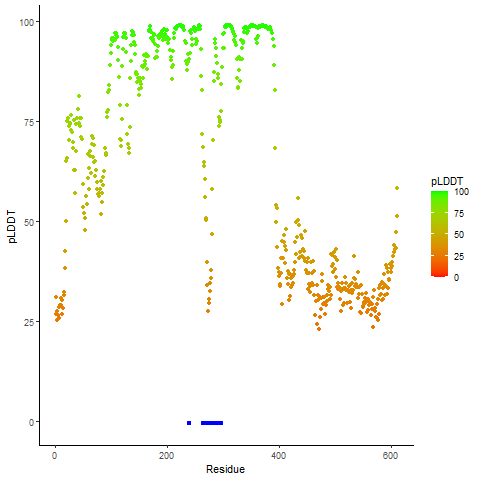
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42472827 | + | ALK | chr2 | 29446394 | - | 0.006545215 | 0.99345475 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42472827 | + | ALK | chr2 | 29446394 | - | 0.006617044 | 0.99338293 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42472827 | + | ALK | chr2 | 29446394 | - | 0.006665675 | 0.9933343 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29446394 | - | 0.001540824 | 0.9984592 |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446394 | - | 0.001366047 | 0.9986339 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446394 | - | 0.003780024 | 0.99622005 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446394 | - | 0.001331759 | 0.9986682 |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29446394 | - | 0.000663732 | 0.9993363 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29446394 | - | 0.000589323 | 0.9994106 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29446394 | - | 0.000660481 | 0.9993395 |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - | 0.001225178 | 0.9987748 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - | 0.001370742 | 0.99862933 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - | 0.001045523 | 0.9989544 |
| ENST00000453191 | ENST00000389048 | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - | 0.005144339 | 0.99485564 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29449940 | - | 0.001402658 | 0.9985973 |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29449940 | - | 0.001344333 | 0.9986557 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29449940 | - | 0.00200867 | 0.9979913 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29449940 | - | 0.001325497 | 0.9986745 |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29450538 | - | 0.000940817 | 0.99905914 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29450538 | - | 0.00193692 | 0.998063 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29450538 | - | 0.000931634 | 0.99906844 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29450538 | - | 0.000994171 | 0.99900585 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42492090 | + | ALK | chr2 | 29446393 | - | 0.001540824 | 0.9984592 |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42491870 | + | ALK | chr2 | 29446393 | - | 0.001366047 | 0.9986339 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42491870 | + | ALK | chr2 | 29446393 | - | 0.003780024 | 0.99622005 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42491870 | + | ALK | chr2 | 29446393 | - | 0.001331759 | 0.9986682 |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42522655 | + | ALK | chr2 | 29446393 | - | 0.000663732 | 0.9993363 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42522655 | + | ALK | chr2 | 29446393 | - | 0.000589323 | 0.9994106 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42522655 | + | ALK | chr2 | 29446393 | - | 0.000660481 | 0.9993395 |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42522660 | + | ALK | chr2 | 29446396 | - | 0.000663732 | 0.9993363 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42522660 | + | ALK | chr2 | 29446396 | - | 0.000589323 | 0.9994106 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42522660 | + | ALK | chr2 | 29446396 | - | 0.000660481 | 0.9993395 |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446396 | - | 0.001366047 | 0.9986339 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446396 | - | 0.003780024 | 0.99622005 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42491871 | + | ALK | chr2 | 29446396 | - | 0.001331759 | 0.9986682 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42492091 | + | ALK | chr2 | 29446396 | - | 0.001540824 | 0.9984592 |
| ENST00000318522 | ENST00000389048 | EML4 | chr2 | 42472827 | + | ALK | chr2 | 29446396 | - | 0.006545215 | 0.99345475 |
| ENST00000402711 | ENST00000389048 | EML4 | chr2 | 42472827 | + | ALK | chr2 | 29446396 | - | 0.006617044 | 0.99338293 |
| ENST00000401738 | ENST00000389048 | EML4 | chr2 | 42472827 | + | ALK | chr2 | 29446396 | - | 0.006665675 | 0.9933343 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >26454_26454_1_EML4-ALK_EML4_chr2_42472827_ENST00000318522_ALK_chr2_29446394_ENST00000389048_length(amino acids)=632AA_BP=69 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKVYRRKHQELQAMQMELQSPEY KLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEA LIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLT CPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRM DPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSP AAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPH DRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQP -------------------------------------------------------------- >26454_26454_2_EML4-ALK_EML4_chr2_42472827_ENST00000318522_ALK_chr2_29446396_ENST00000389048_length(amino acids)=632AA_BP=69 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKVYRRKHQELQAMQMELQSPEY KLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEA LIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLT CPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRM DPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSP AAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPH DRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQP -------------------------------------------------------------- >26454_26454_3_EML4-ALK_EML4_chr2_42472827_ENST00000401738_ALK_chr2_29446394_ENST00000389048_length(amino acids)=632AA_BP=69 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKVYRRKHQELQAMQMELQSPEY KLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEA LIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLT CPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRM DPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSP AAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPH DRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQP -------------------------------------------------------------- >26454_26454_4_EML4-ALK_EML4_chr2_42472827_ENST00000401738_ALK_chr2_29446396_ENST00000389048_length(amino acids)=632AA_BP=69 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKVYRRKHQELQAMQMELQSPEY KLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEA LIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLT CPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRM DPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSP AAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPH DRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQP -------------------------------------------------------------- >26454_26454_5_EML4-ALK_EML4_chr2_42472827_ENST00000402711_ALK_chr2_29446394_ENST00000389048_length(amino acids)=632AA_BP=69 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKVYRRKHQELQAMQMELQSPEY KLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEA LIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLT CPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRM DPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSP AAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPH DRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQP -------------------------------------------------------------- >26454_26454_6_EML4-ALK_EML4_chr2_42472827_ENST00000402711_ALK_chr2_29446396_ENST00000389048_length(amino acids)=632AA_BP=69 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKVYRRKHQELQAMQMELQSPEY KLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEA LIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLT CPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRM DPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSP AAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPH DRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQP -------------------------------------------------------------- >26454_26454_7_EML4-ALK_EML4_chr2_42491870_ENST00000318522_ALK_chr2_29446393_ENST00000389048_length(amino acids)=785AA_BP=222 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSI SDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFIL LELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRK GGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPN FAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRV PRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASL -------------------------------------------------------------- >26454_26454_8_EML4-ALK_EML4_chr2_42491870_ENST00000401738_ALK_chr2_29446393_ENST00000389048_length(amino acids)=785AA_BP=222 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSI SDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFIL LELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRK GGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPN FAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRV PRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASL -------------------------------------------------------------- >26454_26454_9_EML4-ALK_EML4_chr2_42491870_ENST00000402711_ALK_chr2_29446393_ENST00000389048_length(amino acids)=727AA_BP=164 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVYRRKHQELQAMQMEL QSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELD FLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAAR NCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVT SGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKRE EERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIA KKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKN -------------------------------------------------------------- >26454_26454_10_EML4-ALK_EML4_chr2_42491871_ENST00000318522_ALK_chr2_29446394_ENST00000389048_length(amino acids)=785AA_BP=222 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSI SDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFIL LELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRK GGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPN FAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRV PRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASL -------------------------------------------------------------- >26454_26454_11_EML4-ALK_EML4_chr2_42491871_ENST00000318522_ALK_chr2_29446396_ENST00000389048_length(amino acids)=785AA_BP=222 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSI SDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFIL LELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRK GGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPN FAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRV PRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASL -------------------------------------------------------------- >26454_26454_12_EML4-ALK_EML4_chr2_42491871_ENST00000318522_ALK_chr2_29449940_ENST00000389048_length(amino acids)=871AA_BP=222 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVMEGHGEVNIKHYLNCSHCEVDECHMDPESHKVICFCDHGTVLAEDGV SCIVSPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLK EVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELM AGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCA MLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAII LERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGP AVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEP -------------------------------------------------------------- >26454_26454_13_EML4-ALK_EML4_chr2_42491871_ENST00000318522_ALK_chr2_29450538_ENST00000389048_length(amino acids)=904AA_BP=222 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQGGNAASNNDPEMDGEDGVSFISPLGILYTPALKVMEGHGEVNIKHYLN CSHCEVDECHMDPESHKVICFCDHGTVLAEDGVSCIVSPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSP EYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLM EALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCL LTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGG RMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEER SPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKE PHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMN -------------------------------------------------------------- >26454_26454_14_EML4-ALK_EML4_chr2_42491871_ENST00000401738_ALK_chr2_29446394_ENST00000389048_length(amino acids)=785AA_BP=222 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSI SDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFIL LELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRK GGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPN FAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRV PRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASL -------------------------------------------------------------- >26454_26454_15_EML4-ALK_EML4_chr2_42491871_ENST00000401738_ALK_chr2_29446396_ENST00000389048_length(amino acids)=785AA_BP=222 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSI SDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFIL LELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRK GGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPN FAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRV PRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASL -------------------------------------------------------------- >26454_26454_16_EML4-ALK_EML4_chr2_42491871_ENST00000401738_ALK_chr2_29449940_ENST00000389048_length(amino acids)=871AA_BP=222 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVMEGHGEVNIKHYLNCSHCEVDECHMDPESHKVICFCDHGTVLAEDGV SCIVSPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLK EVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELM AGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCA MLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAII LERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGP AVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEP -------------------------------------------------------------- >26454_26454_17_EML4-ALK_EML4_chr2_42491871_ENST00000401738_ALK_chr2_29450538_ENST00000389048_length(amino acids)=904AA_BP=222 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQGGNAASNNDPEMDGEDGVSFISPLGILYTPALKVMEGHGEVNIKHYLN CSHCEVDECHMDPESHKVICFCDHGTVLAEDGVSCIVSPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSP EYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLM EALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCL LTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGG RMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEER SPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKE PHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMN -------------------------------------------------------------- >26454_26454_18_EML4-ALK_EML4_chr2_42491871_ENST00000402711_ALK_chr2_29446394_ENST00000389048_length(amino acids)=727AA_BP=164 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVYRRKHQELQAMQMEL QSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELD FLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAAR NCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVT SGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKRE EERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIA KKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKN -------------------------------------------------------------- >26454_26454_19_EML4-ALK_EML4_chr2_42491871_ENST00000402711_ALK_chr2_29446396_ENST00000389048_length(amino acids)=727AA_BP=164 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVYRRKHQELQAMQMEL QSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELD FLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAAR NCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVT SGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKRE EERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIA KKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKN -------------------------------------------------------------- >26454_26454_20_EML4-ALK_EML4_chr2_42491871_ENST00000402711_ALK_chr2_29449940_ENST00000389048_length(amino acids)=813AA_BP=164 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQVMEGHGEVNIKHYLNC SHCEVDECHMDPESHKVICFCDHGTVLAEDGVSCIVSPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSPE YKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLME ALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLL TCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGR MDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERS PAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEP HDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQ -------------------------------------------------------------- >26454_26454_21_EML4-ALK_EML4_chr2_42491871_ENST00000402711_ALK_chr2_29450538_ENST00000389048_length(amino acids)=846AA_BP=164 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQGGNAASNNDPEMDGED GVSFISPLGILYTPALKVMEGHGEVNIKHYLNCSHCEVDECHMDPESHKVICFCDHGTVLAEDGVSCIVSPTPEPHLPLSLILSVVTSAL VAALVLAFSGIMIVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQ VSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAML DLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWS FGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPL VEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHG SRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNY -------------------------------------------------------------- >26454_26454_22_EML4-ALK_EML4_chr2_42492090_ENST00000401738_ALK_chr2_29446393_ENST00000389048_length(amino acids)=796AA_BP=233 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNP NYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIG VSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMA RDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQ CWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAK KPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPN -------------------------------------------------------------- >26454_26454_23_EML4-ALK_EML4_chr2_42492091_ENST00000401738_ALK_chr2_29446394_ENST00000389048_length(amino acids)=796AA_BP=233 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNP NYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIG VSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMA RDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQ CWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAK KPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPN -------------------------------------------------------------- >26454_26454_24_EML4-ALK_EML4_chr2_42492091_ENST00000401738_ALK_chr2_29446396_ENST00000389048_length(amino acids)=796AA_BP=233 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNP NYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIG VSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMA RDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQ CWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAK KPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPN -------------------------------------------------------------- >26454_26454_25_EML4-ALK_EML4_chr2_42492091_ENST00000401738_ALK_chr2_29449940_ENST00000389048_length(amino acids)=882AA_BP=231 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQVMEGHGEVNIKHYLNCSHCEVDECHMDPESHKVICFC DHGTVLAEDGVSCIVSPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCF AGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQ SLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIY RASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQH QPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTA AEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATG -------------------------------------------------------------- >26454_26454_26_EML4-ALK_EML4_chr2_42492091_ENST00000401738_ALK_chr2_29450538_ENST00000389048_length(amino acids)=915AA_BP=233 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQGGNAASNNDPEMDGEDGVSFISPLGILYTPALKVMEG HGEVNIKHYLNCSHCEVDECHMDPESHKVICFCDHGTVLAEDGVSCIVSPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQE LQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEV CSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHF IHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSN QEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLL VSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKP TKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYE -------------------------------------------------------------- >26454_26454_27_EML4-ALK_EML4_chr2_42522655_ENST00000318522_ALK_chr2_29446393_ENST00000389048_length(amino acids)=1059AA_BP=494 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKLKLEWAYGYRGK DCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLG TFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGK TSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLP RFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRAS YYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPE DRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEI SVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLP -------------------------------------------------------------- >26454_26454_28_EML4-ALK_EML4_chr2_42522655_ENST00000401738_ALK_chr2_29446393_ENST00000389048_length(amino acids)=1070AA_BP=505 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKL KLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSV TLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMT DYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIV RCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGD FGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYR IMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSG KAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCT -------------------------------------------------------------- >26454_26454_29_EML4-ALK_EML4_chr2_42522655_ENST00000402711_ALK_chr2_29446393_ENST00000389048_length(amino acids)=1001AA_BP=436 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITM FIPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQ IAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFH PTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAM QMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQ DELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRD IAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVL EFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQ AKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKN NPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTIL -------------------------------------------------------------- >26454_26454_30_EML4-ALK_EML4_chr2_42522656_ENST00000318522_ALK_chr2_29446393_ENST00000389048_length(amino acids)=1059AA_BP=494 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKLKLEWAYGYRGK DCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLG TFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGK TSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLP RFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRAS YYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPE DRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEI SVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLP -------------------------------------------------------------- >26454_26454_31_EML4-ALK_EML4_chr2_42522656_ENST00000318522_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1059AA_BP=494 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKLKLEWAYGYRGK DCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLG TFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGK TSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLP RFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRAS YYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPE DRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEI SVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLP -------------------------------------------------------------- >26454_26454_32_EML4-ALK_EML4_chr2_42522656_ENST00000401738_ALK_chr2_29446393_ENST00000389048_length(amino acids)=1070AA_BP=505 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKL KLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSV TLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMT DYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIV RCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGD FGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYR IMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSG KAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCT -------------------------------------------------------------- >26454_26454_33_EML4-ALK_EML4_chr2_42522656_ENST00000401738_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1070AA_BP=505 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKL KLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSV TLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMT DYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIV RCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGD FGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYR IMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSG KAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCT -------------------------------------------------------------- >26454_26454_34_EML4-ALK_EML4_chr2_42522656_ENST00000402711_ALK_chr2_29446393_ENST00000389048_length(amino acids)=1001AA_BP=436 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITM FIPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQ IAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFH PTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAM QMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQ DELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRD IAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVL EFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQ AKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKN NPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTIL -------------------------------------------------------------- >26454_26454_35_EML4-ALK_EML4_chr2_42522656_ENST00000402711_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1001AA_BP=436 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITM FIPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQ IAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFH PTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAM QMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQ DELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRD IAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVL EFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQ AKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKN NPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTIL -------------------------------------------------------------- >26454_26454_36_EML4-ALK_EML4_chr2_42522660_ENST00000318522_ALK_chr2_29446319_ENST00000389048_length(amino acids)=526AA_BP= MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKLKLEWAYGYRGK DCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLG TFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF -------------------------------------------------------------- >26454_26454_37_EML4-ALK_EML4_chr2_42522660_ENST00000318522_ALK_chr2_29446396_ENST00000389048_length(amino acids)=1059AA_BP=494 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKLKLEWAYGYRGK DCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLG TFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGK TSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLP RFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRAS YYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPE DRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEI SVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLP -------------------------------------------------------------- >26454_26454_38_EML4-ALK_EML4_chr2_42522660_ENST00000401738_ALK_chr2_29446319_ENST00000389048_length(amino acids)=537AA_BP= MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKL KLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSV TLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS -------------------------------------------------------------- >26454_26454_39_EML4-ALK_EML4_chr2_42522660_ENST00000401738_ALK_chr2_29446396_ENST00000389048_length(amino acids)=1070AA_BP=505 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKL KLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSV TLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMT DYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIV RCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGD FGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYR IMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSG KAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCT -------------------------------------------------------------- >26454_26454_40_EML4-ALK_EML4_chr2_42522660_ENST00000402711_ALK_chr2_29446319_ENST00000389048_length(amino acids)=468AA_BP= MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITM FIPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQ IAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFH PTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGPGCNQHRFADR -------------------------------------------------------------- >26454_26454_41_EML4-ALK_EML4_chr2_42522660_ENST00000402711_ALK_chr2_29446396_ENST00000389048_length(amino acids)=1001AA_BP=436 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITM FIPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQ IAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFH PTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAM QMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQ DELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRD IAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVL EFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQ AKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKN NPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTIL -------------------------------------------------------------- >26454_26454_42_EML4-ALK_EML4_chr2_42528532_ENST00000318522_ALK_chr2_29445266_ENST00000389048_length(amino acids)=779AA_BP= MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKLKLEWAYGYRGK DCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLG TFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHDL NPEREIEDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAV EGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSS -------------------------------------------------------------- >26454_26454_43_EML4-ALK_EML4_chr2_42528532_ENST00000401738_ALK_chr2_29445266_ENST00000389048_length(amino acids)=790AA_BP= MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKL KLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSV TLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGK DRKIILWDHDLNPEREIEDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAE ISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRL -------------------------------------------------------------- >26454_26454_44_EML4-ALK_EML4_chr2_42528532_ENST00000402711_ALK_chr2_29445266_ENST00000389048_length(amino acids)=721AA_BP= MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITM FIPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQ IAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFH PTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAH DGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPA APPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHD RGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQPG -------------------------------------------------------------- >26454_26454_45_EML4-ALK_EML4_chr2_42552694_ENST00000318522_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1310AA_BP=748 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKLKLEWAYGYRGK DCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLG TFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHDL NPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLATHPFKDLLLTCAQDRQVCLWNSMEHRLEWTR LVDEPGHCADFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIYLYVVSENGRKYSRYGRCTGH SSYITHLDWSPDNKYIMSNSGDYEILYLYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIR GLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRE TRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEA FMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDP DVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAF SQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVP -------------------------------------------------------------- >26454_26454_46_EML4-ALK_EML4_chr2_42552694_ENST00000318522_ALK_chr2_29449940_ENST00000389048_length(amino acids)=1396AA_BP=748 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKLKLEWAYGYRGK DCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLG TFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHDL NPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLATHPFKDLLLTCAQDRQVCLWNSMEHRLEWTR LVDEPGHCADFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIYLYVVSENGRKYSRYGRCTGH SSYITHLDWSPDNKYIMSNSGDYEILYLMEGHGEVNIKHYLNCSHCEVDECHMDPESHKVICFCDHGTVLAEDGVSCIVSPTPEPHLPLS LILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGH GAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPR PSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEG IFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVIN TALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSN PPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRL -------------------------------------------------------------- >26454_26454_47_EML4-ALK_EML4_chr2_42552694_ENST00000401738_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1321AA_BP=759 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKL KLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSV TLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGK DRKIILWDHDLNPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLATHPFKDLLLTCAQDRQVCLW NSMEHRLEWTRLVDEPGHCADFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIYLYVVSENGR KYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYLYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLK EVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELM AGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCA MLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAII LERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGP AVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEP -------------------------------------------------------------- >26454_26454_48_EML4-ALK_EML4_chr2_42552694_ENST00000401738_ALK_chr2_29449940_ENST00000389048_length(amino acids)=1407AA_BP=759 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKL KLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSV TLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGK DRKIILWDHDLNPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLATHPFKDLLLTCAQDRQVCLW NSMEHRLEWTRLVDEPGHCADFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIYLYVVSENGR KYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYLMEGHGEVNIKHYLNCSHCEVDECHMDPESHKVICFCDHGTVLAEDGVSCIV SPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPR KNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGD LKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPV KWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERI EYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEG GHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLT -------------------------------------------------------------- >26454_26454_49_EML4-ALK_EML4_chr2_42552694_ENST00000402711_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1252AA_BP=690 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITM FIPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQ IAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFH PTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAH DGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGL ATHPFKDLLLTCAQDRQVCLWNSMEHRLEWTRLVDEPGHCADFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGT FLAVGSHDNFIYLYVVSENGRKYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYLYRRKHQELQAMQMELQSPEYKLSKLRTSTI MTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQN IVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKI GDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPV YRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTS SGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGS -------------------------------------------------------------- >26454_26454_50_EML4-ALK_EML4_chr2_42552694_ENST00000402711_ALK_chr2_29449940_ENST00000389048_length(amino acids)=1338AA_BP=690 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITM FIPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQ IAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFH PTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAH DGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGL ATHPFKDLLLTCAQDRQVCLWNSMEHRLEWTRLVDEPGHCADFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGT FLAVGSHDNFIYLYVVSENGRKYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYLMEGHGEVNIKHYLNCSHCEVDECHMDPESH KVICFCDHGTVLAEDGVSCIVSPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDY NPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRC IGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFG MARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIM TQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKA AKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVP -------------------------------------------------------------- >26454_26454_51_EML4-ALK_EML4_chr2_42552694_ENST00000453191_ALK_chr2_29446394_ENST00000389048_length(amino acids)=611AA_BP=49 MCRFSSKWHSGGHRNALRQGHSSYITHLDWSPDNKYIMSNSGDYEILYLYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFA GKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQS LPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYR ASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQ PEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAA EISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGR -------------------------------------------------------------- >26454_26454_52_EML4-ALK_EML4_chr2_42552694_ENST00000453191_ALK_chr2_29449940_ENST00000389048_length(amino acids)=697AA_BP=49 MCRFSSKWHSGGHRNALRQGHSSYITHLDWSPDNKYIMSNSGDYEILYLMEGHGEVNIKHYLNCSHCEVDECHMDPESHKVICFCDHGTV LAEDGVSCIVSPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTS SISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRF ILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYY RKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDR PNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISV RVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGA -------------------------------------------------------------- >26454_26454_53_EML4-ALK_EML4_chr2_42553392_ENST00000318522_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1343AA_BP=780 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKLKLEWAYGYRGK DCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLG TFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHDL NPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLATHPFKDLLLTCAQDRQVCLWNSMEHRLEWTR LVDEPGHCADFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIYLYVVSENGRKYSRYGRCTGH SSYITHLDWSPDNKYIMSNSGDYEILYWDIPNGCKLIRNRSDCKDIDWTTYTCVLGFQVFVYRRKHQELQAMQMELQSPEYKLSKLRTST IMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQ NIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAK IGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGP VYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTT SSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEG -------------------------------------------------------------- >26454_26454_54_EML4-ALK_EML4_chr2_42553392_ENST00000401738_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1354AA_BP=791 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPSSQPLQIHRQTPESKNATPTKSIKRPSPAEK SHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEKL KLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSV TLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGK DRKIILWDHDLNPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLATHPFKDLLLTCAQDRQVCLW NSMEHRLEWTRLVDEPGHCADFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIYLYVVSENGR KYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYWDIPNGCKLIRNRSDCKDIDWTTYTCVLGFQVFVYRRKHQELQAMQMELQSP EYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLM EALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCL LTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGG RMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEER SPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKE PHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMN -------------------------------------------------------------- >26454_26454_55_EML4-ALK_EML4_chr2_42553392_ENST00000402711_ALK_chr2_29446394_ENST00000389048_length(amino acids)=1285AA_BP=722 MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLRRLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGAN RKPSHTSAVSIAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITM FIPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQ IAGVDKDGRPLQPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTTNEVVLAVEFH PTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAH DGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGL ATHPFKDLLLTCAQDRQVCLWNSMEHRLEWTRLVDEPGHCADFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGT FLAVGSHDNFIYLYVVSENGRKYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYWDIPNGCKLIRNRSDCKDIDWTTYTCVLGFQ VFVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPL QVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIAC GCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSL GYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRP KDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNP TYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEA -------------------------------------------------------------- >26454_26454_56_EML4-ALK_EML4_chr2_42553392_ENST00000453191_ALK_chr2_29446394_ENST00000389048_length(amino acids)=644AA_BP=81 MCRFSSKWHSGGHRNALRQGHSSYITHLDWSPDNKYIMSNSGDYEILYWDIPNGCKLIRNRSDCKDIDWTTYTCVLGFQVFVYRRKHQEL QAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVC SEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFI HRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQ EVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLV SQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPT KKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYED -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:42552694/chr2:29446394) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EML4 | ALK |
| FUNCTION: Essential for the formation and stability of microtubules (MTs) (PubMed:16890222, PubMed:31409757). Required for the organization of the mitotic spindle and for the proper attachment of kinetochores to MTs (PubMed:25789526). Promotes the recruitment of NUDC to the mitotic spindle for mitotic progression (PubMed:25789526). {ECO:0000269|PubMed:16890222, ECO:0000269|PubMed:25789526, ECO:0000269|PubMed:31409757}. | FUNCTION: Catalyzes the methylation of 5-carboxymethyl uridine to 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its methyltransferase domain (PubMed:20123966, PubMed:20308323, PubMed:31079898). Catalyzes the last step in the formation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in target tRNA (PubMed:20123966, PubMed:20308323). Has a preference for tRNA(Arg) and tRNA(Glu), and does not bind tRNA(Lys)(PubMed:20308323). Binds tRNA and catalyzes the iron and alpha-ketoglutarate dependent hydroxylation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its dioxygenase domain, giving rise to 5-(S)-methoxycarbonylhydroxymethyluridine; has a preference for tRNA(Gly) (PubMed:21285950). Required for normal survival after DNA damage (PubMed:20308323). May inhibit apoptosis and promote cell survival and angiogenesis (PubMed:19293182). {ECO:0000269|PubMed:19293182, ECO:0000269|PubMed:20123966, ECO:0000269|PubMed:20308323, ECO:0000269|PubMed:21285950, ECO:0000269|PubMed:31079898}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 1_249 | 496.3333333333333 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 1_249 | 438.3333333333333 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 1_249 | 496.3333333333333 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 1_249 | 438.3333333333333 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 1_249 | 496.3333333333333 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 1_249 | 438.3333333333333 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 1_249 | 747.3333333333334 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 1_249 | 689.3333333333334 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 259_297 | 496.3333333333333 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 301_348 | 496.3333333333333 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 356_396 | 496.3333333333333 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 403_438 | 496.3333333333333 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 445_484 | 496.3333333333333 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 259_297 | 438.3333333333333 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 301_348 | 438.3333333333333 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 356_396 | 438.3333333333333 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 403_438 | 438.3333333333333 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 259_297 | 496.3333333333333 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 301_348 | 496.3333333333333 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 356_396 | 496.3333333333333 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 403_438 | 496.3333333333333 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 445_484 | 496.3333333333333 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 259_297 | 438.3333333333333 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 301_348 | 438.3333333333333 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 356_396 | 438.3333333333333 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 403_438 | 438.3333333333333 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 259_297 | 496.3333333333333 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 301_348 | 496.3333333333333 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 356_396 | 496.3333333333333 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 403_438 | 496.3333333333333 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 445_484 | 496.3333333333333 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 259_297 | 438.3333333333333 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 301_348 | 438.3333333333333 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 356_396 | 438.3333333333333 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 403_438 | 438.3333333333333 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 259_297 | 747.3333333333334 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 301_348 | 747.3333333333334 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 356_396 | 747.3333333333334 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 403_438 | 747.3333333333334 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 445_484 | 747.3333333333334 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 500_538 | 747.3333333333334 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 543_579 | 747.3333333333334 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 582_621 | 747.3333333333334 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 625_662 | 747.3333333333334 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 668_704 | 747.3333333333334 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 711_750 | 747.3333333333334 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 259_297 | 689.3333333333334 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 301_348 | 689.3333333333334 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 356_396 | 689.3333333333334 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 403_438 | 689.3333333333334 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 445_484 | 689.3333333333334 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 500_538 | 689.3333333333334 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 543_579 | 689.3333333333334 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 582_621 | 689.3333333333334 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 625_662 | 689.3333333333334 | 924.0 | Repeat | Note=WD 9 |
| Tgene | ALK | chr2:42472827 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42472827 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42491870 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42491871 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42491871 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42491871 | chr2:29449940 | ENST00000389048 | 16 | 29 | 1116_1392 | 971.3333333333334 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42491871 | chr2:29450538 | ENST00000389048 | 15 | 29 | 1116_1392 | 938.3333333333334 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42492090 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42492091 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42492091 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42492091 | chr2:29449940 | ENST00000389048 | 16 | 29 | 1116_1392 | 971.3333333333334 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42492091 | chr2:29450538 | ENST00000389048 | 15 | 29 | 1116_1392 | 938.3333333333334 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42522655 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42522656 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42522660 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42552694 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr2:42472827 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42472827 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42491870 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42491871 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42491871 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42491871 | chr2:29449940 | ENST00000389048 | 16 | 29 | 1197_1199 | 971.3333333333334 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42491871 | chr2:29450538 | ENST00000389048 | 15 | 29 | 1197_1199 | 938.3333333333334 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42492090 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42492091 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42492091 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42492091 | chr2:29449940 | ENST00000389048 | 16 | 29 | 1197_1199 | 971.3333333333334 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42492091 | chr2:29450538 | ENST00000389048 | 15 | 29 | 1197_1199 | 938.3333333333334 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42522655 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42522656 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42522660 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42552694 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr2:42472827 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42472827 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42491870 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42491871 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42491871 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42491871 | chr2:29449940 | ENST00000389048 | 16 | 29 | 1060_1620 | 971.3333333333334 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42491871 | chr2:29450538 | ENST00000389048 | 15 | 29 | 1060_1620 | 938.3333333333334 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42492090 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42492091 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42492091 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42492091 | chr2:29449940 | ENST00000389048 | 16 | 29 | 1060_1620 | 971.3333333333334 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42492091 | chr2:29450538 | ENST00000389048 | 15 | 29 | 1060_1620 | 938.3333333333334 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42522655 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42522656 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42522660 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42552694 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic | |
| Tgene | ALK | chr2:42491871 | chr2:29449940 | ENST00000389048 | 16 | 29 | 1039_1059 | 971.3333333333334 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42491871 | chr2:29450538 | ENST00000389048 | 15 | 29 | 1039_1059 | 938.3333333333334 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42492091 | chr2:29449940 | ENST00000389048 | 16 | 29 | 1039_1059 | 971.3333333333334 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42492091 | chr2:29450538 | ENST00000389048 | 15 | 29 | 1039_1059 | 938.3333333333334 | 1621.0 | Transmembrane | Helical |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 1_249 | 69.33333333333333 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 1_249 | 69.33333333333333 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 1_249 | 69.33333333333333 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 1_249 | 69.33333333333333 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 1_249 | 222.33333333333334 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 1_249 | 164.33333333333334 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 1_249 | 222.33333333333334 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 1_249 | 164.33333333333334 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 1_249 | 222.33333333333334 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 1_249 | 164.33333333333334 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 1_249 | 222.33333333333334 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 1_249 | 164.33333333333334 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 1_249 | 222.33333333333334 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 1_249 | 164.33333333333334 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 1_249 | 0 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 1_249 | 0 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 1_249 | 0 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 1_249 | 0 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 1_249 | 0 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 1_249 | 0 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 1_249 | 0 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 1_249 | 0 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 1_249 | 0 | 982.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 1_249 | 0 | 924.0 | Region | Microtubule-binding |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 259_297 | 69.33333333333333 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 301_348 | 69.33333333333333 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 356_396 | 69.33333333333333 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 403_438 | 69.33333333333333 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 445_484 | 69.33333333333333 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 500_538 | 69.33333333333333 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 543_579 | 69.33333333333333 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 582_621 | 69.33333333333333 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 625_662 | 69.33333333333333 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 668_704 | 69.33333333333333 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 711_750 | 69.33333333333333 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 760_818 | 69.33333333333333 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000318522 | + | 2 | 23 | 825_864 | 69.33333333333333 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 259_297 | 69.33333333333333 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 301_348 | 69.33333333333333 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 356_396 | 69.33333333333333 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 403_438 | 69.33333333333333 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 445_484 | 69.33333333333333 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 500_538 | 69.33333333333333 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 543_579 | 69.33333333333333 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 582_621 | 69.33333333333333 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 625_662 | 69.33333333333333 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 668_704 | 69.33333333333333 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 711_750 | 69.33333333333333 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 760_818 | 69.33333333333333 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42472827 | chr2:29446394 | ENST00000402711 | + | 2 | 22 | 825_864 | 69.33333333333333 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 259_297 | 69.33333333333333 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 301_348 | 69.33333333333333 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 356_396 | 69.33333333333333 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 403_438 | 69.33333333333333 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 445_484 | 69.33333333333333 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 500_538 | 69.33333333333333 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 543_579 | 69.33333333333333 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 582_621 | 69.33333333333333 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 625_662 | 69.33333333333333 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 668_704 | 69.33333333333333 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 711_750 | 69.33333333333333 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 760_818 | 69.33333333333333 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000318522 | + | 2 | 23 | 825_864 | 69.33333333333333 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 259_297 | 69.33333333333333 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 301_348 | 69.33333333333333 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 356_396 | 69.33333333333333 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 403_438 | 69.33333333333333 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 445_484 | 69.33333333333333 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 500_538 | 69.33333333333333 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 543_579 | 69.33333333333333 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 582_621 | 69.33333333333333 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 625_662 | 69.33333333333333 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 668_704 | 69.33333333333333 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 711_750 | 69.33333333333333 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 760_818 | 69.33333333333333 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42472827 | chr2:29446396 | ENST00000402711 | + | 2 | 22 | 825_864 | 69.33333333333333 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 259_297 | 222.33333333333334 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 301_348 | 222.33333333333334 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 356_396 | 222.33333333333334 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 403_438 | 222.33333333333334 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 445_484 | 222.33333333333334 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 500_538 | 222.33333333333334 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 543_579 | 222.33333333333334 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 582_621 | 222.33333333333334 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 625_662 | 222.33333333333334 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 668_704 | 222.33333333333334 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 711_750 | 222.33333333333334 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 760_818 | 222.33333333333334 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000318522 | + | 6 | 23 | 825_864 | 222.33333333333334 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 259_297 | 164.33333333333334 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 301_348 | 164.33333333333334 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 356_396 | 164.33333333333334 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 403_438 | 164.33333333333334 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 445_484 | 164.33333333333334 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 500_538 | 164.33333333333334 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 543_579 | 164.33333333333334 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 582_621 | 164.33333333333334 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 625_662 | 164.33333333333334 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 668_704 | 164.33333333333334 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 711_750 | 164.33333333333334 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 760_818 | 164.33333333333334 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42491870 | chr2:29446393 | ENST00000402711 | + | 5 | 22 | 825_864 | 164.33333333333334 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 259_297 | 222.33333333333334 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 301_348 | 222.33333333333334 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 356_396 | 222.33333333333334 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 403_438 | 222.33333333333334 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 445_484 | 222.33333333333334 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 500_538 | 222.33333333333334 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 543_579 | 222.33333333333334 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 582_621 | 222.33333333333334 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 625_662 | 222.33333333333334 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 668_704 | 222.33333333333334 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 711_750 | 222.33333333333334 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 760_818 | 222.33333333333334 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000318522 | + | 6 | 23 | 825_864 | 222.33333333333334 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 259_297 | 164.33333333333334 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 301_348 | 164.33333333333334 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 356_396 | 164.33333333333334 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 403_438 | 164.33333333333334 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 445_484 | 164.33333333333334 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 500_538 | 164.33333333333334 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 543_579 | 164.33333333333334 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 582_621 | 164.33333333333334 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 625_662 | 164.33333333333334 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 668_704 | 164.33333333333334 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 711_750 | 164.33333333333334 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 760_818 | 164.33333333333334 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42491871 | chr2:29446394 | ENST00000402711 | + | 5 | 22 | 825_864 | 164.33333333333334 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 259_297 | 222.33333333333334 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 301_348 | 222.33333333333334 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 356_396 | 222.33333333333334 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 403_438 | 222.33333333333334 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 445_484 | 222.33333333333334 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 500_538 | 222.33333333333334 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 543_579 | 222.33333333333334 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 582_621 | 222.33333333333334 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 625_662 | 222.33333333333334 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 668_704 | 222.33333333333334 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 711_750 | 222.33333333333334 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 760_818 | 222.33333333333334 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000318522 | + | 6 | 23 | 825_864 | 222.33333333333334 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 259_297 | 164.33333333333334 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 301_348 | 164.33333333333334 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 356_396 | 164.33333333333334 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 403_438 | 164.33333333333334 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 445_484 | 164.33333333333334 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 500_538 | 164.33333333333334 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 543_579 | 164.33333333333334 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 582_621 | 164.33333333333334 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 625_662 | 164.33333333333334 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 668_704 | 164.33333333333334 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 711_750 | 164.33333333333334 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 760_818 | 164.33333333333334 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42491871 | chr2:29446396 | ENST00000402711 | + | 5 | 22 | 825_864 | 164.33333333333334 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 259_297 | 222.33333333333334 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 301_348 | 222.33333333333334 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 356_396 | 222.33333333333334 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 403_438 | 222.33333333333334 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 445_484 | 222.33333333333334 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 500_538 | 222.33333333333334 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 543_579 | 222.33333333333334 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 582_621 | 222.33333333333334 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 625_662 | 222.33333333333334 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 668_704 | 222.33333333333334 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 711_750 | 222.33333333333334 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 760_818 | 222.33333333333334 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000318522 | + | 6 | 23 | 825_864 | 222.33333333333334 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 259_297 | 164.33333333333334 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 301_348 | 164.33333333333334 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 356_396 | 164.33333333333334 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 403_438 | 164.33333333333334 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 445_484 | 164.33333333333334 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 500_538 | 164.33333333333334 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 543_579 | 164.33333333333334 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 582_621 | 164.33333333333334 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 625_662 | 164.33333333333334 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 668_704 | 164.33333333333334 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 711_750 | 164.33333333333334 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 760_818 | 164.33333333333334 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42491871 | chr2:29449940 | ENST00000402711 | + | 5 | 22 | 825_864 | 164.33333333333334 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 259_297 | 222.33333333333334 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 301_348 | 222.33333333333334 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 356_396 | 222.33333333333334 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 403_438 | 222.33333333333334 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 445_484 | 222.33333333333334 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 500_538 | 222.33333333333334 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 543_579 | 222.33333333333334 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 582_621 | 222.33333333333334 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 625_662 | 222.33333333333334 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 668_704 | 222.33333333333334 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 711_750 | 222.33333333333334 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 760_818 | 222.33333333333334 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000318522 | + | 6 | 23 | 825_864 | 222.33333333333334 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 259_297 | 164.33333333333334 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 301_348 | 164.33333333333334 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 356_396 | 164.33333333333334 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 403_438 | 164.33333333333334 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 445_484 | 164.33333333333334 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 500_538 | 164.33333333333334 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 543_579 | 164.33333333333334 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 582_621 | 164.33333333333334 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 625_662 | 164.33333333333334 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 668_704 | 164.33333333333334 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 711_750 | 164.33333333333334 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 760_818 | 164.33333333333334 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42491871 | chr2:29450538 | ENST00000402711 | + | 5 | 22 | 825_864 | 164.33333333333334 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 259_297 | 0 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 301_348 | 0 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 356_396 | 0 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 403_438 | 0 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 445_484 | 0 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 500_538 | 0 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 543_579 | 0 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 582_621 | 0 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 625_662 | 0 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 668_704 | 0 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 711_750 | 0 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 760_818 | 0 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000318522 | + | 1 | 23 | 825_864 | 0 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 259_297 | 0 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 301_348 | 0 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 356_396 | 0 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 403_438 | 0 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 445_484 | 0 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 500_538 | 0 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 543_579 | 0 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 582_621 | 0 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 625_662 | 0 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 668_704 | 0 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 711_750 | 0 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 760_818 | 0 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42492090 | chr2:29446393 | ENST00000402711 | + | 1 | 22 | 825_864 | 0 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 259_297 | 0 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 301_348 | 0 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 356_396 | 0 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 403_438 | 0 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 445_484 | 0 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 500_538 | 0 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 543_579 | 0 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 582_621 | 0 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 625_662 | 0 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 668_704 | 0 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 