| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:EPS15-KMT2A |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: EPS15-KMT2A | FusionPDB ID: 27061 | FusionGDB2.0 ID: 27061 | Hgene | Tgene | Gene symbol | EPS15 | KMT2A | Gene ID | 2060 | 4297 |
| Gene name | epidermal growth factor receptor pathway substrate 15 | lysine methyltransferase 2A | |
| Synonyms | AF-1P|AF1P|MLLT5 | ALL-1|CXXC7|HRX|HTRX1|MLL|MLL1|MLL1A|TRX1|WDSTS | |
| Cytomap | 1p32.3 | 11q23.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | epidermal growth factor receptor substrate 15ALL1 fused gene from chromosome 1protein AF-1p | histone-lysine N-methyltransferase 2ACXXC-type zinc finger protein 7lysine (K)-specific methyltransferase 2Alysine N-methyltransferase 2Amixed lineage leukemia 1myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)trithorax-like | |
| Modification date | 20200313 | 20200319 | |
| UniProtAcc | Q9UBC2 | Q03164 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000371730, ENST00000371733, ENST00000396122, ENST00000493793, | ENST00000420751, ENST00000354520, ENST00000389506, ENST00000534358, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 17 X 13 X 11=2431 | 31 X 72 X 3=6696 |
| # samples | 17 | 79 | |
| ** MAII score | log2(17/2431*10)=-3.83794324189103 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(79/6696*10)=-3.08337496948588 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: EPS15 [Title/Abstract] AND KMT2A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | KMT2A(118355690)-EPS15(51946988), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor by not retaining the major functional domain in the partially deleted in-frame ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor by not retaining the major functional domain in the partially deleted in-frame ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target by not retaining the major functional domain in the partially deleted in-frame ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target by not retaining the major functional domain in the partially deleted in-frame ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a transcription factor by not retaining the major functional domain in the partially deleted in-frame ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a transcription factor by not retaining the major functional domain in the partially deleted in-frame ORF. KMT2A-EPS15 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KMT2A-EPS15 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. EPS15-KMT2A seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. EPS15-KMT2A seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. EPS15-KMT2A seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. EPS15-KMT2A seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. EPS15-KMT2A seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. EPS15-KMT2A seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. KMT2A-EPS15 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. KMT2A-EPS15 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. KMT2A-EPS15 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. KMT2A-EPS15 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. KMT2A-EPS15 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. KMT2A-EPS15 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. KMT2A-EPS15 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | EPS15 | GO:0048268 | clathrin coat assembly | 12807910 |
| Tgene | KMT2A | GO:0044648 | histone H3-K4 dimethylation | 25561738 |
| Tgene | KMT2A | GO:0045944 | positive regulation of transcription by RNA polymerase II | 20861184 |
| Tgene | KMT2A | GO:0051568 | histone H3-K4 methylation | 19556245 |
| Tgene | KMT2A | GO:0065003 | protein-containing complex assembly | 15199122 |
| Tgene | KMT2A | GO:0080182 | histone H3-K4 trimethylation | 20861184 |
| Tgene | KMT2A | GO:0097692 | histone H3-K4 monomethylation | 25561738|26324722 |
 Fusion gene breakpoints across EPS15 (5'-gene) Fusion gene breakpoints across EPS15 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across KMT2A (3'-gene) Fusion gene breakpoints across KMT2A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | EPS15 | chr1 | 51912631 | - | KMT2A | chr11 | 118353136 | |
| ChimerKB3 | . | . | EPS15 | chr1 | 51913717 | - | KMT2A | chr11 | 118354897 | |
| ChimerKB3 | . | . | EPS15 | chr1 | 51984870 | - | KMT2A | chr11 | 118354897 | |
| ChimerKB3 | . | . | EPS15 | chr1 | 51984870 | - | KMT2A | chr11 | 118354897 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000371730 | EPS15 | chr1 | 51984870 | - | ENST00000534358 | KMT2A | chr11 | 118354897 | + | 12528 | 43 | 10 | 7875 | 2621 |
| ENST00000371730 | EPS15 | chr1 | 51984870 | - | ENST00000389506 | KMT2A | chr11 | 118354897 | + | 9612 | 43 | 10 | 7866 | 2618 |
| ENST00000371730 | EPS15 | chr1 | 51984870 | - | ENST00000354520 | KMT2A | chr11 | 118354897 | + | 10277 | 43 | 10 | 7752 | 2580 |
| ENST00000371733 | EPS15 | chr1 | 51984870 | - | ENST00000534358 | KMT2A | chr11 | 118354897 | + | 12615 | 130 | 13 | 7962 | 2649 |
| ENST00000371733 | EPS15 | chr1 | 51984870 | - | ENST00000389506 | KMT2A | chr11 | 118354897 | + | 9699 | 130 | 13 | 7953 | 2646 |
| ENST00000371733 | EPS15 | chr1 | 51984870 | - | ENST00000354520 | KMT2A | chr11 | 118354897 | + | 10364 | 130 | 13 | 7839 | 2608 |
| ENST00000371730 | EPS15 | chr1 | 51913717 | - | ENST00000534358 | KMT2A | chr11 | 118354897 | 13146 | 661 | 10 | 8493 | 2827 | |
| ENST00000371730 | EPS15 | chr1 | 51913717 | - | ENST00000389506 | KMT2A | chr11 | 118354897 | 10230 | 661 | 10 | 8484 | 2824 | |
| ENST00000371730 | EPS15 | chr1 | 51913717 | - | ENST00000354520 | KMT2A | chr11 | 118354897 | 10895 | 661 | 10 | 8370 | 2786 | |
| ENST00000371733 | EPS15 | chr1 | 51913717 | - | ENST00000534358 | KMT2A | chr11 | 118354897 | 13233 | 748 | 13 | 8580 | 2855 | |
| ENST00000371733 | EPS15 | chr1 | 51913717 | - | ENST00000389506 | KMT2A | chr11 | 118354897 | 10317 | 748 | 13 | 8571 | 2852 | |
| ENST00000371733 | EPS15 | chr1 | 51913717 | - | ENST00000354520 | KMT2A | chr11 | 118354897 | 10982 | 748 | 13 | 8457 | 2814 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >27061_27061_1_EPS15-KMT2A_EPS15_chr1_51913717_ENST00000371730_KMT2A_chr11_118354897_ENST00000354520_length(amino acids)=2786AA_BP=217 MAAAAQLSLTQLSSGNPVYEKYYRQVDTGNTGRVLASDAAAFLKKSGLPDLILGKIWDLADTDGKGILNKQEFFVALRLVACAQNGLEVS LSSLNLAVPPPRFHDTSSPLLISGTSAAELPWAVKPEDKAKYDAIFDSLSPVNGFLSGDKVKPVLLNSKLPVDILGRVWELSDIDHDGML DRDEFAVAMFLVYCALEKEPVPMSLPPALVPPSKRKTEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKFVYCQVCCE PFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGK GWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWR LALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSV LEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSH TEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNC ALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEV FRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDI NSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTH EIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTS SNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAE PSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSS KSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENES QSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARA RSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPK ISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSS LESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGL FEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDP TPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKP ATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLH SFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMT LINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNV VSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQ TTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQ RSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAV LPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSN ARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNP NDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCY -------------------------------------------------------------- >27061_27061_2_EPS15-KMT2A_EPS15_chr1_51913717_ENST00000371730_KMT2A_chr11_118354897_ENST00000389506_length(amino acids)=2824AA_BP=217 MAAAAQLSLTQLSSGNPVYEKYYRQVDTGNTGRVLASDAAAFLKKSGLPDLILGKIWDLADTDGKGILNKQEFFVALRLVACAQNGLEVS LSSLNLAVPPPRFHDTSSPLLISGTSAAELPWAVKPEDKAKYDAIFDSLSPVNGFLSGDKVKPVLLNSKLPVDILGRVWELSDIDHDGML DRDEFAVAMFLVYCALEKEPVPMSLPPALVPPSKRKTEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENVW EMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSY HPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWV HSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSS PEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSR FWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPP GIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYH FMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLF PIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCY YHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVS SVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKS SEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLS GMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMP GVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQS NNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSD EDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKE HVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLK NTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQN PSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPV SPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPM GGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPS SGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQS VGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQ PVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQ TVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETT SLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKP KKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANE PPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFC KRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIY -------------------------------------------------------------- >27061_27061_3_EPS15-KMT2A_EPS15_chr1_51913717_ENST00000371730_KMT2A_chr11_118354897_ENST00000534358_length(amino acids)=2827AA_BP=217 MAAAAQLSLTQLSSGNPVYEKYYRQVDTGNTGRVLASDAAAFLKKSGLPDLILGKIWDLADTDGKGILNKQEFFVALRLVACAQNGLEVS LSSLNLAVPPPRFHDTSSPLLISGTSAAELPWAVKPEDKAKYDAIFDSLSPVNGFLSGDKVKPVLLNSKLPVDILGRVWELSDIDHDGML DRDEFAVAMFLVYCALEKEPVPMSLPPALVPPSKRKTEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENVW EMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSY HPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWV HSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPS RSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVK KSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELN PPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTS NYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCED KLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSG SCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRN NVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLS SKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTL KLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGL SMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQD SQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLS TSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAG EKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDF VLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVP SQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQ VPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVL GPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVA TPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLL PQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGI QDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSN FTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQ ETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITE KKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEE ANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRG LFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMR -------------------------------------------------------------- >27061_27061_4_EPS15-KMT2A_EPS15_chr1_51913717_ENST00000371733_KMT2A_chr11_118354897_ENST00000354520_length(amino acids)=2814AA_BP=245 MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQLSSGNPVYEKYYRQVDTGNTGRVLASDAAAFLKKSGLPDLILGKIWDLADT DGKGILNKQEFFVALRLVACAQNGLEVSLSSLNLAVPPPRFHDTSSPLLISGTSAAELPWAVKPEDKAKYDAIFDSLSPVNGFLSGDKVK PVLLNSKLPVDILGRVWELSDIDHDGMLDRDEFAVAMFLVYCALEKEPVPMSLPPALVPPSKRKTEKPPPVNKQENAGTLNILSTLSNGN SSKQKIPADGVHRIRVDFKFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYP TKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDE MYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLT EVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSN SGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCAL CLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCV FLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVY WSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRI RTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATD LESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAY PGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIIN EHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQV EGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQD RNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFT RTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHK NEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGE SPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDS GEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQK YVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGL NPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISR LQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAG TSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGM FPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGL EQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPG AEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISS DDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSAR AEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMV IEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYK -------------------------------------------------------------- >27061_27061_5_EPS15-KMT2A_EPS15_chr1_51913717_ENST00000371733_KMT2A_chr11_118354897_ENST00000389506_length(amino acids)=2852AA_BP=245 MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQLSSGNPVYEKYYRQVDTGNTGRVLASDAAAFLKKSGLPDLILGKIWDLADT DGKGILNKQEFFVALRLVACAQNGLEVSLSSLNLAVPPPRFHDTSSPLLISGTSAAELPWAVKPEDKAKYDAIFDSLSPVNGFLSGDKVK PVLLNSKLPVDILGRVWELSDIDHDGMLDRDEFAVAMFLVYCALEKEPVPMSLPPALVPPSKRKTEKPPPVNKQENAGTLNILSTLSNGN SSKQKIPADGVHRIRVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCR RCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGN FCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTT SHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEI KKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDS PTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIR GKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPE NIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSS KESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHST SSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSE MKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRN DRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDE VLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTE PISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPF YSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKS SQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTE LLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISA EEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSP PCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNG VTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIG VQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTS SHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNP RLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQ LLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPL DKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEE EESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAV VFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMP MRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINH -------------------------------------------------------------- >27061_27061_6_EPS15-KMT2A_EPS15_chr1_51913717_ENST00000371733_KMT2A_chr11_118354897_ENST00000534358_length(amino acids)=2855AA_BP=245 MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQLSSGNPVYEKYYRQVDTGNTGRVLASDAAAFLKKSGLPDLILGKIWDLADT DGKGILNKQEFFVALRLVACAQNGLEVSLSSLNLAVPPPRFHDTSSPLLISGTSAAELPWAVKPEDKAKYDAIFDSLSPVNGFLSGDKVK PVLLNSKLPVDILGRVWELSDIDHDGMLDRDEFAVAMFLVYCALEKEPVPMSLPPALVPPSKRKTEKPPPVNKQENAGTLNILSTLSNGN SSKQKIPADGVHRIRVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCR RCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGN FCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNS RTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQ PEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGE PDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMA VIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGL EPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTE SSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRR HSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSS SSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRG QRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEF MDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTS PTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEED IPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATT RKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTY NTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSS ISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHI SSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTL PNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGL LIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSL NTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTL TNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQH VNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQ LPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKT VEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILH DAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDL PMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARF -------------------------------------------------------------- >27061_27061_7_EPS15-KMT2A_EPS15_chr1_51984870_ENST00000371730_KMT2A_chr11_118354897_ENST00000354520_length(amino acids)=2580AA_BP=11 MAAAAQLSLTQEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRC KFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFC PLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSH LLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKK ANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPT PLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGK QLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENI HMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKE SQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSS LSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMK QSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDR DQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVL TPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPI SASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYS SSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQ IPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELL KSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEE QFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPC GSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVT QKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQ PPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSH RTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRL LGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLL ASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDK GNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEE SNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVF LIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMR FRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSC -------------------------------------------------------------- >27061_27061_8_EPS15-KMT2A_EPS15_chr1_51984870_ENST00000371730_KMT2A_chr11_118354897_ENST00000389506_length(amino acids)=2618AA_BP=11 MAAAAQLSLTQEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSG HVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCV RCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTC VNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGV KRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHN YAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLL YIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLI KGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKI VECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRP LPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLN SSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSR ELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKK SCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVK VTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGS FKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHN LFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKT QGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGL GLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSE SDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISN AAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASK GLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPS NIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSV SGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAIT AASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTS PGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQ KECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWK SLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFL ASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKR EKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCNCGA -------------------------------------------------------------- >27061_27061_9_EPS15-KMT2A_EPS15_chr1_51984870_ENST00000371730_KMT2A_chr11_118354897_ENST00000534358_length(amino acids)=2621AA_BP=11 MAAAAQLSLTQEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSG HVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCV RCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVA YTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDL EGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSL DHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAG RLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHR DLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYT CKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPG CRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLG PLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNT TSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQK GKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKR TVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQE DGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLA SHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSR VKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLG EGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVA SSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQ ISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPS ASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSS TPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTS GSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTA AITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQAS PTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQ SSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIED AWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMF NFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQT DKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCN -------------------------------------------------------------- >27061_27061_10_EPS15-KMT2A_EPS15_chr1_51984870_ENST00000371733_KMT2A_chr11_118354897_ENST00000354520_length(amino acids)=2608AA_BP=39 MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKFVYCQVC CEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTP GKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAE WRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYT SVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREEN SHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHV NCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGF EVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEP DINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPT THEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTS TSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSF AEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKH SSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMEN ESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSA RARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDL PKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQL SSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDM GLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGV DPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHL KPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQH LHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSN MTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVL NVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPS TQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSS GQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQV AVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEAR SNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEY NPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIG -------------------------------------------------------------- >27061_27061_11_EPS15-KMT2A_EPS15_chr1_51984870_ENST00000371733_KMT2A_chr11_118354897_ENST00000389506_length(amino acids)=2646AA_BP=39 MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAEN VWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRN SYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDR WVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSR SSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKK SRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNP PPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSN YHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDK LFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGS CYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNN VSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSS KSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLK LSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLS MPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDS QSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLST SDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGE KEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFV LKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPS QNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQV PVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLG PMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVAT PSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLP QSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQ DQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNF TQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQE TTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEK KPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEA NEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGL FCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRK -------------------------------------------------------------- >27061_27061_12_EPS15-KMT2A_EPS15_chr1_51984870_ENST00000371733_KMT2A_chr11_118354897_ENST00000534358_length(amino acids)=2649AA_BP=39 MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAEN VWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRN SYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDR WVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESI PSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFS VKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPE LNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSC TSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDC EDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQT SGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYS RNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKV LSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVK TLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDK GLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSC QDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADD LSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPED AGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIM DFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPT VPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPG LQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTS VLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTT VATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAP LLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSL GIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQV SNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIP DQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESI TEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKP EEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHG RGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFA -------------------------------------------------------------- |
Top |
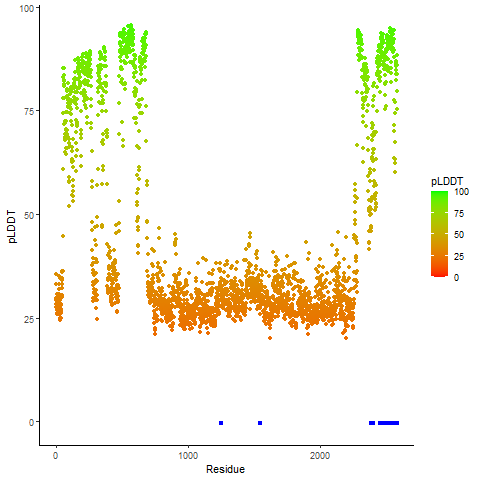
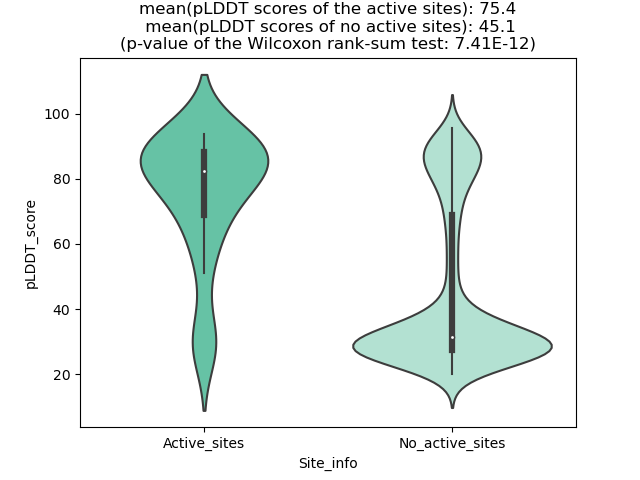
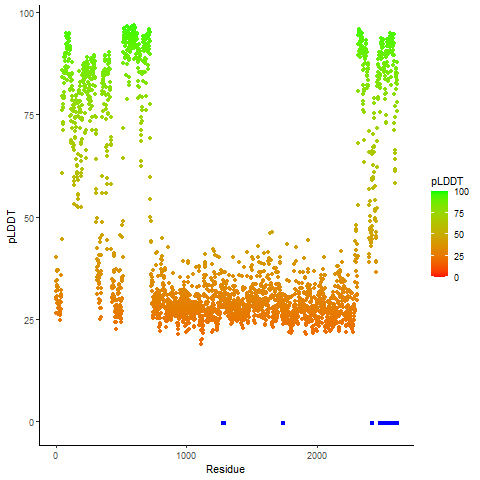
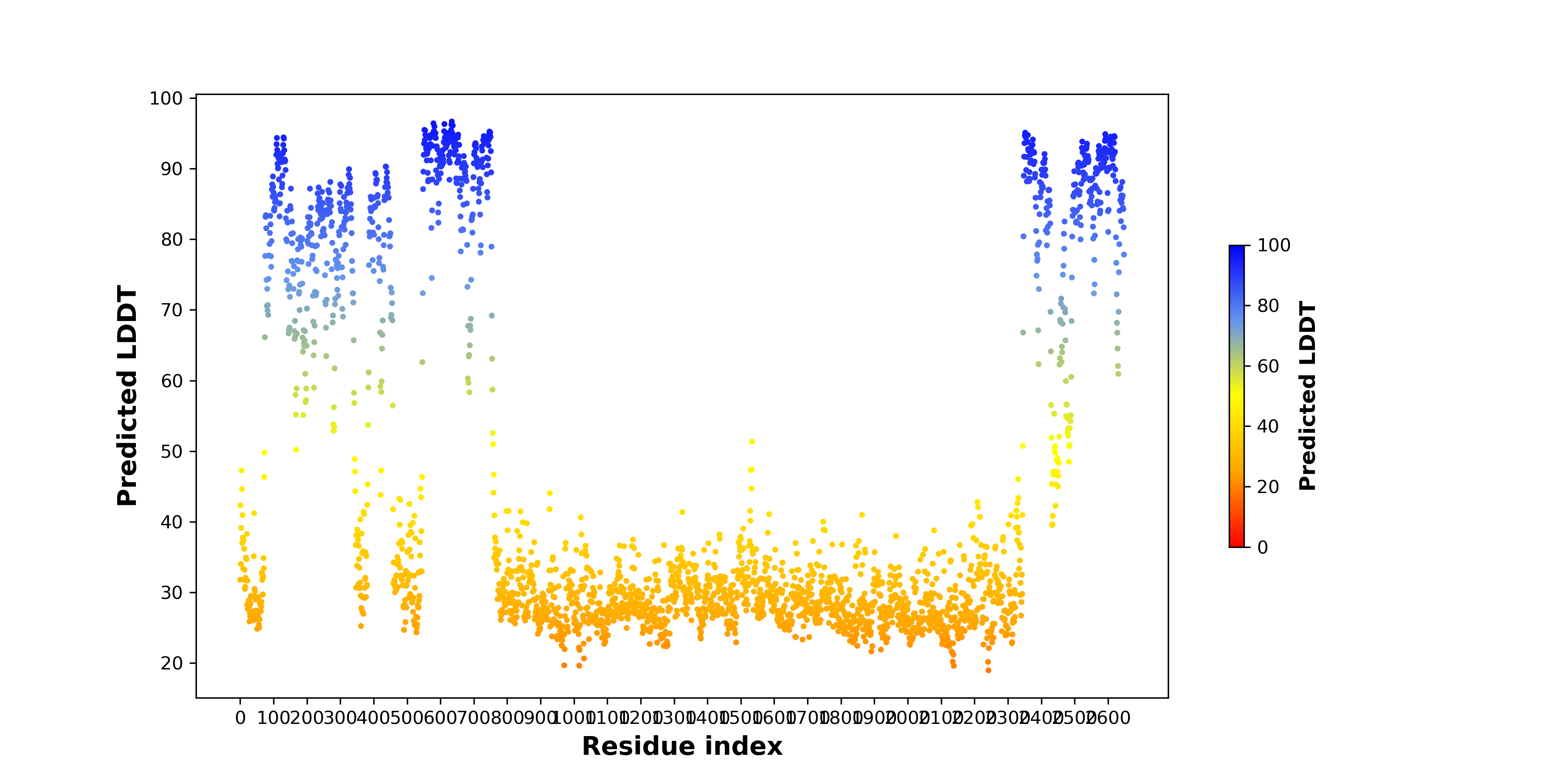
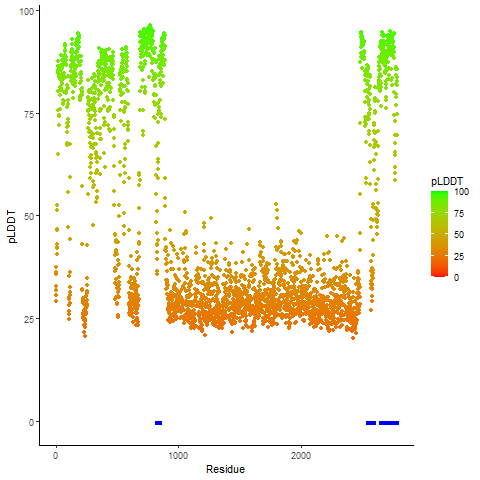
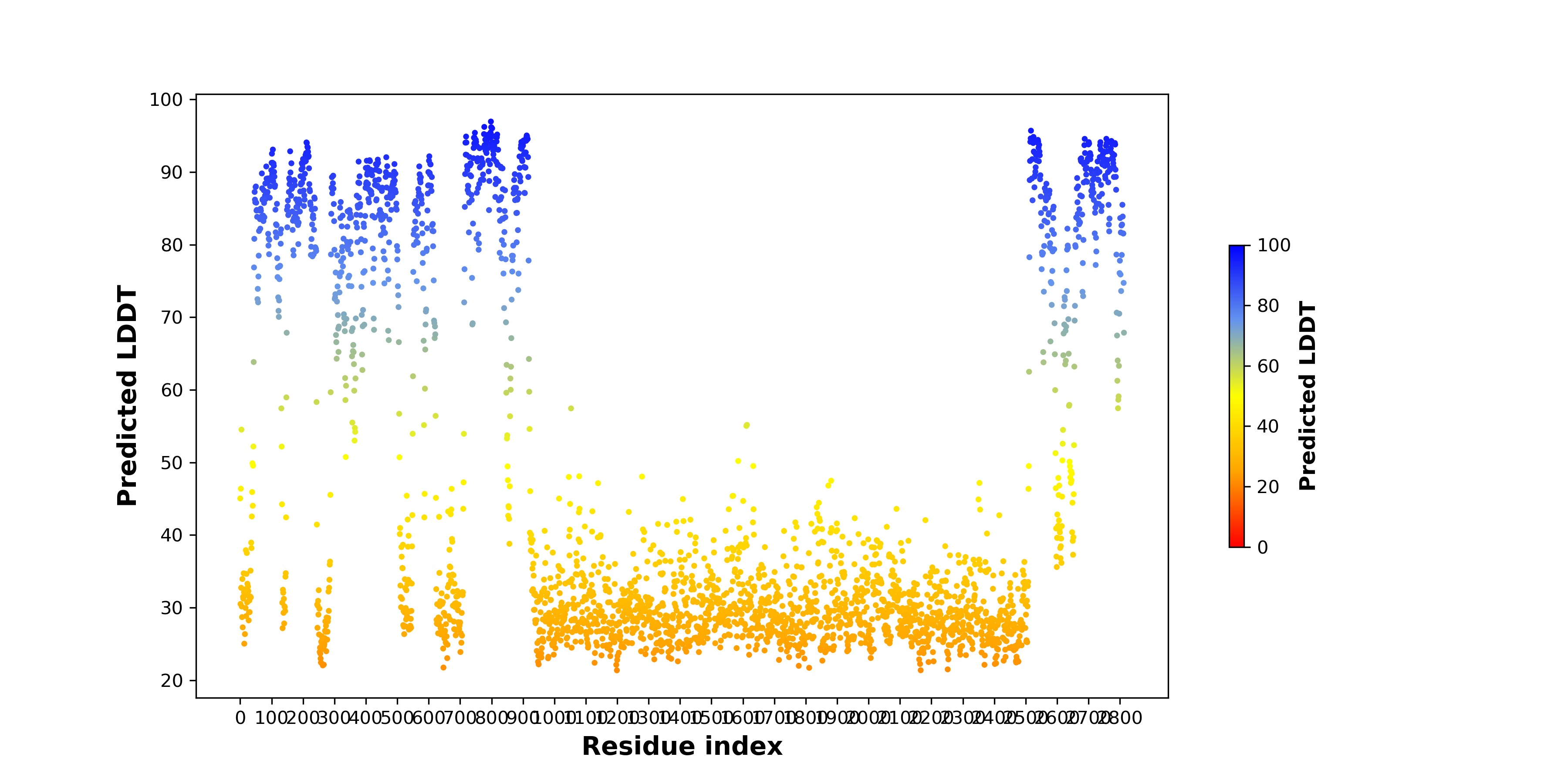
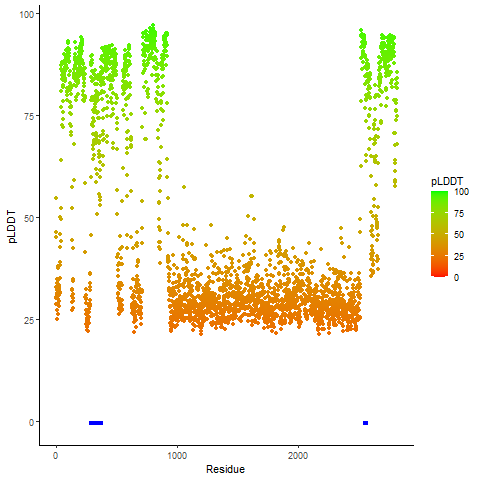
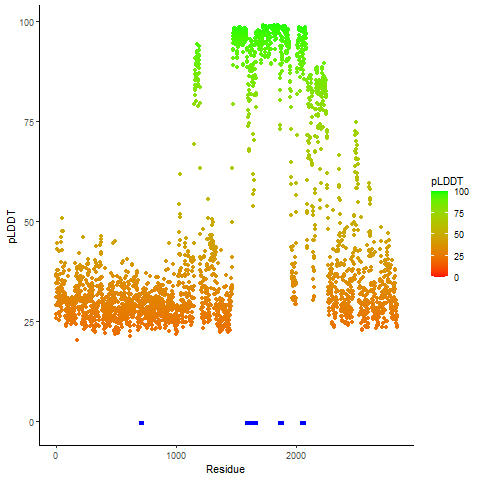
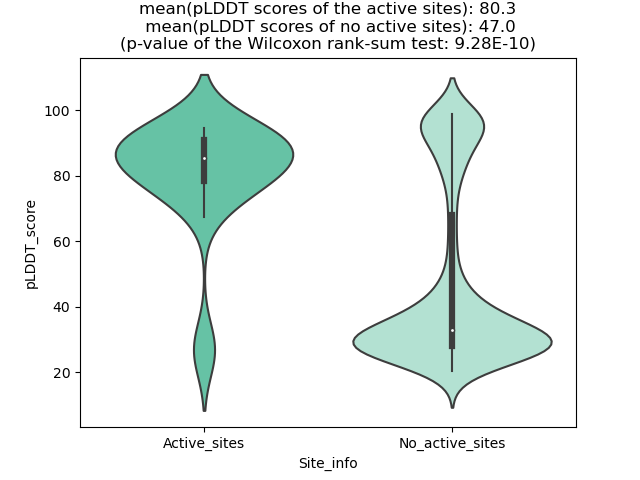
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:118355690/chr11:51946988) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| EPS15 | KMT2A |
| FUNCTION: Seems to be a constitutive component of clathrin-coated pits that is required for receptor-mediated endocytosis. Involved in endocytosis of integrin beta-1 (ITGB1) and transferrin receptor (TFR); internalization of ITGB1 as DAB2-dependent cargo but not TFR seems to require association with DAB2. {ECO:0000269|PubMed:22648170, ECO:0000269|PubMed:9407958}. | FUNCTION: Histone methyltransferase that plays an essential role in early development and hematopoiesis (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalytic subunit of the MLL1/MLL complex, a multiprotein complex that mediates both methylation of 'Lys-4' of histone H3 (H3K4me) complex and acetylation of 'Lys-16' of histone H4 (H4K16ac) (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:24235145, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalyzes methyl group transfer from S-adenosyl-L-methionine to the epsilon-amino group of 'Lys-4' of histone H3 (H3K4) via a non-processive mechanism. Part of chromatin remodeling machinery predominantly forms H3K4me1 and H3K4me2 methylation marks at active chromatin sites where transcription and DNA repair take place (PubMed:25561738, PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Has weak methyltransferase activity by itself, and requires other component of the MLL1/MLL complex to obtain full methyltransferase activity (PubMed:19187761, PubMed:26886794). Has no activity toward histone H3 phosphorylated on 'Thr-3', less activity toward H3 dimethylated on 'Arg-8' or 'Lys-9', while it has higher activity toward H3 acetylated on 'Lys-9' (PubMed:19187761). Binds to unmethylated CpG elements in the promoter of target genes and helps maintain them in the nonmethylated state (PubMed:20010842). Required for transcriptional activation of HOXA9 (PubMed:12453419, PubMed:20677832, PubMed:20010842). Promotes PPP1R15A-induced apoptosis (PubMed:10490642). Plays a critical role in the control of circadian gene expression and is essential for the transcriptional activation mediated by the CLOCK-ARNTL/BMAL1 heterodimer (By similarity). Establishes a permissive chromatin state for circadian transcription by mediating a rhythmic methylation of 'Lys-4' of histone H3 (H3K4me) and this histone modification directs the circadian acetylation at H3K9 and H3K14 allowing the recruitment of CLOCK-ARNTL/BMAL1 to chromatin (By similarity). Also has auto-methylation activity on Cys-3882 in absence of histone H3 substrate (PubMed:24235145). {ECO:0000250|UniProtKB:P55200, ECO:0000269|PubMed:10490642, ECO:0000269|PubMed:12453419, ECO:0000269|PubMed:15960975, ECO:0000269|PubMed:19187761, ECO:0000269|PubMed:19556245, ECO:0000269|PubMed:20010842, ECO:0000269|PubMed:21220120, ECO:0000269|PubMed:24235145, ECO:0000269|PubMed:26886794, ECO:0000305|PubMed:20677832}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 173_184 | 217.0 | 897.0 | Calcium binding | Note=1 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 128_216 | 217.0 | 897.0 | Domain | EH 2 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 15_104 | 217.0 | 897.0 | Domain | EH 1 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 160_195 | 217.0 | 897.0 | Domain | EF-hand 1 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1822_1847 | 1362.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2096_2118 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2146_2174 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2189_2232 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2275_2320 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2402_2418 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2419_2445 | 1362.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2526_2594 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2721_2746 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2747_2784 | 1362.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2785_2821 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3011_3064 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3166_3183 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3195_3222 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3230_3244 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3464_3530 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3563_3606 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1822_1847 | 1362.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2096_2118 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2146_2174 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2189_2232 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2275_2320 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2402_2418 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2419_2445 | 1362.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2526_2594 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2721_2746 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2747_2784 | 1362.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2785_2821 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3011_3064 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3166_3183 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3195_3222 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3230_3244 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3464_3530 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3563_3606 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1822_1847 | 1362.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2096_2118 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2146_2174 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2189_2232 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2275_2320 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2402_2418 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2419_2445 | 1362.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2526_2594 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2721_2746 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2747_2784 | 1362.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2785_2821 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3011_3064 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3166_3183 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3195_3222 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3230_3244 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3464_3530 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3563_3606 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1703_1748 | 1362.0 | 3932.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2018_2074 | 1362.0 | 3932.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3666_3747 | 1362.0 | 3932.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3829_3945 | 1362.0 | 3932.0 | Domain | SET | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3953_3969 | 1362.0 | 3932.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1703_1748 | 1362.0 | 3970.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2018_2074 | 1362.0 | 3970.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3666_3747 | 1362.0 | 3970.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3829_3945 | 1362.0 | 3970.0 | Domain | SET | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3953_3969 | 1362.0 | 3970.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1703_1748 | 1362.0 | 3973.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2018_2074 | 1362.0 | 3973.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3666_3747 | 1362.0 | 3973.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3829_3945 | 1362.0 | 3973.0 | Domain | SET | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3953_3969 | 1362.0 | 3973.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1663_1713 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1806_1869 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2081_2133 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2145_2232 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2275_2333 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2373_2460 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2475_2618 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2647_2675 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2713_2821 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 2961_3064 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3166_3244 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3464_3608 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3620_3643 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3785_3808 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 3906_3907 | 1362.0 | 3932.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1663_1713 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1806_1869 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2081_2133 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2145_2232 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2275_2333 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2373_2460 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2475_2618 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2647_2675 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2713_2821 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 2961_3064 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3166_3244 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3464_3608 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3620_3643 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3785_3808 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 3906_3907 | 1362.0 | 3970.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1663_1713 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1806_1869 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2081_2133 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2145_2232 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2275_2333 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2373_2460 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2475_2618 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2647_2675 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2713_2821 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 2961_3064 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3166_3244 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3464_3608 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3620_3643 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3785_3808 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 3906_3907 | 1362.0 | 3973.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1431_1482 | 1362.0 | 3932.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1479_1533 | 1362.0 | 3932.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1566_1627 | 1362.0 | 3932.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1870_1910 | 1362.0 | 3932.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1931_1978 | 1362.0 | 3932.