711_750 | 0 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 760_818 | 0 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000318522 | + | 1 | 23 | 825_864 | 0 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 259_297 | 0 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 301_348 | 0 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 356_396 | 0 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 403_438 | 0 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 445_484 | 0 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 500_538 | 0 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 543_579 | 0 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 582_621 | 0 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 625_662 | 0 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 668_704 | 0 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 711_750 | 0 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 760_818 | 0 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42492091 | chr2:29446394 | ENST00000402711 | + | 1 | 22 | 825_864 | 0 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 259_297 | 0 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 301_348 | 0 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 356_396 | 0 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 403_438 | 0 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 445_484 | 0 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 500_538 | 0 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 543_579 | 0 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 582_621 | 0 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 625_662 | 0 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 668_704 | 0 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 711_750 | 0 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 760_818 | 0 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000318522 | + | 1 | 23 | 825_864 | 0 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 259_297 | 0 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 301_348 | 0 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 356_396 | 0 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 403_438 | 0 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 445_484 | 0 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 500_538 | 0 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 543_579 | 0 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 582_621 | 0 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 625_662 | 0 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 668_704 | 0 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 711_750 | 0 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 760_818 | 0 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42492091 | chr2:29446396 | ENST00000402711 | + | 1 | 22 | 825_864 | 0 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 259_297 | 0 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 301_348 | 0 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 356_396 | 0 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 403_438 | 0 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 445_484 | 0 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 500_538 | 0 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 543_579 | 0 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 582_621 | 0 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 625_662 | 0 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 668_704 | 0 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 711_750 | 0 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 760_818 | 0 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000318522 | + | 1 | 23 | 825_864 | 0 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 259_297 | 0 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 301_348 | 0 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 356_396 | 0 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 403_438 | 0 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 445_484 | 0 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 500_538 | 0 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 543_579 | 0 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 582_621 | 0 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 625_662 | 0 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 668_704 | 0 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 711_750 | 0 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 760_818 | 0 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42492091 | chr2:29449940 | ENST00000402711 | + | 1 | 22 | 825_864 | 0 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 259_297 | 0 | 982.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 301_348 | 0 | 982.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 356_396 | 0 | 982.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 403_438 | 0 | 982.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 445_484 | 0 | 982.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 500_538 | 0 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 543_579 | 0 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 582_621 | 0 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 625_662 | 0 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 668_704 | 0 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 711_750 | 0 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 760_818 | 0 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000318522 | + | 1 | 23 | 825_864 | 0 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 259_297 | 0 | 924.0 | Repeat | Note=WD 1 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 301_348 | 0 | 924.0 | Repeat | Note=WD 2 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 356_396 | 0 | 924.0 | Repeat | Note=WD 3 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 403_438 | 0 | 924.0 | Repeat | Note=WD 4 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 445_484 | 0 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 500_538 | 0 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 543_579 | 0 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 582_621 | 0 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 625_662 | 0 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 668_704 | 0 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 711_750 | 0 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 760_818 | 0 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42492091 | chr2:29450538 | ENST00000402711 | + | 1 | 22 | 825_864 | 0 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 500_538 | 496.3333333333333 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 543_579 | 496.3333333333333 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 582_621 | 496.3333333333333 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 625_662 | 496.3333333333333 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 668_704 | 496.3333333333333 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 711_750 | 496.3333333333333 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 760_818 | 496.3333333333333 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000318522 | + | 13 | 23 | 825_864 | 496.3333333333333 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 445_484 | 438.3333333333333 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 500_538 | 438.3333333333333 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 543_579 | 438.3333333333333 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 582_621 | 438.3333333333333 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 625_662 | 438.3333333333333 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 668_704 | 438.3333333333333 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 711_750 | 438.3333333333333 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 760_818 | 438.3333333333333 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42522655 | chr2:29446393 | ENST00000402711 | + | 12 | 22 | 825_864 | 438.3333333333333 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 500_538 | 496.3333333333333 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 543_579 | 496.3333333333333 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 582_621 | 496.3333333333333 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 625_662 | 496.3333333333333 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 668_704 | 496.3333333333333 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 711_750 | 496.3333333333333 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 760_818 | 496.3333333333333 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000318522 | + | 13 | 23 | 825_864 | 496.3333333333333 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 445_484 | 438.3333333333333 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 500_538 | 438.3333333333333 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 543_579 | 438.3333333333333 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 582_621 | 438.3333333333333 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 625_662 | 438.3333333333333 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 668_704 | 438.3333333333333 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 711_750 | 438.3333333333333 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 760_818 | 438.3333333333333 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42522656 | chr2:29446394 | ENST00000402711 | + | 12 | 22 | 825_864 | 438.3333333333333 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 500_538 | 496.3333333333333 | 982.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 543_579 | 496.3333333333333 | 982.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 582_621 | 496.3333333333333 | 982.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 625_662 | 496.3333333333333 | 982.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 668_704 | 496.3333333333333 | 982.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 711_750 | 496.3333333333333 | 982.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 760_818 | 496.3333333333333 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000318522 | + | 13 | 23 | 825_864 | 496.3333333333333 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 445_484 | 438.3333333333333 | 924.0 | Repeat | Note=WD 5 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 500_538 | 438.3333333333333 | 924.0 | Repeat | Note=WD 6 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 543_579 | 438.3333333333333 | 924.0 | Repeat | Note=WD 7 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 582_621 | 438.3333333333333 | 924.0 | Repeat | Note=WD 8 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 625_662 | 438.3333333333333 | 924.0 | Repeat | Note=WD 9 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 668_704 | 438.3333333333333 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 711_750 | 438.3333333333333 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 760_818 | 438.3333333333333 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42522660 | chr2:29446396 | ENST00000402711 | + | 12 | 22 | 825_864 | 438.3333333333333 | 924.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 760_818 | 747.3333333333334 | 982.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000318522 | + | 20 | 23 | 825_864 | 747.3333333333334 | 982.0 | Repeat | Note=WD 13 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 668_704 | 689.3333333333334 | 924.0 | Repeat | Note=WD 10 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 711_750 | 689.3333333333334 | 924.0 | Repeat | Note=WD 11 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 760_818 | 689.3333333333334 | 924.0 | Repeat | Note=WD 12 |
| Hgene | EML4 | chr2:42552694 | chr2:29446394 | ENST00000402711 | + | 19 | 22 | 825_864 | 689.3333333333334 | 924.0 | Repeat | Note=WD 13 |
| Tgene | ALK | chr2:42472827 | chr2:29446394 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42472827 | chr2:29446396 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42491870 | chr2:29446393 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42491871 | chr2:29446394 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42491871 | chr2:29446396 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42491871 | chr2:29449940 | ENST00000389048 | 16 | 29 | 816_940 | 971.3333333333334 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42491871 | chr2:29450538 | ENST00000389048 | 15 | 29 | 816_940 | 938.3333333333334 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42492090 | chr2:29446393 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42492091 | chr2:29446394 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42492091 | chr2:29446396 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42492091 | chr2:29449940 | ENST00000389048 | 16 | 29 | 816_940 | 971.3333333333334 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42492091 | chr2:29450538 | ENST00000389048 | 15 | 29 | 816_940 | 938.3333333333334 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42522655 | chr2:29446393 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42522656 | chr2:29446394 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42522660 | chr2:29446396 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42552694 | chr2:29446394 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr2:42472827 | chr2:29446394 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42472827 | chr2:29446394 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42472827 | chr2:29446394 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42472827 | chr2:29446396 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42472827 | chr2:29446396 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42472827 | chr2:29446396 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42491870 | chr2:29446393 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42491870 | chr2:29446393 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42491870 | chr2:29446393 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42491871 | chr2:29446394 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42491871 | chr2:29446394 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42491871 | chr2:29446394 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42491871 | chr2:29446396 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42491871 | chr2:29446396 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42491871 | chr2:29446396 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42491871 | chr2:29449940 | ENST00000389048 | 16 | 29 | 264_427 | 971.3333333333334 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42491871 | chr2:29449940 | ENST00000389048 | 16 | 29 | 437_473 | 971.3333333333334 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42491871 | chr2:29449940 | ENST00000389048 | 16 | 29 | 478_636 | 971.3333333333334 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42491871 | chr2:29450538 | ENST00000389048 | 15 | 29 | 264_427 | 938.3333333333334 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42491871 | chr2:29450538 | ENST00000389048 | 15 | 29 | 437_473 | 938.3333333333334 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42491871 | chr2:29450538 | ENST00000389048 | 15 | 29 | 478_636 | 938.3333333333334 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42492090 | chr2:29446393 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42492090 | chr2:29446393 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42492090 | chr2:29446393 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42492091 | chr2:29446394 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42492091 | chr2:29446394 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42492091 | chr2:29446394 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42492091 | chr2:29446396 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42492091 | chr2:29446396 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42492091 | chr2:29446396 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42492091 | chr2:29449940 | ENST00000389048 | 16 | 29 | 264_427 | 971.3333333333334 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42492091 | chr2:29449940 | ENST00000389048 | 16 | 29 | 437_473 | 971.3333333333334 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42492091 | chr2:29449940 | ENST00000389048 | 16 | 29 | 478_636 | 971.3333333333334 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42492091 | chr2:29450538 | ENST00000389048 | 15 | 29 | 264_427 | 938.3333333333334 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42492091 | chr2:29450538 | ENST00000389048 | 15 | 29 | 437_473 | 938.3333333333334 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42492091 | chr2:29450538 | ENST00000389048 | 15 | 29 | 478_636 | 938.3333333333334 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42522655 | chr2:29446393 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42522655 | chr2:29446393 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42522655 | chr2:29446393 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42522656 | chr2:29446394 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42522656 | chr2:29446394 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42522656 | chr2:29446394 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42522660 | chr2:29446396 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42522660 | chr2:29446396 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42522660 | chr2:29446396 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42552694 | chr2:29446394 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr2:42552694 | chr2:29446394 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr2:42552694 | chr2:29446394 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr2:42472827 | chr2:29446394 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42472827 | chr2:29446396 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42491870 | chr2:29446393 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42491871 | chr2:29446394 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42491871 | chr2:29446396 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42491871 | chr2:29449940 | ENST00000389048 | 16 | 29 | 19_1038 | 971.3333333333334 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42491871 | chr2:29450538 | ENST00000389048 | 15 | 29 | 19_1038 | 938.3333333333334 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42492090 | chr2:29446393 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42492091 | chr2:29446394 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42492091 | chr2:29446396 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42492091 | chr2:29449940 | ENST00000389048 | 16 | 29 | 19_1038 | 971.3333333333334 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42492091 | chr2:29450538 | ENST00000389048 | 15 | 29 | 19_1038 | 938.3333333333334 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42522655 | chr2:29446393 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42522656 | chr2:29446394 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42522660 | chr2:29446396 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42552694 | chr2:29446394 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr2:42472827 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42472827 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42491870 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42491871 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42491871 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42492090 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42492091 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42492091 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42522655 | chr2:29446393 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42522656 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42522660 | chr2:29446396 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical | |
| Tgene | ALK | chr2:42552694 | chr2:29446394 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (245) >>>245.pdbFusion protein BP residue: CIF file (245) >>>245.cif | EML4 | chr2 | 42522660 | + | ALK | chr2 | 29446319 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKV TKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEK LKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGH TDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIG LGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTT NEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKF VQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGPGCNQHRFADR | 468 |
| 3D view using mol* of 245 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (352) >>>352.pdbFusion protein BP residue: CIF file (352) >>>352.cif | EML4 | chr2 | 42522660 | + | ALK | chr2 | 29446319 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDI RTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEER TQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVT LSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKA KGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGPGC | 526 |
| 3D view using mol* of 352 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (370) >>>370.pdbFusion protein BP residue: CIF file (370) >>>370.cif | EML4 | chr2 | 42522660 | + | ALK | chr2 | 29446319 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMF IPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIA SVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPL QPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEH MLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEP | 537 |
| 3D view using mol* of 370 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (481) >>>481.pdbFusion protein BP residue: 49 CIF file (481) >>>481.cif | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - | MCRFSSKWHSGGHRNALRQGHSSYITHLDWSPDNKYIMSNSGDYEILYLY RRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLK EVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQ DELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLR ETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTC PGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSK TDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVY RIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEE KVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAA EISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGS WFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEP SSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTIL | 611 |
| 3D view using mol* of 481 (AA BP:49) | ||||||||||
 | ||||||||||
| PDB file (518) >>>518.pdbFusion protein BP residue: 81 CIF file (518) >>>518.cif | EML4 | chr2 | 42553392 | + | ALK | chr2 | 29446394 | - | MCRFSSKWHSGGHRNALRQGHSSYITHLDWSPDNKYIMSNSGDYEILYWD IPNGCKLIRNRSDCKDIDWTTYTCVLGFQVFVYRRKHQELQAMQMELQSP EYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGA FGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNH QNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDL LHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARD IYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLG YMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNF AIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLV SQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHV NMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKE PHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRH | 644 |
| 3D view using mol* of 518 (AA BP:81) | ||||||||||
 | ||||||||||
| PDB file (567) >>>567.pdbFusion protein BP residue: 49 CIF file (567) >>>567.cif | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29449940 | - | MCRFSSKWHSGGHRNALRQGHSSYITHLDWSPDNKYIMSNSGDYEILYLM EGHGEVNIKHYLNCSHCEVDECHMDPESHKVICFCDHGTVLAEDGVSCIV SPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMEL QSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLG HGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISK FNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAM LDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGM ARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIF SLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDR PNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPP LLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEG GHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIA KKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFR | 697 |
| 3D view using mol* of 567 (AA BP:49) | ||||||||||
 | ||||||||||
| PDB file (590) >>>590.pdbFusion protein BP residue: CIF file (590) >>>590.cif | EML4 | chr2 | 42528532 | + | ALK | chr2 | 29445266 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKV TKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEK LKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGH TDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIG LGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTT NEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKF VQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAH DGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEDPDVINTALPI EYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSS GKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKP TSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGR LPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAP | 721 |
| 3D view using mol* of 590 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (595) >>>595.pdbFusion protein BP residue: 164 CIF file (595) >>>595.cif | EML4 | chr2 | 42491870 | + | ALK | chr2 | 29446393 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKV TKTADKHKDVIINQVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYN PNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPS PLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRF ILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEEN HFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPV KWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVT SGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVI NTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPP LPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVH GSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPP NVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPL | 727 |
| 3D view using mol* of 595 (AA BP:164) | ||||||||||
 | ||||||||||
| PDB file (639) >>>639.pdbFusion protein BP residue: CIF file (639) >>>639.cif | EML4 | chr2 | 42528532 | + | ALK | chr2 | 29445266 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDI RTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEER TQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVT LSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKA KGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQ ISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEDPD VINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAP PPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHK VHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTV PPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGL | 779 |
| 3D view using mol* of 639 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (642) >>>642.pdbFusion protein BP residue: 222 CIF file (642) >>>642.cif | EML4 | chr2 | 42491870 | + | ALK | chr2 | 29446393 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQVYRRKHQELQAMQMELQSPEYKLSKLRT STIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQV SGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGV SLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIAC GCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRK GGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSN QEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEY CTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEE RSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNP PSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGL EGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYG | 785 |
| 3D view using mol* of 642 (AA BP:222) | ||||||||||
 | ||||||||||
| PDB file (644) >>>644.pdbFusion protein BP residue: CIF file (644) >>>644.cif | EML4 | chr2 | 42528532 | + | ALK | chr2 | 29445266 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMF IPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIA SVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPL QPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEH MLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEP TPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHD LNPEREIEDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQA KREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAF SQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDR GNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCG | 790 |
| 3D view using mol* of 644 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (649) >>>649.pdbFusion protein BP residue: 233 CIF file (649) >>>649.cif | EML4 | chr2 | 42492090 | + | ALK | chr2 | 29446393 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQAKMSTREKNSQVYRRKHQELQAMQMELQ SPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGH GAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKF NHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAML DLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMA RDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFS LGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRP NFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPL LVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGG HVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAK KEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRL | 796 |
| 3D view using mol* of 649 (AA BP:233) | ||||||||||
 | ||||||||||
| PDB file (752) >>>752.pdbFusion protein BP residue: 436 CIF file (752) >>>752.cif | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29446393 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKV TKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEK LKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGH TDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIG LGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTT NEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKF VQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRRKHQELQAM QMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLI RGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEAL IISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPS SLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIG DFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLL WEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQ PEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPE GVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGP AVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKN NPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEV PLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQPG | 1001 |
| 3D view using mol* of 752 (AA BP:436) | ||||||||||
 | ||||||||||
| PDB file (785) >>>785.pdbFusion protein BP residue: 494 CIF file (785) >>>785.cif | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29446393 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDI RTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEER TQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVT LSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKA KGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKVYRR KHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEV PRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDE LDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRET RPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPG PGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTD TWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRI MTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKV PVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEI SVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWF TEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSS LTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKS | 1059 |
| 3D view using mol* of 785 (AA BP:494) | ||||||||||
 | ||||||||||
| PDB file (788) >>>788.pdbFusion protein BP residue: 505 CIF file (788) >>>788.cif | EML4 | chr2 | 42522656 | + | ALK | chr2 | 29446393 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMF IPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIA SVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPL QPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEH MLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEP TPGKGPKVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAG KTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVK TLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMA GGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDI AARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEA FMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDP PKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIE YGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSG KAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPT SLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRL PGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPG | 1070 |
| 3D view using mol* of 788 (AA BP:505) | ||||||||||
 | ||||||||||
| PDB file (867)CIF file (867) >>>867.cif | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKV TKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEK LKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGH TDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIG LGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTT NEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKF VQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAH DGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEVPDQYGTIRAV AEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLATHPFKDLLL TCAQDRQVCLWNSMEHRLEWTRLVDEPGHCADFHPSGTVVAIGTHSGRWF VLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIYLYVVSENG RKYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYLYRRKHQELQA MQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITL IRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEA LIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQP SSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKI GDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVL LWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQH QPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDP EGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRG PAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKK NNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKE VPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMNQP | 1252 |
| 3D view using mol* of 867 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (878)CIF file (878) >>>878.cif | EML4 | chr2 | 42553392 | + | ALK | chr2 | 29446394 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKV TKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEK LKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGH TDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIG LGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTT NEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKF VQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAH DGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEVPDQYGTIRAV AEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLATHPFKDLLL TCAQDRQVCLWNSMEHRLEWTRLVDEPGHCADFHPSGTVVAIGTHSGRWF VLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIYLYVVSENG RKYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYWDIPNGCKLIR NRSDCKDIDWTTYTCVLGFQVFVYRRKHQELQAMQMELQSPEYKLSKLRT STIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQV SGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGV SLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIAC GCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRK GGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSN QEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEY CTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEE RSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNP PSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGL EGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYG | 1285 |