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1431_1482 | 1362.0 | 3970.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1479_1533 | 1362.0 | 3970.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1566_1627 | 1362.0 | 3970.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1870_1910 | 1362.0 | 3970.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1931_1978 | 1362.0 | 3970.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1431_1482 | 1362.0 | 3973.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1479_1533 | 1362.0 | 3973.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1566_1627 | 1362.0 | 3973.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1870_1910 | 1362.0 | 3973.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1931_1978 | 1362.0 | 3973.0 | Zinc finger | PHD-type 4 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 236_247 | 217.0 | 897.0 | Calcium binding | Note=2 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 542_586 | 217.0 | 897.0 | Compositional bias | Polar residues | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 587_601 | 217.0 | 897.0 | Compositional bias | Basic and acidic residues | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 771_785 | 217.0 | 897.0 | Compositional bias | Pro residues | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 826_848 | 217.0 | 897.0 | Compositional bias | Polar residues | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 223_258 | 217.0 | 897.0 | Domain | EF-hand 2 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 224_314 | 217.0 | 897.0 | Domain | EH 3 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 851_870 | 217.0 | 897.0 | Domain | UIM 1 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 877_896 | 217.0 | 897.0 | Domain | UIM 2 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 768_774 | 217.0 | 897.0 | Motif | Note=SH3-binding | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 542_606 | 217.0 | 897.0 | Region | Disordered | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 599_827 | 217.0 | 897.0 | Region | Note=15 X 3 AA repeats of D-P-F | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 760_848 | 217.0 | 897.0 | Region | Disordered | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 599_601 | 217.0 | 897.0 | Repeat | Note=1 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 623_625 | 217.0 | 897.0 | Repeat | Note=2 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 629_631 | 217.0 | 897.0 | Repeat | Note=3 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 634_636 | 217.0 | 897.0 | Repeat | Note=4 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 640_642 | 217.0 | 897.0 | Repeat | Note=5 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 645_647 | 217.0 | 897.0 | Repeat | Note=6 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 651_653 | 217.0 | 897.0 | Repeat | Note=7 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 664_666 | 217.0 | 897.0 | Repeat | Note=8 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 672_674 | 217.0 | 897.0 | Repeat | Note=9 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 692_694 | 217.0 | 897.0 | Repeat | Note=10 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 709_711 | 217.0 | 897.0 | Repeat | Note=11 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 737_739 | 217.0 | 897.0 | Repeat | Note=12 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 798_800 | 217.0 | 897.0 | Repeat | Note=13 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 804_806 | 217.0 | 897.0 | Repeat | Note=14 | |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 825_827 | 217.0 | 897.0 | Repeat | Note=15 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1046_1061 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1249_1272 | 1362.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1278_1302 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1303_1317 | 1362.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 184_203 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 204_226 | 1362.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 236_253 | 1362.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 325_351 | 1362.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 37_58 | 1362.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 445_495 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 535_556 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 557_576 | 1362.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 713_731 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 747_780 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 798_845 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 81_103 | 1362.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 846_898 | 1362.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1046_1061 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1249_1272 | 1362.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1278_1302 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1303_1317 | 1362.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 184_203 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 204_226 | 1362.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 236_253 | 1362.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 325_351 | 1362.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 37_58 | 1362.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 445_495 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 535_556 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 557_576 | 1362.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 713_731 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 747_780 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 798_845 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 81_103 | 1362.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 846_898 | 1362.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1046_1061 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1249_1272 | 1362.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1278_1302 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1303_1317 | 1362.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 184_203 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 204_226 | 1362.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 236_253 | 1362.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 325_351 | 1362.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 37_58 | 1362.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 445_495 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 535_556 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 557_576 | 1362.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 713_731 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 747_780 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 798_845 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 81_103 | 1362.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 846_898 | 1362.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 169_180 | 1362.0 | 3932.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 217_227 | 1362.0 | 3932.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 301_309 | 1362.0 | 3932.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 169_180 | 1362.0 | 3970.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 217_227 | 1362.0 | 3970.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 301_309 | 1362.0 | 3970.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 169_180 | 1362.0 | 3973.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 217_227 | 1362.0 | 3973.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 301_309 | 1362.0 | 3973.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 123_134 | 1362.0 | 3932.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 147_152 | 1362.0 | 3932.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 6_25 | 1362.0 | 3932.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 123_134 | 1362.0 | 3970.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 147_152 | 1362.0 | 3970.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 6_25 | 1362.0 | 3970.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 123_134 | 1362.0 | 3973.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 147_152 | 1362.0 | 3973.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 6_25 | 1362.0 | 3973.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1038_1066 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1106_1166 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1200_1375 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 132_253 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1_108 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 301_352 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 445_585 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 713_780 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 798_949 | 1362.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1038_1066 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1106_1166 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1200_1375 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 132_253 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1_108 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 301_352 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 445_585 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 713_780 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 798_949 | 1362.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1038_1066 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1106_1166 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1200_1375 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 132_253 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1_108 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 301_352 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 445_585 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 713_780 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 798_949 | 1362.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000354520 | 7 | 35 | 1147_1195 | 1362.0 | 3932.0 | Zinc finger | CXXC-type | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000389506 | 7 | 36 | 1147_1195 | 1362.0 | 3970.0 | Zinc finger | CXXC-type | |
| Tgene | KMT2A | chr1:51913717 | chr11:118354897 | ENST00000534358 | 7 | 36 | 1147_1195 | 1362.0 | 3973.0 | Zinc finger | CXXC-type |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (1149) >>>1149.pdbFusion protein BP residue: 11 CIF file (1149) >>>1149.cif | EPS15 | chr1 | 51984870 | - | KMT2A | chr11 | 118354897 | + | MAAAAQLSLTQEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRI RVDFKFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQH QATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGST TPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCG KCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKE LQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGP DPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAIN SDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGML PNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPT PLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGR LLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKP GATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPEN GFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSD CEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEH DENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVI SKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRS IGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESS AKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMK QSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTT SRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSA NSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKE KHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGD KGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNAL KESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQ NLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRS YGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVI SSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQ IPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRV KTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSD DCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREK DMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSP TVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHM TPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTV PIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLY VLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSL PTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQ PPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSST PSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRS KSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSS VLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQS LGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITA ASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDA PNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPK TKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAE QQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEE SNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDA WKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKH CRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPP EYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIH GRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDD SEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEE | 2580 |
| 3D view using mol* of 1149 (AA BP:11) | ||||||||||
 | ||||||||||
| PDB file (1155) >>>1155.pdbFusion protein BP residue: 39 CIF file (1155) >>>1155.cif | EPS15 | chr1 | 51984870 | - | KMT2A | chr11 | 118354897 | + | MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQEKPPPVNKQEN AGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKFVYCQVCCEPFHKFCLE ENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLG PNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAK LFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILS NLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLL RYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGV KRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQ MERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREEN SHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPEL NPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVF EDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRA KNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFL NGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDA RKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQ NTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRP LPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMS PMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTS TSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVL SSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSS KEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRS SIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLK PEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKE LQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISA SENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGS FKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSA EGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDL PKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDA GEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVH TSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQ ALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSV SSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVT ITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGS VEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISN AAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSST PSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQH LHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTV ATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQ LPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAA TAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGH VTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGT SQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLAS KTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTS PGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLR TSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQV AVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESIT EKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSF AGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPH GSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATS MDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGN VIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEP NCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCNCGA | 2608 |
| 3D view using mol* of 1155 (AA BP:39) | ||||||||||
 | ||||||||||
| PDB file (1160) >>>1160.pdbFusion protein BP residue: 11 CIF file (1160) >>>1160.cif | EPS15 | chr1 | 51984870 | - | KMT2A | chr11 | 118354897 | + | MAAAAQLSLTQEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRI RVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVC CEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKC RNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSH DFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCEN LSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTAL LNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQD DQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKAN SMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHN YAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILST DRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHV NCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCT SNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDF EGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQC SRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTS FTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYS PTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLS PQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLN SSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSS SLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSF AEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKT LKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQ VTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAP PMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIE STSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLP DGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSS TGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHN LFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTE NLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQL SSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMD FVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQL PTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAV ISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDA DHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNST DSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQK IQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASK GLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSES SQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSN MTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPL LPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTT PTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPP SSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQ LQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKAL SSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGN GKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQ KECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQ QEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEAR SNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPE EANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEV QLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDA GEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNA ARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDA | 2618 |
| 3D view using mol* of 1160 (AA BP:11) | ||||||||||
 | ||||||||||
| PDB file (1161) >>>1161.pdbFusion protein BP residue: 11 CIF file (1161) >>>1161.cif | EPS15 | chr1 | 51984870 | - | KMT2A | chr11 | 118354897 | + | MAAAAQLSLTQEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPADGVHRI RVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVC CEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKC RNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSH DFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCEN LSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVL TALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVS KQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIK KANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSL DHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPI LSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEW THVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLT SCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVF VDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIG YQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHS PTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTP SYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTS SLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLG PLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVS KSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKI GSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTE VKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLE PGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVP KAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPL QIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNL MLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFY SSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLA SHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKEN GTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLE AQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSD IMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFS QQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSR LAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGH MDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVP NSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGV TQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPS ASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLV SESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDV VSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEP APLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQT TTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVA LPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADE HYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQN KALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLD KGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQ SSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLML WLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQ EARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFH KPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEE EEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRN IDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMH GNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPI | 2621 |
| 3D view using mol* of 1161 (AA BP:11) | ||||||||||
 | ||||||||||
| PDB file (1170) >>>1170.pdbFusion protein BP residue: 39 CIF file (1170) >>>1170.cif | EPS15 | chr1 | 51984870 | - | KMT2A | chr11 | 118354897 | + | MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQEKPPPVNKQEN AGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENVWEMGGLGIL TSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLEN WCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKK VWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCD KCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVN CTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLN PETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSV LEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKK SRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKI IPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQC ALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHM AVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVY CQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMI GSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVE CRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPD RPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTH EIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNN VSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTV GNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNV AYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLR GQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSR DRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEY MGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVT LTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQP SPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARA RSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLST SDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDG TESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVG HKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDT YNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSE LLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFEL PLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESD PALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLT RNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKP ATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLG PMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSF PPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPI SRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGS LNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDT SHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTP DIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAA SSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLD SASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQ RSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQE TTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNP ANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEI SSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGIL HDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKS AFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHL KKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREK YYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQ | 2646 |
| 3D view using mol* of 1170 (AA BP:39) | ||||||||||
 | ||||||||||
| PDB file (1171) >>>1171.pdbFusion protein BP residue: 39 CIF file (1171) >>>1171.cif | EPS15 | chr1 | 51984870 | - | KMT2A | chr11 | 118354897 | + | MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQEKPPPVNKQEN AGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENVWEMGGLGIL TSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLEN WCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKK VWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCD KCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYT CVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPP DLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNY TSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFS VKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLM KKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDN RQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKN VHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDK KVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIH MMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCK IVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPP SPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTP TTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYS RNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTV VTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSA HNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHL HLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGS SSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLT PEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTV KVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPV AQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRS ARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADD LSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGV DDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKS SVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNL LDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESS SSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQ FELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASS ESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQ DLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPH LKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTS VLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQ SSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKK RPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLD LGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTIS QDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLL GTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAI TAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQR DLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPS SGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIP DQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVT QNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLV FEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRML GILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHL RKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRF RHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDK REKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINI | 2649 |
| 3D view using mol* of 1171 (AA BP:39) | ||||||||||
 | ||||||||||
| PDB file (1202) >>>1202.pdbFusion protein BP residue: 217 CIF file (1202) >>>1202.cif | EPS15 | chr1 | 51913717 | - | KMT2A | chr11 | 118354897 | MAAAAQLSLTQLSSGNPVYEKYYRQVDTGNTGRVLASDAAAFLKKSGLPD LILGKIWDLADTDGKGILNKQEFFVALRLVACAQNGLEVSLSSLNLAVPP PRFHDTSSPLLISGTSAAELPWAVKPEDKAKYDAIFDSLSPVNGFLSGDK VKPVLLNSKLPVDILGRVWELSDIDHDGMLDRDEFAVAMFLVYCALEKEP VPMSLPPALVPPSKRKTEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPA DGVHRIRVDFKFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCH VCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRC KSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYES KMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWR LALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSR SSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKI IQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVS SNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPG EPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDS ANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRC EFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKG EVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGI LNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDI NSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGS CYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLL SSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTA TDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGS SSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAP QVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHT DSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSC KETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVS SDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENES QSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDS QSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTP LYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYN FTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTAT TRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNC HSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDS DNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGL DSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSV LTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDP TPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQV PVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQ NMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTT GLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSG LLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKK LAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVP NIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSG LASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLI KASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQ TTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNF TQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGP TKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGT PGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPK TVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICA ESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQ LSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLAS KHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGV YRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCY MFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRK | 2786 | |
| 3D view using mol* of 1202 (AA BP:217) | ||||||||||
 | ||||||||||
| PDB file (1206) >>>1206.pdbFusion protein BP residue: 245 CIF file (1206) >>>1206.cif | EPS15 | chr1 | 51913717 | - | KMT2A | chr11 | 118354897 | MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQLSSGNPVYEKY YRQVDTGNTGRVLASDAAAFLKKSGLPDLILGKIWDLADTDGKGILNKQE FFVALRLVACAQNGLEVSLSSLNLAVPPPRFHDTSSPLLISGTSAAELPW AVKPEDKAKYDAIFDSLSPVNGFLSGDKVKPVLLNSKLPVDILGRVWELS DIDHDGMLDRDEFAVAMFLVYCALEKEPVPMSLPPALVPPSKRKTEKPPP VNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKFVYCQVCCEPF HKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSY HPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSL CHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDE MYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSR TTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQP LDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVK SFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQW QEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSR EDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCAL WSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYH FMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGIS LRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVY WSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTES SSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQR SPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRS KLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTS LGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKG EKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPS SVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLS GMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTG EEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQV EGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSP TEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPK PQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKK RGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFRE EEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKI DRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLE SSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLK NTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTE PVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDS GEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHIS SPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPG PSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLT SSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLP MSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRT DLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLI NFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQS VGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSS ASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGM FPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHV NQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAV QASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKH KVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECG QPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQK RKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNAR LKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANE PPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKS ARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMV IEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFI NHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKL | 2814 | |
| 3D view using mol* of 1206 (AA BP:245) | ||||||||||
 | ||||||||||
| PDB file (1208) >>>1208.pdbFusion protein BP residue: 217 CIF file (1208) >>>1208.cif | EPS15 | chr1 | 51913717 | - | KMT2A | chr11 | 118354897 | MAAAAQLSLTQLSSGNPVYEKYYRQVDTGNTGRVLASDAAAFLKKSGLPD LILGKIWDLADTDGKGILNKQEFFVALRLVACAQNGLEVSLSSLNLAVPP PRFHDTSSPLLISGTSAAELPWAVKPEDKAKYDAIFDSLSPVNGFLSGDK VKPVLLNSKLPVDILGRVWELSDIDHDGMLDRDEFAVAMFLVYCALEKEP VPMSLPPALVPPSKRKTEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPA DGVHRIRVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEF VYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQL LECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGW DAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWV HSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLK QVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLT EVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQP EIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLP PSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPT PPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQ NEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGC CLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFR RVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLF PIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTI AHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRI RTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRH STSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDH VLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASD LVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNV SKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDE DTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKS FLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMP GVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPA SPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQD RNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDI PFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEE RLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNG KENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQD SLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNIL PSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFE VFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQN PSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFI QGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQK YVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLP NGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSL FPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQ LLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAP SDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMY FEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVS MQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQ PVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGE ADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGL EQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQL PLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTAS VEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSP LMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTD KVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKF RFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPND EEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFC KRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDA TMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYK | 2824 | |
| 3D view using mol* of 1208 (AA BP:217) | ||||||||||
 | ||||||||||
| PDB file (1209) >>>1209.pdbFusion protein BP residue: 217 CIF file (1209) >>>1209.cif | EPS15 | chr1 | 51913717 | - | KMT2A | chr11 | 118354897 | MAAAAQLSLTQLSSGNPVYEKYYRQVDTGNTGRVLASDAAAFLKKSGLPD LILGKIWDLADTDGKGILNKQEFFVALRLVACAQNGLEVSLSSLNLAVPP PRFHDTSSPLLISGTSAAELPWAVKPEDKAKYDAIFDSLSPVNGFLSGDK VKPVLLNSKLPVDILGRVWELSDIDHDGMLDRDEFAVAMFLVYCALEKEP VPMSLPPALVPPSKRKTEKPPPVNKQENAGTLNILSTLSNGNSSKQKIPA DGVHRIRVDFKEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEF VYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQL LECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGW DAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWV HSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQI SLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPP VLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDG GQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNA VLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLH PPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLY IGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGAT VGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFE VFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCED KLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDEN RTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKV PRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGS RRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKV VDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSS ASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRE LNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSS PDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHS SKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGL SMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKES SPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLP VQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGE EDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSG GEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPK RNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQ GQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCG NILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMG LFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVP SQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPD HFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQ NQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQ TLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTS QSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPP DPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSN IAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSS IMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLN VVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGI QDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASP SGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNS MGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKR FQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQD TASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNF SSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKS LTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRN YKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYN PNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRG LFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEV VDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTY | 2827 | |
| 3D view using mol* of 1209 (AA BP:217) | ||||||||||
 | ||||||||||
| PDB file (1213) >>>1213.pdbFusion protein BP residue: 245 CIF file (1213) >>>1213.cif | EPS15 | chr1 | 51913717 | - | KMT2A | chr11 | 118354897 | MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQLSSGNPVYEKY YRQVDTGNTGRVLASDAAAFLKKSGLPDLILGKIWDLADTDGKGILNKQE FFVALRLVACAQNGLEVSLSSLNLAVPPPRFHDTSSPLLISGTSAAELPW AVKPEDKAKYDAIFDSLSPVNGFLSGDKVKPVLLNSKLPVDILGRVWELS DIDHDGMLDRDEFAVAMFLVYCALEKEPVPMSLPPALVPPSKRKTEKPPP VNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENVWEM GGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPL EDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTK PTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGN FCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESV AYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAA KPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQ GNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFP WFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQP PLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGI EDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGS LKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFL DDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPE NIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVY TCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEII SPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGS PTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGN TYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQ RTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSE GSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSF PHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEH MGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDE VLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRK RTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGD GPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYP RRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDG ADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQL DGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHV TKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSD KNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESP ESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISA EEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSV ASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHG NNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTT PPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMET NTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPA ATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSG LKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPS LLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTS TISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNP RLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPST AAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHS SQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPS SPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEA HIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEV QVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKK GLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGL RMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAE VHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMP MRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQ TDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRV INIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCNCGAKKCRKF | 2852 | |
| 3D view using mol* of 1213 (AA BP:245) | ||||||||||
 | ||||||||||
| PDB file (1214) | EPS15 | chr1 | 51913717 | - | KMT2A | chr11 | 118354897 | MRPLPPPPPRPEPQSARLPSPPLHDGNTMAAAAQLSLTQLSSGNPVYEKY YRQVDTGNTGRVLASDAAAFLKKSGLPDLILGKIWDLADTDGKGILNKQE FFVALRLVACAQNGLEVSLSSLNLAVPPPRFHDTSSPLLISGTSAAELPW AVKPEDKAKYDAIFDSLSPVNGFLSGDKVKPVLLNSKLPVDILGRVWELS DIDHDGMLDRDEFAVAMFLVYCALEKEPVPMSLPPALVPPSKRKTEKPPP VNKQENAGTLNILSTLSNGNSSKQKIPADGVHRIRVDFKEDCEAENVWEM GGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPL EDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTK PTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGN FCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLP ESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYR QAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRK MDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMER VFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHT EQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPP PGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDD DGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNC VFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGL EPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKR CVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTA EIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPS AGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMR TGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSS NLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSK SSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEA LSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSII NEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEF MDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQA PRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASEN PGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKR RYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQ VDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKI SQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEK EHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTST PSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALG ESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSS ISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITE KSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQ GHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAV QTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSV METNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHS FPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATP SSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPN HPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAA GTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTL TNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQT PSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTG IHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGG SPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSS SEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVL PEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKK PKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGV NGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSA RAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDL PMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIR SIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCY SRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCNCGAKKC | 2855 | |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
EPS15_pLDDT.png |
KMT2A_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 1149 | 1.037 | 200 | 1.053 | 525.133 | 0.569 | 0.753 | 0.996 | 0.637 | 1.04 | 0.613 | 0.738 | Chain A: 1249,1250,1251,1544,1546,2384,2386,2387,2 388,2389,2390,2393,2446,2447,2448,2449,2450,2451,2 452,2469,2487,2490,2491,2492,2493,2494,2495,2496,2 508,2513,2514,2515,2516,2517,2518,2539,2553,2555,2 557,2566,2567,2568,2569,2579 |
| 1160 | 1.044 | 460 | 1.061 | 1450.547 | 0.495 | 0.76 | 0.971 | 0.693 | 1.028 | 0.674 | 1.002 | Chain A: 1282,1283,1284,1285,1286,1287,1288,1289,1 290,1291,1740,1741,1742,2422,2424,2425,2426,2427,2 487,2488,2489,2490,2507,2517,2521,2524,2525,2528,2 529,2530,2531,2532,2533,2534,2535,2541,2545,2546,2 551,2552,2553,2554,2555,2556,2563,2564,2565,2566,2 577,2591,2593,2594,2595,2596,2598,2602,2604,2605,2 606,2607,2608,2617,2618 |
| 1161 | 1.092 | 155 | 1.088 | 232.554 | 0.351 | 0.836 | 1.112 | 0.907 | 1.083 | 0.837 | 0.802 | Chain A: 52,53,54,55,56,58,69,70,72,86,87,88,543,5 44,546,547,574,650,651,653,654,655,656,658,662,673 ,2378,2381,2382 |
| 1171 | 1.115 | 149 | 1.096 | 240.1 | 0.382 | 0.87 | 1.141 | 1.029 | 1.123 | 0.916 | 0.985 | Chain A: 80,82,83,84,86,97,98,100,114,115,571,572, 573,574,575,602,678,679,681,682,683,684,685,686,69 0,701,2406,2409,2410 |
| 1202 | 1.0116 | 508 | 1.0348 | 1408.015 | 0.5369 | 0.7058 | 0.9414 | 0.7037 | 1.0084 | 0.6979 | 0.8727 | Chain A: 823,824,825,826,827,828,830,831,833,836,8 48,2547,2549,2550,2551,2552,2554,2555,2556,2557,25 58,2560,2561,2562,2564,2565,2566,2567,2568,2569,25 70,2582,2583,2584,2585,2586,2587,2588,2589,2590,25 92,2593,2594,2595,2596,2599,2652,2653,2654,2655,26 56,2657,2658,2675,2696,2697,2698,2699,2700,2701,27 02,2703,2714,2719,2720,2721,2722,2723,2724,2745,27 59,2761,2763,2772,2773,2774,2775,2776,2785 |
| 1206 | 1.0626 | 229 | 1.1096 | 551.887 | 0.5492 | 0.7277 | 0.9402 | 0.7432 | 0.819 | 0.9074 | 0.5531 | Chain A: 295,308,309,310,311,312,315,318,319,320,3 22,323,324,325,326,328,333,334,339,340,341,342,344 ,346,347,348,349,363,368,369,370,2552,2553,2554,25 55 |
| 1208 | 0.9936 | 479 | 0.9923 | 1060.213 | 0.5581 | 0.6884 | 0.9224 | 0.2321 | 1.1066 | 0.2097 | 0.6394 | Chain A: 267,270,271,304,305,316,318,319,320,321,3 22,325,327,328,329,330,332,333,334,335,336,338,341 ,342,343,344,345,346,347,349,350,351,352,354,356,3 57,358,359,362,363,371,372,373,378,379,380,2328,23 29,2330,2331,2381,2383,2384,2386,2542,2545,2546,25 49,2550,2553,2554,2556,2557,2562,2563,2564,2565,25 66,2567,2568,2569,2570,2571,2573 |
| 1209 | 1.0187 | 257 | 1.0516 | 791.987 | 0.6058 | 0.6968 | 0.8513 | 0.3466 | 0.9402 | 0.3687 | 0.9556 | Chain A: 710,711,713,714,1593,1594,1595,1596,1597, 1598,1599,1605,1606,1607,1609,1610,1614,1616,1619, 1622,1657,1658,1659,1660,1661,1662,1663,1864,1865, 1866,1867,1868,1872,1873,1874,1875,1876,1878,1879, 1880,2050,2051,2054,2058,2059,2061,2063,2065 |
| 1213 | 1.046 | 229 | 1.088 | 599.564 | 0.609 | 0.717 | 0.919 | 0.772 | 0.867 | 0.89 | 0.695 | Chain A: 298,344,345,346,347,348,349,350,352,353,3 55,356,357,358,360,363,364,366,372,377,378,379,380 ,382,383,384,385,386,387,390,401,406,407,408,2590, 2591,2592,2593 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
| 1161 | ZINC000000121541 | DB00643 | Mebendazole | -7.29933 | -7.95753 |
| 1161 | ZINC000000000469 | DB00816 | Orciprenaline | -7.47712 | -7.49152 |
| 1161 | ZINC000000643046 | DB00975 | Dipyridamole | -7.43608 | -7.43688 |
| 1161 | ZINC000003830943 | DB01362 | Iohexol | -7.42868 | -7.42868 |
| 1161 | ZINC000000025958 | DB12332 | Rucaparib | -7.39136 | -7.40636 |
| 1161 | ZINC000002169830 | DB00194 | Vidarabine | -7.3961 | -7.3961 |
| 1161 | ZINC000000004166 | DB00489 | Sotalol | -7.36645 | -7.38765 |
| 1161 | ZINC000002036848 | DB00140 | Riboflavin | -7.35402 | -7.35442 |
| 1161 | ZINC000043206370 | DB11793 | Niraparib | -7.28448 | -7.28448 |
| 1161 | ZINC000000007601 | DB01001 | Salbutamol | -7.24878 | -7.24878 |
| 1161 | ZINC000034220093 | DB15058 | Flutemetamol | -7.22405 | -7.23815 |
| 1161 | ZINC000003977952 | DB00581 | Lactulose | -7.18746 | -7.18746 |
| 1161 | ZINC000000056652 | DB01064 | Isoprenaline | -7.17912 | -7.17912 |
| 1161 | ZINC000030691797 | DB08883 | Perampanel | -7.15909 | -7.15909 |
| 1161 | ZINC000004214955 | DB06769 | Bendamustine | -6.67472 | -7.11862 |
| 1161 | Netarsudil | DB06769 | -7.09715 | -7.10825 | |
| 1161 | ZINC000003830990 | DB11868 | Etiracetam | -7.07817 | -7.07817 |
| 1161 | ZINC000012414057 | DB08167 | Methylthioninium | -7.05314 | -7.05614 |
| 1161 | ZINC000000897258 | DB00983 | Formoterol | -7.03826 | -7.05046 |
| 1161 | ZINC000000034157 | DB06707 | Levonordefrin | -7.02248 | -7.03998 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
| ZINC000000121541 | DB00643 | Mebendazole | Small molecule | COC(=O)NC1=NC2=C(N1)C=C(C=C2)C(=O)C1=CC=CC=C1 | Approved|Vet_approved |
| ZINC000000000469 | DB00816 | Orciprenaline | Small molecule | CC(C)NCC(O)C1=CC(O)=CC(O)=C1 | Approved |
| ZINC000000643046 | DB00975 | Dipyridamole | Small molecule | OCCN(CCO)C1=NC2=C(N=C(N=C2N2CCCCC2)N(CCO)CCO)C(=N1)N1CCCCC1 | Approved |
| ZINC000000025958 | DB12332 | Rucaparib | Small molecule | CNCC1=CC=C(C=C1)C1=C2CCNC(=O)C3=C2C(N1)=CC(F)=C3 | Approved|Investigational |
| ZINC000002169830 | DB00194 | Vidarabine | Small molecule | NC1=NC=NC2=C1N=CN2[C@@H]1O[C@H](CO)[C@@H](O)[C@@H]1O | Approved|Investigational |
| ZINC000002036848 | DB00140 | Riboflavin | Small molecule | CC1=C(C)C=C2N(C[C@H](O)[C@H](O)[C@H](O)CO)C3=NC(=O)NC(=O)C3=NC2=C1 | Approved|Investigational|Nutraceutical|Vet_approved |
| ZINC000043206370 | DB11793 | Niraparib | Small molecule | NC(=O)C1=CC=CC2=CN(N=C12)C1=CC=C(C=C1)[C@@H]1CCCNC1 | Approved|Investigational |
| ZINC000034220093 | DB15058 | Flutemetamol | Small molecule | CNC1=C(F)C=C(C=C1)C1=NC2=C(S1)C=C(O)C=C2 | Investigational |
| ZINC000003977952 | DB00581 | Lactulose | Small molecule | OC[C@H]1O[C@](O)(CO)[C@@H](O)[C@@H]1O[C@@H]1O[C@H](CO)[C@H](O)[C@H](O)[C@H]1O | Approved |
| ZINC000030691797 | DB08883 | Perampanel | Small molecule | O=C1N(C=C(C=C1C1=CC=CC=C1C#N)C1=NC=CC=C1)C1=CC=CC=C1 | Approved |
| ZINC000004214955 | DB06769 | Bendamustine | Small molecule | CN1C(CCCC(O)=O)=NC2=CC(=CC=C12)N(CCCl)CCCl | Approved|Investigational |
| ZINC000003830990 | DB11868 | Etiracetam | Small molecule | CCC(N1CCCC1=O)C(N)=O | Investigational |
| ZINC000012414057 | DB08167 | Methylthioninium | Small molecule | CN(C)C1=CC2=[S+]C3=CC(=CC=C3N=C2C=C1)N(C)C | Experimental|Investigational |
| ZINC000000897258 | DB00983 | Formoterol | Small molecule | COC1=CC=C(CC(C)NCC(O)C2=CC(NC=O)=C(O)C=C2)C=C1 | Approved|Investigational |
| ZINC000000034157 | DB06707 | Levonordefrin | Small molecule | C[C@H](N)[C@H](O)C1=CC(O)=C(O)C=C1 | Approved |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
| ZINC000000121541 | 295.297 | 6.562 | 576.161 | 98.468 | 169.387 | 308.306 | 0 | 957.18 | 1 | 5 | 8.911 | 3 | 83.641 | 0 | 0 |
| ZINC000000121541 | 295.297 | 2.873 | 577.033 | 96.773 | 172.83 | 307.43 | 0 | 959.022 | 0 | 3 | 8.668 | 3 | 88.187 | 0 | 0 |
| ZINC000000121541 | 295.297 | 10.919 | 575.199 | 98.469 | 170.072 | 306.658 | 0 | 956.419 | 1 | 5 | 8.861 | 3 | 83.461 | 0 | 0 |
| ZINC000000000469 | 211.26 | 2.192 | 479.658 | 212.457 | 159.848 | 107.353 | 0 | 782.094 | 4 | 4.7 | 9.205 | 2 | 61.391 | 0 | 0 |
| ZINC000000000469 | 211.26 | 3.668 | 477.852 | 212.113 | 158.053 | 107.685 | 0 | 779.059 | 4 | 4.7 | 8.929 | 2 | 61.69 | 0 | 0 |
| ZINC000000643046 | 504.631 | 1.586 | 815.307 | 592.733 | 195.941 | 26.633 | 0 | 1582.633 | 4 | 12.8 | 7.716 | 2 | 52.494 | 2 | 0 |
| ZINC000003830943 | 821.143 | 2.11 | 797.152 | 329.149 | 341.979 | 8.796 | 117.227 | 1489.14 | 8 | 18.2 | 9.356 | 1 | 0 | 3 | 1 |
| ZINC000000025958 | 323.369 | 9.363 | 601.398 | 214.232 | 112.926 | 227.819 | 46.421 | 1032.273 | 3 | 4 | 8.52 | 3 | 85.064 | 0 | 0 |
| ZINC000000025958 | 323.369 | 9.466 | 597.964 | 212.386 | 112.007 | 227.152 | 46.419 | 1026.229 | 3 | 4 | 8.513 | 3 | 85.039 | 0 | 0 |
| ZINC000002169830 | 267.244 | 3.396 | 469.36 | 95.245 | 259.374 | 114.741 | 0 | 784.03 | 5 | 10.8 | 8.527 | 2 | 44.327 | 0 | 0 |
| ZINC000000004166 | 272.362 | 5.095 | 568.775 | 304.129 | 140.425 | 123.778 | 0.442 | 948.347 | 3 | 7.7 | 9.15 | 3 | 65.285 | 0 | 0 |
| ZINC000000004166 | 272.362 | 6.328 | 565.576 | 303.726 | 137.239 | 124.569 | 0.042 | 941.283 | 3 | 7.7 | 9.164 | 3 | 65.814 | 0 | 0 |
| ZINC000002036848 | 376.368 | 8.767 | 609.496 | 261.237 | 282.582 | 65.677 | 0 | 1115.69 | 5 | 12.3 | 9.092 | 2 | 45.86 | 0 | 1 |
| ZINC000043206370 | 320.393 | 3.754 | 607.688 | 185.794 | 143.417 | 278.476 | 0 | 1055.805 | 3 | 5 | 8.546 | 3 | 76.72 | 0 | 0 |
| ZINC000000007601 | 239.314 | 3.133 | 507.069 | 275.176 | 145.075 | 86.817 | 0 | 875.643 | 4 | 5.15 | 9.068 | 2 | 66.522 | 0 | 0 |
| ZINC000034220093 | 274.312 | 1.949 | 511.177 | 89.833 | 86.339 | 254.76 | 80.245 | 847.322 | 2 | 3.25 | 8.207 | 3 | 100 | 0 | 0 |
| ZINC000034220093 | 274.312 | 1.174 | 509.609 | 89.83 | 84.419 | 255.176 | 80.184 | 844.623 | 2 | 3.25 | 8.146 | 3 | 100 | 0 | 0 |
| ZINC000003977952 | 342.299 | 7.257 | 545.281 | 230.326 | 314.956 | 0 | 0 | 978.368 | 8 | 16.8 | 10.609 | 1 | 0 | 2 | 2 |
| ZINC000000056652 | 211.26 | 3.507 | 478.577 | 212.52 | 152.49 | 113.567 | 0 | 781.059 | 4 | 4.7 | 8.941 | 2 | 63.01 | 0 | 0 |
| ZINC000030691797 | 349.391 | 5.825 | 656.081 | 0 | 102.198 | 553.883 | 0 | 1144.654 | 0 | 5.5 | 8.69 | 1 | 100 | 0 | 1 |
| ZINC000004214955 | 358.267 | 3.446 | 660.401 | 298.857 | 122.78 | 92.485 | 146.279 | 1135.219 | 1 | 4.5 | 8.979 | 3 | 95.316 | 0 | 1 |
| ZINC000004214955 | 358.267 | 5.922 | 650.52 | 300.031 | 115.164 | 97.2 | 138.125 | 1124.979 | 1 | 4.5 | 8.29 | 3 | 96.403 | 0 | 1 |
| ZINC000003830990 | 170.211 | 3.555 | 386.854 | 254.13 | 132.723 | 0 | 0 | 627.855 | 2 | 5.5 | 9.607 | 2 | 59.075 | 0 | 0 |
| ZINC000000897258 | 344.41 | 7.083 | 674.489 | 258.736 | 187.926 | 227.826 | 0 | 1168.666 | 4 | 7.2 | 8.855 | 2 | 65.222 | 0 | 0 |
| ZINC000000897258 | 344.41 | 4.982 | 586.764 | 251.201 | 138.136 | 197.426 | 0 | 1091.391 | 4 | 7.2 | 8.677 | 3 | 73.278 | 0 | 0 |
| ZINC000000034157 | 183.207 | 1.584 | 399.847 | 106.772 | 186.848 | 106.227 | 0 | 646.199 | 5 | 4.2 | 8.948 | 2 | 52.109 | 0 | 0 |
| ZINC000000034157 | 183.207 | 2.284 | 398.836 | 105.841 | 186.106 | 106.889 | 0 | 643.154 | 5 | 4.2 | 8.781 | 2 | 52.19 | 0 | 0 |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
| ZINC000000121541 | COC(=O)/N=c1[nH]c2ccc(C(=O)c3ccccc3)cc2[nH]1 | 0.209696342 | 0.592445172 |
| ZINC000000000469 | CC(C)NC[C@@H](O)c1cc(O)cc(O)c1 | 0.170221255 | 0.19797014 |
| ZINC000000643046 | OCCN(CCO)c1nc(N2CCCCC2)c2nc(N(CCO)CCO)nc(N3CCCCC3)c2n1 | 0.137158563 | 0.379972524 |
| ZINC000003830943 | CC(=O)N(C[C@H](O)CO)c1c(I)c(C(=O)NC[C@H](O)CO)c(I)c(C(=O)NC[C@H](O)CO)c1I | 0.042332243 | 0.211666342 |
| ZINC000000025958 | CNCc1ccc(-c2[nH]c3cc(F)cc4c3c2CCNC4=O)cc1 | 0.204259799 | 0.179994505 |
| ZINC000002169830 | Nc1ncnc2c1ncn2[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O | 0.079554768 | 0.347763769 |
| ZINC000000004166 | CC(C)NC[C@@H](O)c1ccc(NS(C)(=O)=O)cc1 | 0.233835874 | 0.174875168 |
| ZINC000002036848 | Cc1cc2nc3c(=O)[nH]c(=O)nc-3n(C[C@H](O)[C@H](O)[C@H](O)CO)c2cc1C | 0.061390494 | 0.448129596 |
| ZINC000043206370 | NC(=O)c1cccc2cn(-c3ccc([C@@H]4CCCNC4)cc3)nc12 | 0.15810306 | 0.243905197 |
| ZINC000000007601 | CC(C)(C)NC[C@H](O)c1ccc(O)c(CO)c1 | 0.176238455 | 0.331044881 |
| ZINC000034220093 | CNc1ccc(-c2nc3ccc(O)cc3s2)cc1F | 0.301158621 | 0.239116152 |
| ZINC000003977952 | OC[C@H]1O[C@](O)(CO)[C@@H](O)[C@@H]1O[C@@H]1O[C@H](CO)[C@H](O)[C@H](O)[C@H]1O | 0.029403356 | 0.491913174 |
| ZINC000000056652 | CC(C)NC[C@H](O)c1ccc(O)c(O)c1 | 0.202150594 | 0.209218132 |
| ZINC000030691797 | N#Cc1ccccc1-c1cc(-c2ccccn2)cn(-c2ccccc2)c1=O | 0.292416362 | 0.331128937 |
| ZINC000004214955 | Cn1c(CCCC(=O)O)nc2cc(N(CCCl)CCCl)ccc21 | 0.20795331 | 0.394281261 |
| ZINC000003830990 | CC[C@H](C(N)=O)N1CCCC1=O | 0.157335861 | 0.410047131 |
| ZINC000012414057 | CN(C)c1ccc2nc3ccc(=[N+](C)C)cc-3sc2c1 | 0.17581235 | 0.481215426 |
| ZINC000000897258 | COc1ccc(C[C@@H](C)NC[C@@H](O)c2ccc(O)c(NC=O)c2)cc1 | 0.133863078 | 0.246482573 |
| ZINC000000034157 | C[C@H](N)[C@H](O)c1ccc(O)c(O)c1 | 0.135079192 | 0.302260037 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| EPS15 | RNF11, SNAP91, AGFG1, NAGPA, STON2, HGS, AP2A2, DNM2, AP2A1, EGFR, CLINT1, EPN1, SCAMP1, STAM2, GRB2, CRK, RAB11FIP2, Synj1, REPS2, UBE2D1, UBE2D2, UBE2D3, UBE2H, UBE2E1, SLC6A3, PARK2, USP8, CD4, EPS15, OCLN, UBQLN1, UBC, NEDD4, GJA1, TGFB1, Zwint, FCHO2, ITSN1, ITSN2, ELAVL1, RAD23A, UBQLN2, EPN3, SH3BP4, AP1B1, NUMB, SPG20, TFAP2A, ABL1, AP2B1, IQGAP1, KRT85, KRT1, KRT10, KRT2, AP2M1, KRT14, KRT86, KRT5, KRT6A, KRT16, KRT9, HSPA8, KRT35, HSPA9, KRT17, KRT33B, KRT83, BMP2K, KRT34, KRT82, NONO, KRT38, KRT75, KRT79, PRDX1, KRT36, KRT32, MAP4, HSPA1L, HSPA1B, KRT37, HSPA5, KRT73, AP2S1, NECAP2, TRAF2, MCCC2, KRT4, ACTB, KRT3, VPRBP, TUBB, HIST4H4, PCCB, KRTAP9-3, NECAP1, VSIG8, CCT3, CRKL, Dab2, FN1, VCAM1, Fcho2, LAPTM5, ITGA4, OPTN, RABL6, EPHA2, CORO7, UBI4, AGFG2, SLC25A41, AURKA, SPC25, Numb, NTRK1, TMEM17, XPO1, CAPZA2, CLTB, CLTC, DBN1, FLNA, GAK, MYH9, PPP1CB, PICALM, PDLIM7, SYNPO, MAPRE1, LIMA1, GTSE1, ANLN, MYO19, Actb, Flot1, Myh9, Myo1c, Tpm1, Lima1, Calml3, Myh10, Sec24c, Flnb, MAPK14, AP1G1, MYO6, SPOPL, SGIP1, FCHO1, CDH1, RUNDC3A, LSR, STAMBPL1, CCDC172, OXCT1, MTNR1A, RNF26, fcho1, HNRNPL, STK40, DYRK1A, PLK1, DVL2, MIB1, ORF7a, ESR1, TRIM56, KIF14, Rnf183, DNAJC5, DDX58, ACBD5, CLTA, VASP, NAA40, RNPS1, MACC1, SYCE1, RARRES1, POLR2J3, EPS15L1, C15orf59, ZCCHC10, SYNC, INO80E, DNAJC6, RUFY4, |
| KMT2A | PPIE, PPP1R15A, KMT2A, ASH2L, HCFC1, HCFC2, MEN1, RBBP5, WDR5, AVP, INS, OXT, MAP3K5, HDAC1, CTBP1, CBX4, BMI1, CREBBP, SMARCB1, CXXC1, MYB, CTNNB1, SNW1, E2F2, E2F4, E2F6, PSIP1, MLLT4, POLR2A, KAT8, RNF2, TP53, tat, SBF1, MTM1, SET, HIST1H3A, HIST1H4A, KAT6A, ELL, AFF1, AFF4, CDK9, CCNT1, CTR9, LEO1, PAF1, CDC73, WDR61, MLLT3, DOT1L, SKP2, HIST3H3, SVIL, HIST2H3C, SIN3A, MLLT1, RUNX1, CBFB, H3F3A, SIRT7, ASB2, TCEB1, TCEB2, CBX8, TOP1, TAF6, NCL, HECW2, LGR4, CSNK2A2, SENP3, SYMPK, PKN1, PIH1D1, KRAS, TAF1, CHD3, SMARCA2, SMARCC2, SMARCC1, HDAC2, RBBP4, RBBP7, TBP, MBD3, SAP30, RAN, TAF9, TASP1, HIST1H2BG, EWSR1, DYNLT1, KIF11, ING4, Cbx1, Set, ZNF131, ASB7, AR, CSNK2A1, OGT, BRD4, KDM5B, KDM5D, KDM6B, CKS1B, CKS2, CHD4, ESR2, AGR2, EZH2, FBXW7, SOX2, MED17, WDR82, ACTR8, GEMIN5, RUVBL1, MCM2, RTF1, MED15, MED8, MRGBP, MED14, HIST1H2BB, HIST1H2AB, KIAA1429, SP1, MYC, NR2C2, IRF2, PLEKHA4, DPY30, PTEN, NPM1, RPL13, CCT6A, TMCO1, ITGB1, ESR1, MYCN, SUPT5H, CIT, Pik3r2, FASN, LDLR, CDC27, KMT2B, SETD1A, KDM6A, KANSL2, BUB3, SETD1B, KANSL1, RERE, MCRS1, KMT2C, ZZZ3, MBIP, YEATS2, PAXIP1, KANSL3, KMT2D, BOD1L1, CSRP2BP, ZNF608, ATN1, PHF20L1, PHF20, NCOA6, TADA3, APEX1, ASF1A, CBX3, CD3EAP, CENPA, NUP50, POLR1E, TERF2IP, ZNF330, NAA40, PRKRA, SRSF4, KRR1, RPL13A, ABT1, CSNK2B, OASL, SRSF5, SRSF6, RPS9, FGFBP1, RPLP0, RPL36, KAL1, RPL3, PML, ELF1, ELF2, FEV, NHLH1, EN1, KLF12, KLF3, NFIX, NFYC, YY1, BRD3, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| EPS15 |  |
| KMT2A |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | EPS15 | chr1:51913717 | chr11:118354897 | ENST00000371733 | 9 | 25 | 2_330 | 217.0 | 897.0 | DAB2 |
Top |
Related Drugs to EPS15-KMT2A |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to EPS15-KMT2A |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | KMT2A | C2826025 | Mixed phenotype acute leukemia | 3 | ORPHANET |
| Tgene | KMT2A | C0023418 | leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023452 | Childhood Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023453 | L2 Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023466 | Leukemia, Monocytic, Chronic | 2 | CTD_human |
| Tgene | KMT2A | C0023467 | Leukemia, Myelocytic, Acute | 2 | CTD_human |
| Tgene | KMT2A | C0023470 | Myeloid Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0026998 | Acute Myeloid Leukemia, M1 | 2 | CTD_human |
| Tgene | KMT2A | C1854630 | Growth Deficiency and Mental Retardation with Facial Dysmorphism | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | KMT2A | C1879321 | Acute Myeloid Leukemia (AML-M2) | 2 | CTD_human |
| Tgene | KMT2A | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 2 | CTD_human |
| Tgene | KMT2A | C0001418 | Adenocarcinoma | 1 | CTD_human |
| Tgene | KMT2A | C0004403 | Autosome Abnormalities | 1 | CTD_human |
| Tgene | KMT2A | C0005684 | Malignant neoplasm of urinary bladder | 1 | CTD_human |
| Tgene | KMT2A | C0005695 | Bladder Neoplasm | 1 | CTD_human |
| Tgene | KMT2A | C0007138 | Carcinoma, Transitional Cell | 1 | CTD_human |
| Tgene | KMT2A | C0008625 | Chromosome Aberrations | 1 | CTD_human |
| Tgene | KMT2A | C0023448 | Lymphoid leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0023465 | Acute monocytic leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0023479 | Acute myelomonocytic leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human |
| Tgene | KMT2A | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Tgene | KMT2A | C0036341 | Schizophrenia | 1 | PSYGENET |
| Tgene | KMT2A | C0038356 | Stomach Neoplasms | 1 | CTD_human |
| Tgene | KMT2A | C0149925 | Small cell carcinoma of lung | 1 | CTD_human |
| Tgene | KMT2A | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| Tgene | KMT2A | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| Tgene | KMT2A | C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| Tgene | KMT2A | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| Tgene | KMT2A | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| Tgene | KMT2A | C0270972 | Cornelia De Lange Syndrome | 1 | ORPHANET |
| Tgene | KMT2A | C0280141 | Acute Undifferentiated Leukemia | 1 | ORPHANET |
| Tgene | KMT2A | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| Tgene | KMT2A | C0856823 | Undifferentiated type acute leukemia | 1 | ORPHANET |
| Tgene | KMT2A | C1535926 | Neurodevelopmental Disorders | 1 | CTD_human |
| Tgene | KMT2A | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human |
| Tgene | KMT2A | C2239176 | Liver carcinoma | 1 | CTD_human |
| Tgene | KMT2A | C2930974 | Acute erythroleukemia | 1 | CTD_human |
| Tgene | KMT2A | C2930975 | Acute erythroleukemia - M6a subtype | 1 | CTD_human |
| Tgene | KMT2A | C2930976 | Acute myeloid leukemia FAB-M6 | 1 | CTD_human |
| Tgene | KMT2A | C2930977 | Acute erythroleukemia - M6b subtype | 1 | CTD_human |