| 3D view using mol* of 878 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (882)CIF file (882) >>>882.cif | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDI RTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEER TQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVT LSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKA KGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQ ISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEVPD QYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLAT HPFKDLLLTCAQDRQVCLWNSMEHRLEWTRLVDEPGHCADFHPSGTVVAI GTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIY LYVVSENGRKYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYLYR RKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKE VPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQD ELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRE TRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCP GPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKT DTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYR IMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEK VPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAE ISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSW FTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPS SLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILK | 1310 |
| 3D view using mol* of 882 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (884)CIF file (884) >>>884.cif | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29446394 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMF IPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIA SVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPL QPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEH MLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEP TPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHD LNPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQ GHTDELWGLATHPFKDLLLTCAQDRQVCLWNSMEHRLEWTRLVDEPGHCA DFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTF LAVGSHDNFIYLYVVSENGRKYSRYGRCTGHSSYITHLDWSPDNKYIMSN SGDYEILYLYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFA GKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAV KTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELM AGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRD IAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPE AFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMD PPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPI EYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSS GKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKP TSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGR LPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAP | 1321 |
| 3D view using mol* of 884 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (891)CIF file (891) >>>891.cif | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29449940 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSIKRPSPAEKSHNSWENSDDSRNKLSKIPSTPKLIPKV TKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDIRTELPPEK LKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEERTQRHYLGH TDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVTLSTLQIIG LGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKAKGAEIKTT NEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIFGKYEKPKF VQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQISKQIKAH DGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEVPDQYGTIRAV AEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLATHPFKDLLL TCAQDRQVCLWNSMEHRLEWTRLVDEPGHCADFHPSGTVVAIGTHSGRWF VLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIYLYVVSENG RKYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYLMEGHGEVNIK HYLNCSHCEVDECHMDPESHKVICFCDHGTVLAEDGVSCIVSPTPEPHLP LSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQSPEYKLSK LRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYE GQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRC IGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARD IACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASY YRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPS KSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILER IEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKR EEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQ SNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGN LGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNV | 1338 |
| 3D view using mol* of 891 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (892)CIF file (892) >>>892.cif | EML4 | chr2 | 42553392 | + | ALK | chr2 | 29446394 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDI RTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEER TQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVT LSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKA KGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQ ISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEVPD QYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLAT HPFKDLLLTCAQDRQVCLWNSMEHRLEWTRLVDEPGHCADFHPSGTVVAI GTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIY LYVVSENGRKYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYWDI PNGCKLIRNRSDCKDIDWTTYTCVLGFQVFVYRRKHQELQAMQMELQSPE YKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAF GEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQ NIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLL HVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDI YRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGY MPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFA IILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVS QQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVN MAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEP HDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHF | 1343 |
| 3D view using mol* of 892 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (896)CIF file (896) >>>896.cif | EML4 | chr2 | 42553392 | + | ALK | chr2 | 29446394 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMF IPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIA SVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPL QPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEH MLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEP TPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHD LNPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQ GHTDELWGLATHPFKDLLLTCAQDRQVCLWNSMEHRLEWTRLVDEPGHCA DFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTF LAVGSHDNFIYLYVVSENGRKYSRYGRCTGHSSYITHLDWSPDNKYIMSN SGDYEILYWDIPNGCKLIRNRSDCKDIDWTTYTCVLGFQVFVYRRKHQEL QAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNI TLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLM EALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPS QPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVA KIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFG VLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCW QHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPK DPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVP RGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPT KKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANM KEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMN | 1354 |
| 3D view using mol* of 896 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (908) >>>908.pdbFusion protein BP residue: 748 CIF file (908) >>>908.cif | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29449940 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQEGEYIKMFMRGRPITMFIPSDVDNYDDI RTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIASVVVLFNYEER TQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPLQPHVRVWDSVT LSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEHMLTVWDWQKKA KGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWSGNSLTRKQGIF GKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEPTPGKGPKGVYQ ISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHDLNPEREIEVPD QYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQGHTDELWGLAT HPFKDLLLTCAQDRQVCLWNSMEHRLEWTRLVDEPGHCADFHPSGTVVAI GTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTFLAVGSHDNFIY LYVVSENGRKYSRYGRCTGHSSYITHLDWSPDNKYIMSNSGDYEILYLME GHGEVNIKHYLNCSHCEVDECHMDPESHKVICFCDHGTVLAEDGVSCIVS PTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKHQELQAMQMELQ SPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGH GAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKF NHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAML DLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMA RDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFS LGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRP NFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPL LVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGG HVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAK KEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRL | 1396 |
| 3D view using mol* of 908 (AA BP:748) | ||||||||||
 | ||||||||||
| PDB file (912)CIF file (912) >>>912.cif | EML4 | chr2 | 42552694 | + | ALK | chr2 | 29449940 | - | MDGFAGSLDDSISAASTSDVQDRLSALESRVQQQEDEITVLKAALADVLR RLAISEDHVASVKKSVSSKGQPSPRAVIPMSCITNGSGANRKPSHTSAVS IAGKETLSSAAKSGTEKKKEKPQGQREKKEESHSNDQSPQIRASPSPQPS SQPLQIHRQTPESKNATPTKSIKRPSPAEKSHNSWENSDDSRNKLSKIPS TPKLIPKVTKTADKHKDVIINQAKMSTREKNSQEGEYIKMFMRGRPITMF IPSDVDNYDDIRTELPPEKLKLEWAYGYRGKDCRANVYLLPTGKIVYFIA SVVVLFNYEERTQRHYLGHTDCVKCLAIHPDKIRIATGQIAGVDKDGRPL QPHVRVWDSVTLSTLQIIGLGTFERGVGCLDFSKADSGVHLCIIDDSNEH MLTVWDWQKKAKGAEIKTTNEVVLAVEFHPTDANTIITCGKSHIFFWTWS GNSLTRKQGIFGKYEKPKFVQCLAFLGNGDVLTGDSGGVMLIWSKTTVEP TPGKGPKGVYQISKQIKAHDGSVFTLCQMRNGMLLTGGGKDRKIILWDHD LNPEREIEVPDQYGTIRAVAEGKADQFLVGTSRNFILRGTFNDGFQIEVQ GHTDELWGLATHPFKDLLLTCAQDRQVCLWNSMEHRLEWTRLVDEPGHCA DFHPSGTVVAIGTHSGRWFVLDAETRDLVSIHTDGNEQLSVMRYSIDGTF LAVGSHDNFIYLYVVSENGRKYSRYGRCTGHSSYITHLDWSPDNKYIMSN SGDYEILYLMEGHGEVNIKHYLNCSHCEVDECHMDPESHKVICFCDHGTV LAEDGVSCIVSPTPEPHLPLSLILSVVTSALVAALVLAFSGIMIVYRRKH QELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPR KNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELD FLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRP RPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPG RVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTW SFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMT QCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPV RPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISV RVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTE KPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLT ANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKN | 1407 |
| 3D view using mol* of 912 (AA BP:) | ||||||||||
 | ||||||||||
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
EML4_pLDDT.png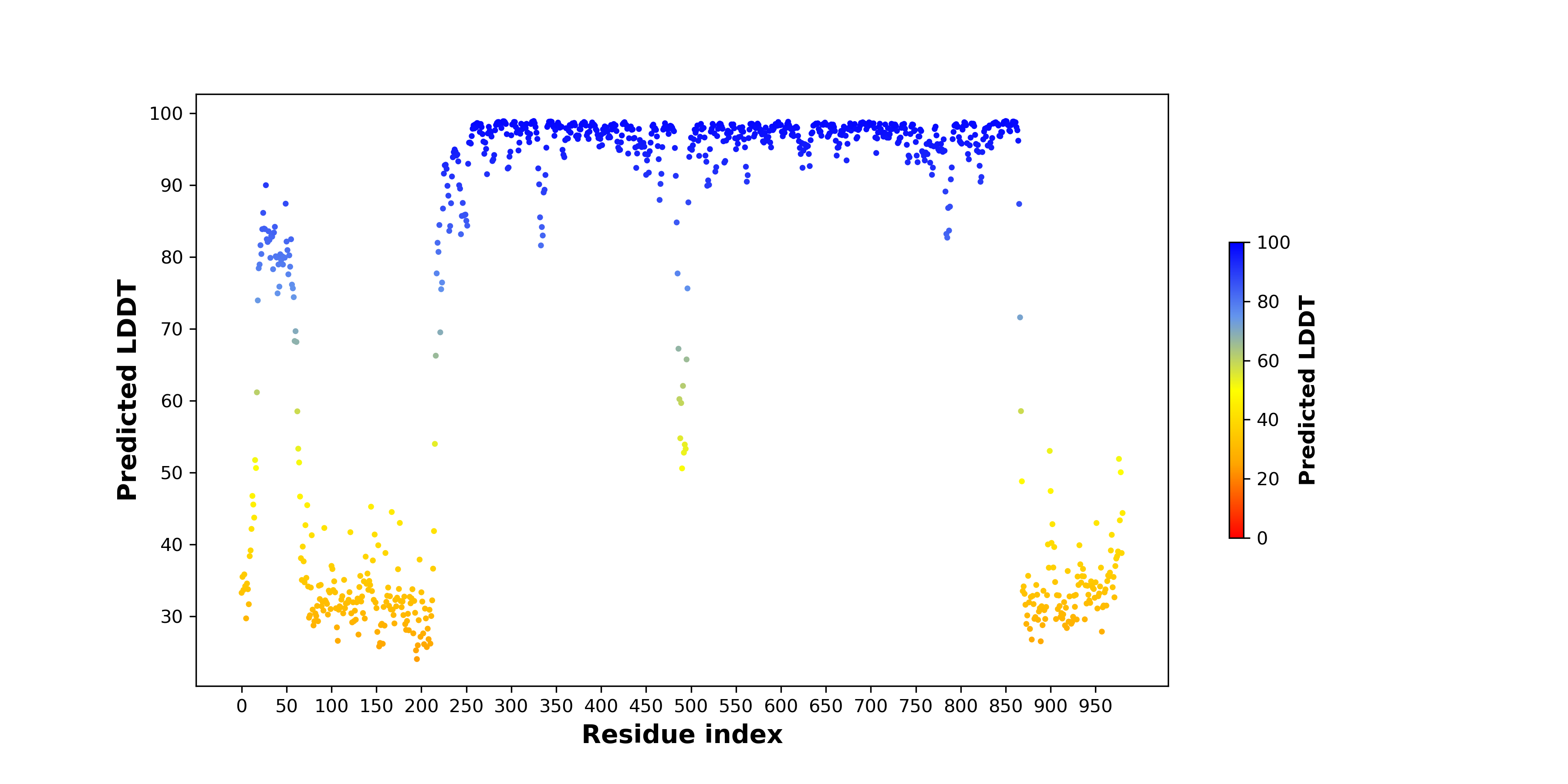 |
ALK_pLDDT.png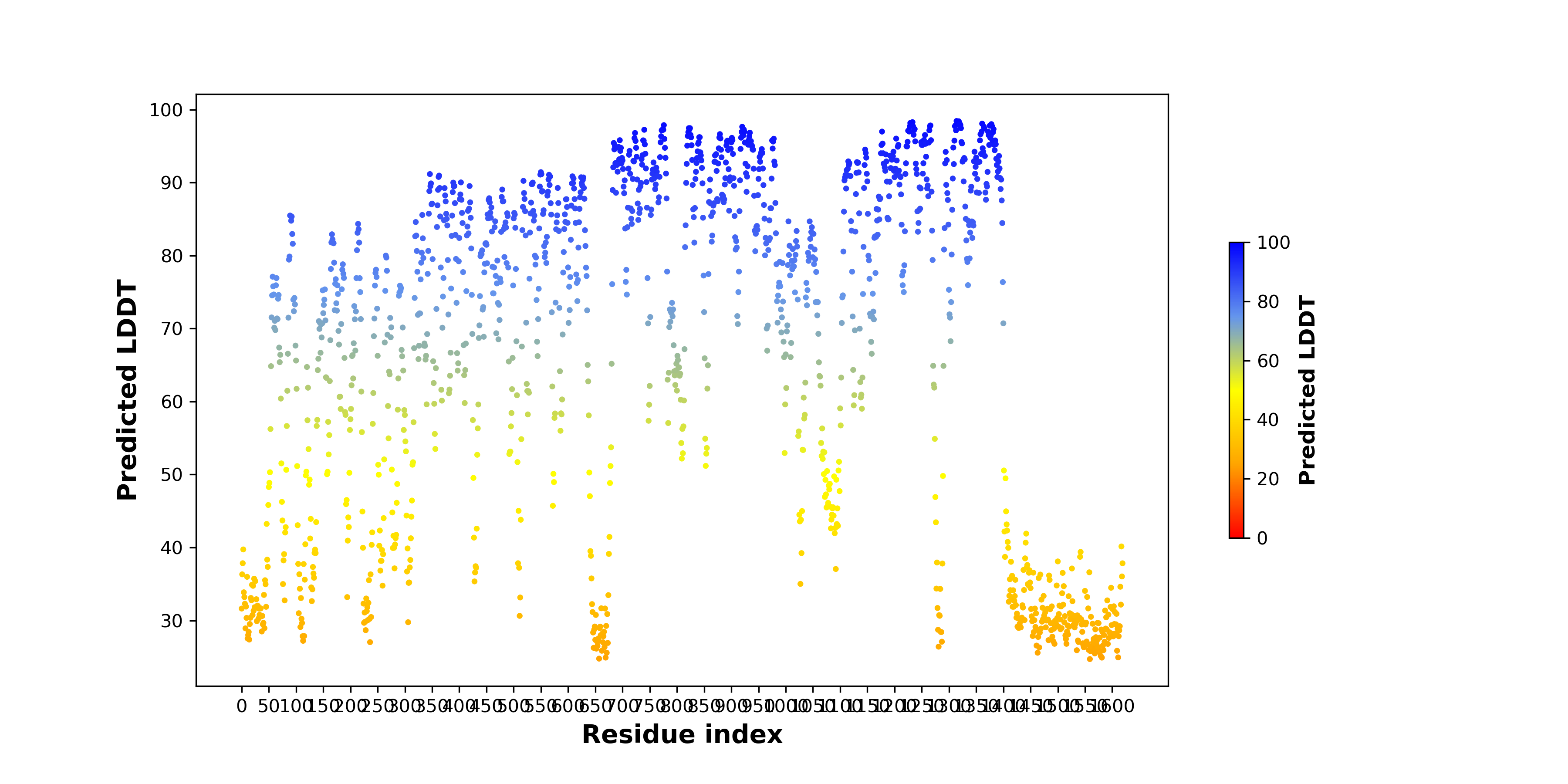 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 245 | 1.032 | 350 | 1.074 | 921.984 | 0.563 | 0.697 | 0.929 | 0.745 | 0.871 | 0.855 | 0.86 | Chain A: 33,36,37,40,41,43,44,46,47,49,50,211,212, 213,214,215,216,217,219,255,256,257,258,259,260,26 1,263,309,310,311,312,315,325,327,331,353,354,355, 356,357,358,400,402,403,404,405,413,415,416,417,41 8,419,420,421,452,453,454,455,456,457,458,459,460, 461 |
| 352 | 1.053 | 394 | 1.095 | 1233.428 | 0.531 | 0.723 | 0.947 | 0.96 | 0.854 | 1.124 | 0.753 | Chain A: 33,37,40,41,43,44,45,47,48,49,50,51,52,54 ,55,267,268,269,270,271,272,273,274,275,276,277,27 8,279,280,282,283,284,286,295,296,297,298,313,314, 315,316,317,318,319,321,367,368,369,370,373,383,38 5,389,411,412,413,414,415,416,419,460,461,462,463, 464,471,472,473,479 |
| 370 | 0.924 | 2827 | 1.0373 | 1578.486 | 0.7292 | 0.4233 | 0.3729 | 0.2356 | 0.4807 | 0.4902 | 0.9445 | Chain A: 15,19,20,23,24,26,27,30,31,33,34,35,36,37 ,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,5 4,55,56,58,59,270,271,272,273,274,275,276,277,278, 279,280,281,282,283,284,285,286,287,288,289,290,29 1,292,293,295,297,299,300,301,302,306,307,308,309, 310,311,313,317,319,320,321,324,325,326,327,328,32 9,330,332,340,341,343,344,345,349,350,351,352,353, 355,367,368,369,370,371,372,373,374,375,378,379,38 0,381,384,387,393,394,395,396,397,398,399,400,401, 403,405,410,411,412,415,417,420,421,422,423,424,42 5,426,427,428,430,431,464,465,466,467,468,469,471, 472,473,474,475,476,477,479,482,483,484,485,486,48 7,488,489,490,491,492,515,516,517,518,519,520,521, 522,523,524,525,526,527,528,529,530,531,532,533,53 4,536 |
| 481 | 1.027 | 100 | 1.066 | 358.778 | 0.598 | 0.695 | 0.952 | 0.729 | 0.891 | 0.818 | 0.783 | Chain A: 239,264,267,268,271,272,273,274,275,276,2 77,278,279,280,282,283,287,291,292,294,295,297 |
| 518 | 1.041 | 352 | 1.068 | 963.83 | 0.536 | 0.736 | 0.942 | 0.697 | 0.962 | 0.725 | 0.938 | Chain A: 145,146,147,148,149,150,151,153,154,155,1 72,174,191,195,204,220,221,222,223,224,225,226,227 ,229,230,231,233,234,271,273,277,278,280,293,294,2 95,296,297,298,299,300,313,314,315,316,317,318,319 ,352,358,359,523,524,525,527,529,530,531,532,533,5 34,535,536,538 |
| 567 | 1.021 | 292 | 1.035 | 866.761 | 0.521 | 0.73 | 0.981 | 0.734 | 1.051 | 0.699 | 0.554 | Chain A: 197,199,200,201,202,203,204,207,209,225,2 27,244,248,257,273,274,275,276,277,279,280,282,283 ,286,289,324,326,330,331,333,346,347,348,349,350,3 51,352,366,367,368,369,370,371,372,403,404,405,407 ,408,409,410,412,579,580,581,582,583,584,585,586,5 87,588 |
| 590 | 1.047 | 218 | 1.078 | 552.916 | 0.491 | 0.738 | 0.962 | 1.055 | 0.935 | 1.128 | 0.66 | Chain A: 557,558,559,560,561,562,563,564,565,582,5 84,586,593,596,597,599,600,604,613,629,631,633,634 ,636,637,638,641,642,645,646,677,678,682,683,685,6 95,696,697,698,699,700,701 |
| 595 | 1.035 | 135 | 1.095 | 368.725 | 0.591 | 0.666 | 0.859 | 0.92 | 0.749 | 1.229 | 0.997 | Chain A: 28,29,32,33,36,37,39,40,41,397,604,605,60 6,607,609,610,611,612,613,614,616,618,620,622 |
| 639 | 1.081 | 141 | 1.097 | 329.966 | 0.502 | 0.809 | 1.049 | 0.745 | 1.008 | 0.739 | 0.746 | Chain A: 273,274,277,314,315,316,317,321,368,369,3 70,373,413,414,415,416,419,460,461,462,463,464,465 ,471,513,515,516,517,518,519,523,631,632,633,634,6 36,637,638 |
| 642 | 1.016 | 198 | 1.049 | 605.738 | 0.609 | 0.692 | 0.853 | 0.654 | 0.939 | 0.697 | 0.605 | Chain A: 40,47,48,51,262,263,264,265,408,409,410,4 11,413,438,441,442,443,445,446,447,448,449,450,451 ,452,453,454,456,457,458,461,465,466,469,471,475,4 79,504 |
| 644 | 1.1 | 155 | 1.169 | 268.912 | 0.362 | 0.735 | 1.026 | 1.698 | 0.65 | 2.61 | 0.89 | Chain A: 281,524,525,527,535,536,537,538,539,540,5 42,544,557,559,562,564,646,647,648,649,650,651,652 ,653 |
| 649 | 1.024 | 112 | 1.045 | 296.695 | 0.539 | 0.727 | 0.941 | 0.659 | 1.018 | 0.648 | 0.675 | Chain A: 424,449,452,453,456,457,458,459,460,461,4 62,463,465,467,468,469,476,477,480,482,515 |
| 752 | 1.099 | 112 | 1.159 | 409.542 | 0.582 | 0.751 | 0.989 | 1.468 | 0.711 | 2.064 | 0.872 | Chain A: 503,504,505,506,507,508,511,529,530,531,5 48,552,561,577,578,580,581,583,584,587,628,630,634 ,635,637,650,651,652,653,654,655,656,670,671,672,6 73 |
| 785 | 1.041 | 386 | 1.043 | 814.625 | 0.459 | 0.759 | 0.994 | 0.629 | 1.081 | 0.582 | 0.724 | Chain A: 268,270,271,272,273,274,275,276,277,278,2 79,280,282,287,313,314,315,316,317,319,321,329,330 ,364,367,368,369,370,371,373,383,384,385,389,411,4 12,413,414,415,416,419,458,460,461,462,463,464,471 ,472,473,475,479,943,944,945,946,1011,1012,1013,10 14,1015,1016,1017,1018,1019,1020,1022 |
| 788 | 1.056 | 307 | 1.045 | 582.757 | 0.404 | 0.782 | 1.074 | 0.664 | 1.118 | 0.594 | 0.857 | Chain A: 282,283,284,285,286,287,288,289,290,291,2 98,324,325,326,327,328,330,332,378,379,380,381,382 ,384,394,396,400,422,423,424,425,426,427,471,472,4 73,474,475,482,483,484,490,1022,1023,1025,1026,102 7,1028,1029,1030 |
| 867 | 1.069 | 450 | 0.978 | 1105.146 | 0.366 | 0.802 | 1.1 | 0.384 | 1.359 | 0.283 | 0.949 | Chain A: 213,214,215,216,217,218,219,220,221,222,2 29,255,256,257,258,259,260,261,263,306,309,310,311 ,312,313,315,325,327,328,329,331,351,352,353,354,3 55,356,357,358,402,403,404,405,406,455,456,457,458 ,459,460,461,498,499,500,501,502,503,504,505,1134, 1135,1136,1137,1142,1143,1145 |
| 878 | 1.07 | 169 | 1.11 | 583.443 | 0.624 | 0.749 | 0.87 | 0.738 | 0.856 | 0.862 | 0.805 | Chain A: 39,206,207,208,210,212,213,230,231,253,51 5,532,533,534,535,536,537,538,540,549,551,561,1167 ,1168,1170,1171,1172,1244,1245,1246,1247,1248 |
| 882 | 1.063 | 266 | 1.094 | 776.552 | 0.528 | 0.757 | 0.937 | 0.692 | 0.919 | 0.753 | 1.372 | Chain A: 595,596,597,598,599,600,601,602,603,604,6 05,606,607,621,638,639,640,641,643,679,680,682,683 ,685,725,726,727,728,729,731,1261,1262,1264,1265,1 266,1267,1268,1269,1270,1271,1272,1273 |
| 884 | 1.048 | 436 | 1.038 | 1265.67 | 0.469 | 0.769 | 1.008 | 0.511 | 1.113 | 0.459 | 1.072 | Chain A: 29,30,32,33,35,36,37,38,39,40,42,282,283, 284,285,286,287,288,289,290,291,293,298,324,325,32 6,327,328,329,330,331,332,378,379,380,381,382,384, 387,394,396,400,422,423,424,425,426,427,430,431,47 1,472,473,474,475,476,477,479,524,525,526,527,528, 529,530,567,568,569,570,571,572,573 |
| 891 | 1.091 | 320 | 0.967 | 790.615 | 0.351 | 0.834 | 1.153 | 0.323 | 1.454 | 0.222 | 0.827 | Chain A: 213,214,215,216,217,218,219,220,221,222,2 24,229,255,256,257,258,259,261,263,309,310,311,312 ,313,315,325,327,331,354,355,356,357,358,402,403,4 04,405,406,455,456,457,458,459,460,461,498,499,500 ,501,502,504,1224,1225 |
| 892 | 1.052 | 393 | 1.04 | 845.152 | 0.458 | 0.776 | 0.998 | 0.41 | 1.122 | 0.366 | 0.958 | Chain A: 231,232,310,331,332,333,334,595,596,597,5 98,599,600,602,637,638,639,640,641,643,677,679,680 ,681,682,683,684,685,695,723,724,725,726,728,729,7 39,740,741,743,769,770,772,774,775,779,783,1295,12 96,1298,1299,1300,1301,1302,1303,1304,1305,1306,13 07,1308,1309 |
| 896 | 1.094 | 164 | 1.121 | 395.136 | 0.452 | 0.806 | 1.052 | 0.705 | 0.927 | 0.761 | 2.214 | Chain A: 610,611,613,614,651,652,654,693,694,695,6 96,739,740,780,781,782,783,1306,1307,1308,1309,131 0,1311,1312,1313 |
| 908 | 1.074 | 385 | 1.026 | 1171.002 | 0.44 | 0.809 | 1.076 | 0.508 | 1.227 | 0.414 | 0.789 | Chain A: 40,43,44,46,47,50,51,268,270,271,272,273, 274,275,276,277,278,279,280,287,313,314,315,316,31 7,318,319,321,329,367,368,369,370,371,373,383,384, 385,389,411,412,413,414,415,416,458,460,461,462,46 3,464,513,514,515,516,517,518,519,556,557,558,559, 560,561,562,563 |
| 912 | 1.061 | 161 | 1.088 | 575.554 | 0.559 | 0.761 | 0.971 | 0.865 | 0.946 | 0.914 | 0.83 | Chain A: 946,950,1035,1060,1062,1063,1064,1066,106 7,1068,1069,1070,1071,1072,1073,1074,1075,1076,107 8,1079,1080,1083,1087,1088,1089,1091,1093,1123,112 6,1378,1379,1380,1381 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
| 352 | ZINC000150588351 | DB11574 | Elbasvir | -8.443 | -9.8345 |
| 352 | ZINC000001550477 | DB01259 | Lapatinib | -7.18653 | -8.35493 |
| 352 | ZINC000003943279 | DB06695 | Dabigatran etexilate | -5.96896 | -7.44016 |
| 352 | ZINC000043207851 | DB08877 | Ruxolitinib | -7.38566 | -7.38566 |
| 352 | ZINC000000896731 | DB00744 | Zileuton | -7.32304 | -7.34164 |
| 352 | ZINC000003775644 | DB01123 | Proflavine | -7.26336 | -7.29416 |
| 352 | ZINC000096006020 | DB01229 | Paclitaxel | -7.16034 | -7.16184 |
| 352 | ZINC000003872994 | DB01026 | Ketoconazole | -6.81185 | -7.13655 |
| 352 | ZINC000068204830 | DB09102 | Daclatasvir | -6.22856 | -7.06876 |
| 352 | ZINC000029571072 | DB06636 | Isavuconazonium | -5.2512 | -7.0235 |
| 352 | ZINC000008577218 | DB00158 | Folic acid | -5.61894 | -6.96364 |
| 352 | ZINC000000033882 | DB00988 | Dopamine | -6.90882 | -6.90882 |
| 352 | ZINC000000000850 | DB00744 | Zileuton | -6.88987 | -6.90847 |
| 352 | Baricitinib | DB00744 | -6.88438 | -6.88438 | |
| 352 | ZINC000022059930 | DB01155 | Gemifloxacin | -6.80622 | -6.84712 |
| 352 | ZINC000150601177 | DB09296 | Ombitasvir | -6.83123 | -6.83123 |
| 352 | ZINC000000002055 | DB08797 | Salicylamide | -5.16212 | -6.80912 |
| 352 | ZINC000008035395 | DB09134 | Ioversol | -6.8074 | -6.8074 |
| 352 | ZINC000004235575 | DB11279 | Brilliant green cation | -6.06717 | -6.77387 |
| 352 | ZINC000000643138 | DB01026 | Ketoconazole | -6.19845 | -6.74715 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
| ZINC000150588351 | DB11574 | Elbasvir | Small molecule | [H][C@]1(CCCN1C(=O)[C@@H](NC(=O)OC)C(C)C)C1=NC=C(N1)C1=CC2=C(C=C1)N1[C@@H](OC3=C(C=CC(=C3)C3=CN=C(N3)[C@]3([H])CCCN3C(=O)[C@@H](NC(=O)OC)C(C)C)C1=C2)C1=CC=CC=C1 | Approved |
| ZINC000001550477 | DB01259 | Lapatinib | Small molecule | CS(=O)(=O)CCNCC1=CC=C(O1)C1=CC2=C(C=C1)N=CN=C2NC1=CC(Cl)=C(OCC2=CC(F)=CC=C2)C=C1 | Approved|Investigational |
| ZINC000003943279 | DB06695 | Dabigatran etexilate | Small molecule | CCCCCCOC(=O)N=C(N)C1=CC=C(NCC2=NC3=C(C=CC(=C3)C(=O)N(CCC(=O)OCC)C3=NC=CC=C3)N2C)C=C1 | Approved |
| ZINC000043207851 | DB08877 | Ruxolitinib | Small molecule | N#CC[C@H](C1CCCC1)N1C=C(C=N1)C1=C2C=CNC2=NC=N1 | Approved |
| ZINC000000896731 | DB00744 | Zileuton | Small molecule | CC(N(O)C(N)=O)C1=CC2=CC=CC=C2S1 | Approved|Investigational|Withdrawn |
| ZINC000003775644 | DB01123 | Proflavine | Small molecule | NC1=CC2=NC3=C(C=CC(N)=C3)C=C2C=C1 | Approved |
| ZINC000096006020 | DB01229 | Paclitaxel | Small molecule | [H][C@]12[C@H](OC(=O)C3=CC=CC=C3)[C@]3(O)C[C@H](OC(=O)[C@H](O)[C@@H](NC(=O)C4=CC=CC=C4)C4=CC=CC=C4)C(C)=C([C@@H](OC(C)=O)C(=O)[C@]1(C)[C@@H](O)C[C@H]1OC[C@@]21OC(C)=O)C3(C)C | Approved|Vet_approved |
| ZINC000068204830 | DB09102 | Daclatasvir | Small molecule | COC(=O)N[C@@H](C(C)C)C(=O)N1CCC[C@H]1C1=NC=C(N1)C1=CC=C(C=C1)C1=CC=C(C=C1)C1=CN=C(N1)[C@@H]1CCCN1C(=O)[C@@H](NC(=O)OC)C(C)C | Approved|Investigational |
| ZINC000008577218 | DB00158 | Folic acid | Small molecule | NC1=NC(=O)C2=NC(CNC3=CC=C(C=C3)C(=O)N[C@@H](CCC(O)=O)C(O)=O)=CN=C2N1 | Approved|Nutraceutical|Vet_approved |
| ZINC000000033882 | DB00988 | Dopamine | Small molecule | NCCC1=CC(O)=C(O)C=C1 | Approved |
| ZINC000022059930 | DB01155 | Gemifloxacin | Small molecule | CON=C1/CN(CC1CN)C1=NC2=C(C=C1F)C(=O)C(=CN2C1CC1)C(O)=O | Approved|Investigational |
| ZINC000150601177 | DB09296 | Ombitasvir | Small molecule | COC(=O)N[C@@H](C(C)C)C(=O)N1CCC[C@H]1C(=O)NC1=CC=C(C=C1)[C@@H]1CC[C@H](N1C1=CC=C(C=C1)C(C)(C)C)C1=CC=C(NC(=O)[C@@H]2CCCN2C(=O)[C@@H](NC(=O)OC)C(C)C)C=C1 | Approved|Investigational |
| ZINC000000002055 | DB08797 | Salicylamide | Small molecule | NC(=O)C1=CC=CC=C1O | Approved |
| ZINC000004235575 | DB11279 | Brilliant green cation | Small molecule | CCN(CC)C1=CC=C(C=C1)C(C1=CC=CC=C1)=C1C=CC(C=C1)=[N+](CC)CC | Approved|Vet_approved |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
| ZINC000150588351 | 882.03 | 6.572 | 1345.74 | 717.517 | 186.345 | 441.878 | 0 | 2635.972 | 2.5 | 13.25 | 8.001 | 1 | 68.973 | 3 | 1 |
| ZINC000150588351 | 882.03 | 15.581 | 1298.014 | 670.728 | 196.39 | 430.896 | 0 | 2598.655 | 2.5 | 13.25 | 8.107 | 1 | 65.891 | 3 | 1 |
| ZINC000150588351 | 882.03 | 7.694 | 1389.327 | 750.107 | 173.462 | 465.758 | 0 | 2673.848 | 2.5 | 13.25 | 8.124 | 1 | 73.085 | 3 | 1 |
| ZINC000150588351 | 882.03 | 2.219 | 1373.023 | 721.483 | 195.055 | 456.485 | 0 | 2655.192 | 2.5 | 13.25 | 8.091 | 1 | 68.497 | 3 | 1 |
| ZINC000150588351 | 882.03 | 13.761 | 1347.364 | 706.26 | 205.501 | 435.603 | 0 | 2635.257 | 2.5 | 13.25 | 8.253 | 1 | 64.824 | 3 | 1 |
| ZINC000150588351 | 882.03 | 6.922 | 1403.807 | 740.212 | 182.531 | 481.064 | 0 | 2678.744 | 2.5 | 13.25 | 8.307 | 1 | 71.991 | 3 | 1 |
| ZINC000150588351 | 882.03 | 13.99 | 1367.982 | 729.766 | 188.685 | 449.531 | 0 | 2678.673 | 2.5 | 13.25 | 7.973 | 1 | 70.208 | 3 | 2 |
| ZINC000150588351 | 882.03 | 10.656 | 1391.126 | 719.587 | 194.166 | 477.373 | 0 | 2673.624 | 2.5 | 13.25 | 7.937 | 1 | 69.244 | 3 | 2 |
| ZINC000150588351 | 882.03 | 8.233 | 1375.056 | 718.117 | 197.908 | 459.032 | 0 | 2655.63 | 2.5 | 13.25 | 7.962 | 1 | 67.787 | 3 | 2 |
| ZINC000001550477 | 581.06 | 8.763 | 984.429 | 219.392 | 136.563 | 520.447 | 108.027 | 1738.439 | 1 | 8.25 | 8.353 | 1 | 75.112 | 2 | 1 |
| ZINC000001550477 | 581.06 | 8.228 | 914.704 | 239.853 | 89.107 | 476.75 | 108.994 | 1676.884 | 1 | 8.25 | 8.025 | 1 | 82.627 | 2 | 1 |
| ZINC000003943279 | 627.742 | 4.667 | 1184.331 | 574.306 | 231.253 | 378.772 | 0 | 2103.011 | 2 | 11 | 8.55 | 1 | 58.304 | 3 | 1 |
| ZINC000003943279 | 627.742 | 7.938 | 1170.006 | 572.981 | 237.303 | 359.722 | 0 | 2090.012 | 2 | 11 | 8.404 | 1 | 56.447 | 3 | 1 |
| ZINC000003943279 | 627.742 | 6.121 | 1133.409 | 561.818 | 218.501 | 353.089 | 0 | 2055.355 | 2 | 11 | 8.475 | 1 | 59.143 | 3 | 1 |
| ZINC000003943279 | 627.742 | 10.2 | 1166.266 | 573.103 | 235.814 | 357.349 | 0 | 2084.657 | 2 | 11 | 8.292 | 1 | 56.55 | 3 | 1 |
| ZINC000003943279 | 627.742 | 9.595 | 1158.598 | 580.852 | 244.759 | 332.987 | 0 | 2079.282 | 2 | 11 | 8.313 | 1 | 54.32 | 3 | 1 |
| ZINC000003943279 | 627.742 | 7.327 | 1178.116 | 572.965 | 232.033 | 373.118 | 0 | 2096.733 | 2 | 11 | 8.58 | 1 | 57.88 | 3 | 1 |
| ZINC000003943279 | 627.742 | 9.958 | 1175.042 | 575.778 | 234.38 | 364.884 | 0 | 2092.735 | 2 | 11 | 8.327 | 1 | 57.186 | 3 | 1 |
| ZINC000043207851 | 306.369 | 8.338 | 581.1 | 222.006 | 148.565 | 210.529 | 0 | 1017.193 | 1 | 5 | 8.708 | 3 | 89.622 | 0 | 0 |
| ZINC000000896731 | 236.288 | 3.773 | 444.598 | 84.301 | 131.134 | 195.105 | 34.058 | 746.481 | 3 | 3.7 | 8.644 | 3 | 78.18 | 0 | 0 |
| ZINC000000896731 | 236.288 | 5.297 | 450.816 | 85.528 | 135.414 | 201.881 | 27.993 | 746.812 | 3 | 3.7 | 8.57 | 3 | 76.556 | 0 | 0 |
| ZINC000003775644 | 209.25 | 0.181 | 433.081 | 0 | 137.936 | 295.144 | 0 | 706.541 | 3 | 3 | 7.603 | 3 | 83.637 | 0 | 0 |
| ZINC000003775644 | 209.25 | 0.371 | 432.112 | 0 | 138.151 | 293.961 | 0 | 705.014 | 3 | 3 | 7.627 | 3 | 83.535 | 0 | 0 |
| ZINC000096006020 | 853.918 | 7.222 | 1008.963 | 367.497 | 222.021 | 419.445 | 0 | 2212.853 | 3 | 17.65 | 9.796 | 2 | 61.419 | 2 | 2 |
| ZINC000003872994 | 531.438 | 2.643 | 843.259 | 319.79 | 80.343 | 323.603 | 119.523 | 1551.551 | 0 | 8.25 | 8.241 | 3 | 92.53 | 1 | 0 |
| ZINC000003872994 | 531.438 | 6.575 | 844.887 | 311.956 | 81.254 | 336.082 | 115.594 | 1549.966 | 0 | 8.25 | 8.233 | 3 | 92.405 | 1 | 0 |
| ZINC000003872994 | 531.438 | 5.578 | 860.223 | 333.686 | 78.629 | 331.472 | 116.437 | 1569.565 | 0 | 8.25 | 9.26 | 3 | 92.881 | 1 | 0 |
| ZINC000003872994 | 531.438 | 5.486 | 858.441 | 334.024 | 79.024 | 328.466 | 116.927 | 1566.075 | 0 | 8.25 | 9.254 | 3 | 92.765 | 1 | 0 |
| ZINC000068204830 | 738.885 | 9.186 | 1185.225 | 698.369 | 189.608 | 297.248 | 0 | 2297.481 | 2.5 | 12.5 | 8.424 | 1 | 56.25 | 3 | 1 |
| ZINC000068204830 | 738.885 | 12.728 | 1242.653 | 732.724 | 195.462 | 314.467 | 0 | 2325.851 | 2.5 | 12.5 | 8.586 | 1 | 56.993 | 3 | 1 |
| ZINC000068204830 | 738.885 | 5.659 | 1215.343 | 718.68 | 189.061 | 307.602 | 0 | 2308.854 | 2.5 | 12.5 | 8.456 | 1 | 57.668 | 3 | 1 |
| ZINC000068204830 | 738.885 | 3.036 | 1238.237 | 724.613 | 189.774 | 323.849 | 0 | 2337.465 | 2.5 | 12.5 | 8.529 | 1 | 58.588 | 3 | 1 |
| ZINC000068204830 | 738.885 | 6.612 | 1209.345 | 719.031 | 182.477 | 307.837 | 0 | 2307.347 | 2.5 | 12.5 | 8.288 | 1 | 59.648 | 3 | 1 |
| ZINC000068204830 | 738.885 | 4.849 | 1244.561 | 737.115 | 179.716 | 327.731 | 0 | 2332.759 | 2.5 | 12.5 | 8.571 | 1 | 60.798 | 3 | 1 |
| ZINC000008577218 | 441.402 | 13.51 | 755.446 | 101.238 | 451.657 | 202.551 | 0 | 1308.074 | 6.25 | 12.75 | 8.35 | 1 | 0 | 2 | 1 |
| ZINC000008577218 | 441.402 | 14.14 | 754.923 | 100.926 | 451.592 | 202.405 | 0 | 1306.451 | 6.25 | 12.75 | 8.302 | 1 | 0 | 2 | 1 |
| ZINC000008577218 | 441.402 | 14.323 | 755.349 | 100.67 | 452.288 | 202.391 | 0 | 1307.577 | 6.25 | 12.75 | 8.399 | 1 | 0 | 2 | 1 |
| ZINC000000033882 | 153.18 | 2.708 | 369.386 | 84.168 | 166.575 | 118.644 | 0 | 574.834 | 4 | 2.5 | 8.835 | 2 | 53.663 | 0 | 0 |
| ZINC000000000850 | 236.288 | 5.297 | 457.642 | 85.783 | 137.035 | 198.523 | 36.3 | 755.015 | 3 | 3.7 | 8.91 | 3 | 75.351 | 0 | 0 |
| ZINC000000000850 | 236.288 | 5.297 | 450.837 | 85.532 | 135.419 | 201.893 | 27.994 | 746.839 | 3 | 3.7 | 8.57 | 3 | 76.555 | 0 | 0 |
| ZINC000022059930 | 389.385 | 5.771 | 675.499 | 333.722 | 232.902 | 67.773 | 41.101 | 1199.671 | 2 | 8.7 | 9.049 | 2 | 36.482 | 0 | 1 |
| ZINC000022059930 | 389.385 | 7.083 | 660.215 | 323.57 | 227.182 | 68.271 | 41.191 | 1181.717 | 2 | 8.7 | 9.157 | 2 | 37.113 | 0 | 1 |
| ZINC000150601177 | 894.121 | 2.674 | 1361.773 | 931.062 | 187.357 | 243.353 | 0 | 2751.452 | 2.5 | 15.5 | 7.779 | 1 | 67.605 | 3 | 2 |
| ZINC000000002055 | 137.138 | 6.473 | 320.989 | 0 | 148.306 | 172.683 | 0 | 487.389 | 2 | 2.25 | 9.228 | 3 | 80.822 | 0 | 0 |
| ZINC000000002055 | 137.138 | 5.405 | 320.141 | 0 | 149.757 | 170.384 | 0 | 487.073 | 2 | 2.25 | 9.337 | 3 | 80.506 | 0 | 0 |
| ZINC000008035395 | 807.116 | 12.861 | 789.663 | 274.386 | 383.301 | 10.854 | 121.123 | 1453.819 | 8 | 18.2 | 9.427 | 1 | 0 | 3 | 1 |
| ZINC000000643138 | 531.438 | 3.645 | 839.043 | 328.65 | 81.288 | 309.88 | 119.224 | 1549.678 | 0 | 8.25 | 8.204 | 3 | 92.395 | 1 | 0 |
| ZINC000000643138 | 531.438 | 7.586 | 808.295 | 331.514 | 70.114 | 287.19 | 119.477 | 1511.81 | 0 | 8.25 | 7.964 | 3 | 94.972 | 1 | 0 |
| ZINC000000643138 | 531.438 | 4.172 | 832.524 | 334.374 | 75.085 | 307.237 | 115.829 | 1545.459 | 0 | 8.25 | 9.194 | 3 | 93.862 | 1 | 0 |
| ZINC000000643138 | 531.438 | 6.909 | 811.792 | 344.459 | 66.106 | 282.902 | 118.325 | 1517.814 | 0 | 8.25 | 9.025 | 3 | 95.484 | 1 | 0 |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
| ZINC000150588351 | COC(=O)N[C@H](C(=O)N1CCC[C@H]1c1nc(-c2ccc3c(c2)O[C@@H](c2ccccc2)n2c-3cc3cc(-c4c[nH]c([C@@H]5CCCN5C(=O)[C@@H](NC(=O)OC)C(C)C)n4)ccc32)c[nH]1)C(C)C | 0.015875917 | 0.217012798 |
| ZINC000001550477 | CS(=O)(=O)CCNCc1ccc(-c2ccc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3c2)o1 | 0.18759569 | 0.281219747 |
| ZINC000003943279 | CCCCCCOC(=O)/N=C(N)c1ccc(NCc2nc3cc(C(=O)N(CCC(=O)OCC)c4ccccn4)ccc3n2C)cc1 | 0.136066223 | 0.265677856 |
| ZINC000043207851 | N#CC[C@H](C1CCCC1)n1cc(-c2ncnc3[nH]ccc23)cn1 | 0.090743892 | 0.35666618 |
| ZINC000000896731 | C[C@H](c1cc2ccccc2s1)N(O)C(N)=O | 0.136335882 | 0.223511692 |
| ZINC000003775644 | Nc1ccc2cc3ccc(N)cc3nc2c1 | 0.346762085 | 0.473171062 |
| ZINC000096006020 | CC(=O)O[C@H]1C(=O)[C@]2(C)[C@@H](O)C[C@H]3OC[C@@]3(OC(C)=O)[C@H]2[C@H](OC(=O)c2ccccc2)[C@]2(O)C[C@H](OC(=O)[C@H](O)[C@@H](NC(=O)c3ccccc3)c3ccccc3)C(C)=C1C2(C)C | 0.007331142 | 0.303016135 |
| ZINC000003872994 | CC(=O)N1CCN(c2ccc(OC[C@H]3CO[C@@](Cn4ccnc4)(c4ccc(Cl)cc4Cl)O3)cc2)CC1 | 0.087393272 | 0.387082142 |
| ZINC000068204830 | COC(=O)N[C@H](C(=O)N1CCC[C@H]1c1ncc(-c2ccc(-c3ccc(-c4cnc([C@@H]5CCCN5C(=O)[C@@H](NC(=O)OC)C(C)C)[nH]4)cc3)cc2)[nH]1)C(C)C | 0.039971092 | 0.226481814 |
| ZINC000029571072 | CNCC(=O)OCc1cccnc1N(C)C(=O)O[C@H](C)[n+]1cnn(C[C@](O)(c2cc(F)ccc2F)[C@@H](C)c2nc(-c3ccc(C#N)cc3)cs2)c1 | 0.025416027 | 0.235213823 |
| ZINC000008577218 | Nc1nc(O)c2nc(CNc3ccc(C(=O)N[C@@H](CCC(=O)O)C(=O)O)cc3)cnc2n1 | 0.127688186 | 0.392534973 |
| ZINC000000033882 | NCCc1ccc(O)c(O)c1 | 0.385526059 | 0.415023995 |
| ZINC000000000850 | C[C@@H](c1cc2ccccc2s1)N(O)C(N)=O | 0.136335882 | 0.223511692 |
| ZINC000022059930 | CO/N=C1CN(c2nc3c(cc2F)c(=O)c(C(=O)O)cn3C2CC2)C[C@H]1CN | 0.077611514 | 0.258972331 |
| ZINC000150601177 | COC(=O)N[C@H](C(=O)N1CCC[C@H]1C(=O)Nc1ccc([C@@H]2CC[C@@H](c3ccc(NC(=O)[C@@H]4CCCN4C(=O)[C@@H](NC(=O)OC)C(C)C)cc3)N2c2ccc(C(C)(C)C)cc2)cc1)C(C)C | 0.026337368 | 0.185460331 |
| ZINC000000002055 | NC(=O)c1ccccc1O | 0.561746126 | 0.479667743 |
| ZINC000008035395 | O=C(NC[C@@H](O)CO)c1c(I)c(C(=O)NC[C@@H](O)CO)c(I)c(N(CCO)C(=O)CO)c1I | 0.051780669 | 0.23149478 |
| ZINC000004235575 | CCN(CC)c1ccc(C(=C2C=CC(=[N+](CC)CC)C=C2)c2ccccc2)cc1 | 0.165302956 | 0.522075975 |
| ZINC000000643138 | CC(=O)N1CCN(c2ccc(OC[C@H]3CO[C@](Cn4ccnc4)(c4ccc(Cl)cc4Cl)O3)cc2)CC1 | 0.087393272 | 0.387082142 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| EML4 | NEK6, ARL4D, ARIH2, ELAVL1, EML4, ILK, PRPS1, TUBB2A, TUBB2B, FBXW11, NTRK1, SPICE1, DCTN1, TFEB, Nek9, NUDC, EML1, FBXO28, TUBB3, CTAGE5, TUBB, TKT, PHKG2, NEK7, SKP1, EFTUD2, FGF11, MYC, CDK9, KIAA1429, ATG16L1, Bach1, N, HCVgp1, TUBA1B, DDRGK1, TP53, KIF2A, MAPRE1, MAPRE3, TRIM36, TUBA4A, WDR5, C15orf59, C1QTNF1, PABPN1L, BAG2, CAPSL, PRPS2, HSPA8, TUBB6, KXD1, ZNF174, GPS2, NEK9, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| EML4 | 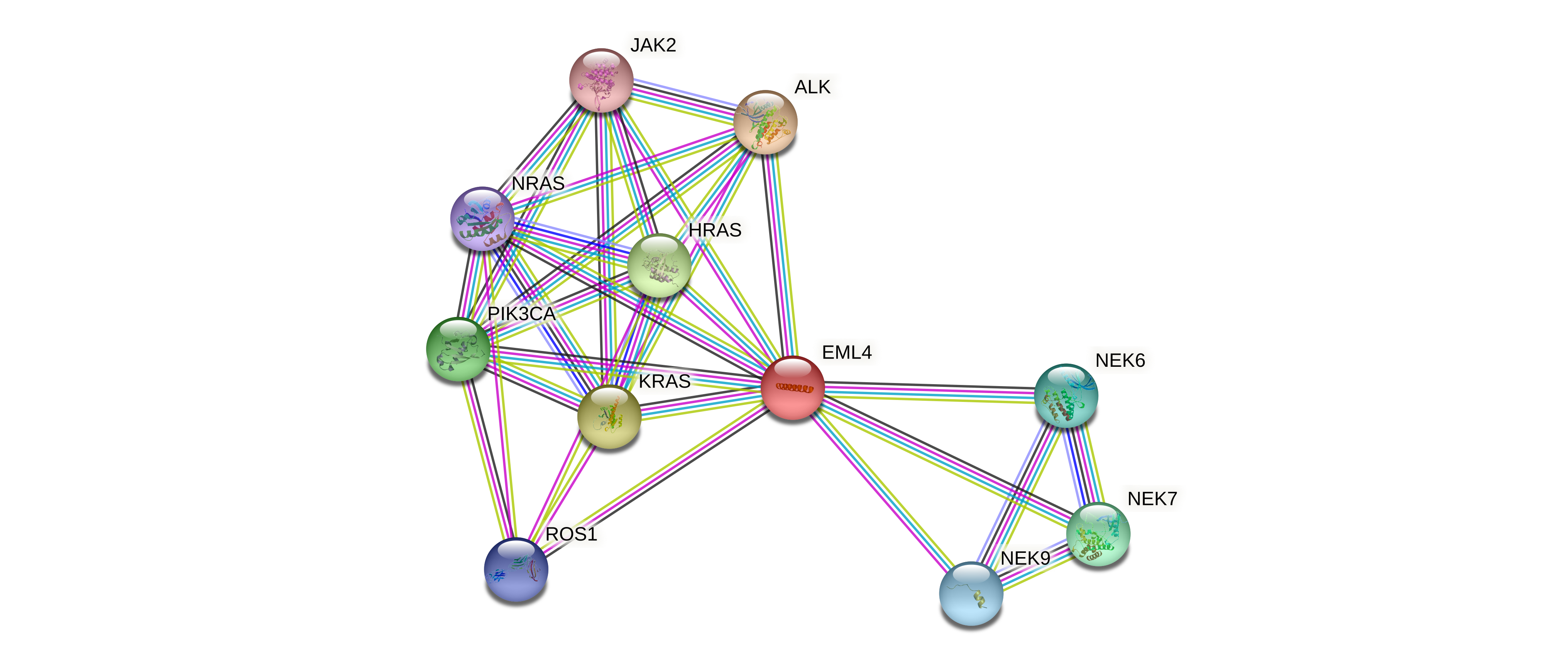 |
| ALK |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to EML4-ALK |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
| EML4 | ALK | Crizotinib | PubMed | 33157918 |
Top |
Related Diseases to EML4-ALK |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| EML4 | ALK | Lung Adenocarcinoma | PubMed | 33157918 |
| EML4 | ALK | Lung Adenocarcinoma | MyCancerGenome | |
| EML4 | ALK | Non-Small Cell Lung Carcinoma | MyCancerGenome | |
| EML4 | ALK | Adenocarcinoma Of Unknown Primary | MyCancerGenome | |
| EML4 | ALK | Unknown | MyCancerGenome | |
| EML4 | ALK | Breast Invasive Ductal Carcinoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | EML4 | C0007131 | Non-Small Cell Lung Carcinoma | 6 | CTD_human |
| Hgene | EML4 | C0027627 | Neoplasm Metastasis | 2 | CTD_human |
| Hgene | EML4 | C0152013 | Adenocarcinoma of lung (disorder) | 2 | CTD_human |
| Hgene | EML4 | C0006118 | Brain Neoplasms | 1 | CTD_human |
| Hgene | EML4 | C0153633 | Malignant neoplasm of brain | 1 | CTD_human |
| Hgene | EML4 | C0496899 | Benign neoplasm of brain, unspecified | 1 | CTD_human |
| Hgene | EML4 | C0750974 | Brain Tumor, Primary | 1 | CTD_human |
| Hgene | EML4 | C0750977 | Recurrent Brain Neoplasm | 1 | CTD_human |
| Hgene | EML4 | C0750979 | Primary malignant neoplasm of brain | 1 | CTD_human |
| Hgene | EML4 | C1527390 | Neoplasms, Intracranial | 1 | CTD_human |
| Tgene | ALK | C0007131 | Non-Small Cell Lung Carcinoma | 28 | CGI;CTD_human |
| Tgene | ALK | C0027819 | Neuroblastoma | 13 | CGI;CTD_human;ORPHANET |
| Tgene | ALK | C0152013 | Adenocarcinoma of lung (disorder) | 8 | CGI;CTD_human |
| Tgene | ALK | C2751681 | NEUROBLASTOMA, SUSCEPTIBILITY TO, 3 | 8 | CLINGEN;UNIPROT |
| Tgene | ALK | C0206180 | Ki-1+ Anaplastic Large Cell Lymphoma | 6 | CGI;CTD_human |
| Tgene | ALK | C0334121 | Inflammatory Myofibroblastic Tumor | 4 | CGI;CTD_human;ORPHANET |
| Tgene | ALK | C0018199 | Granuloma, Plasma Cell | 3 | CTD_human |
| Tgene | ALK | C0007621 | Neoplastic Cell Transformation | 2 | CTD_human |
| Tgene | ALK | C0027627 | Neoplasm Metastasis | 2 | CTD_human |
| Tgene | ALK | C0238463 | Papillary thyroid carcinoma | 2 | ORPHANET |
| Tgene | ALK | C0001973 | Alcoholic Intoxication, Chronic | 1 | PSYGENET |
| Tgene | ALK | C0006118 | Brain Neoplasms | 1 | CGI;CTD_human |
| Tgene | ALK | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | ALK | C0007134 | Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0011570 | Mental Depression | 1 | PSYGENET |
| Tgene | ALK | C0011581 | Depressive disorder | 1 | PSYGENET |
| Tgene | ALK | C0027643 | Neoplasm Recurrence, Local | 1 | CTD_human |
| Tgene | ALK | C0036341 | Schizophrenia | 1 | PSYGENET |
| Tgene | ALK | C0079744 | Diffuse Large B-Cell Lymphoma | 1 | CTD_human |
| Tgene | ALK | C0085269 | Plasma Cell Granuloma, Pulmonary | 1 | CTD_human |
| Tgene | ALK | C0153633 | Malignant neoplasm of brain | 1 | CGI;CTD_human |
| Tgene | ALK | C0278601 | Inflammatory Breast Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0496899 | Benign neoplasm of brain, unspecified | 1 | CTD_human |
| Tgene | ALK | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0750974 | Brain Tumor, Primary | 1 | CTD_human |
| Tgene | ALK | C0750977 | Recurrent Brain Neoplasm | 1 | CTD_human |
| Tgene | ALK | C0750979 | Primary malignant neoplasm of brain | 1 | CTD_human |
| Tgene | ALK | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | ALK | C1266042 | Chromophobe Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1266043 | Sarcomatoid Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1266044 | Collecting Duct Carcinoma of the Kidney | 1 | CTD_human |
| Tgene | ALK | C1306837 | Papillary Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1332079 | Anaplastic Large Cell Lymphoma, ALK-Positive | 1 | ORPHANET |
| Tgene | ALK | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | ALK | C1527390 | Neoplasms, Intracranial | 1 | CTD_human |
| Tgene | ALK | C2931189 | Neural crest tumor | 1 | ORPHANET |
| Tgene | ALK | C3899155 | hereditary neuroblastoma | 1 | GENOMICS_ENGLAND |
| Tgene | ALK | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |