| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:AGRN-MXI1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: AGRN-MXI1 | FusionPDB ID: 3001 | FusionGDB2.0 ID: 3001 | Hgene | Tgene | Gene symbol | AGRN | MXI1 | Gene ID | 375790 | 4601 |
| Gene name | agrin | MAX interactor 1, dimerization protein | |
| Synonyms | AGRIN|CMS8|CMSPPD | MAD2|MXD2|MXI|bHLHc11 | |
| Cytomap | 1p36.33 | 10q25.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | agrinagrin proteoglycan | max-interacting protein 1MAX dimerization protein 2Max-related transcription factorclass C basic helix-loop-helix protein 11 | |
| Modification date | 20200315 | 20200313 | |
| UniProtAcc | O00468 | P50539 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000477585, ENST00000379370, | ENST00000332674, ENST00000485566, ENST00000239007, ENST00000361248, ENST00000369612, ENST00000393134, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 14 X 7 X 10=980 | 17 X 10 X 9=1530 |
| # samples | 18 | 19 | |
| ** MAII score | log2(18/980*10)=-2.4447848426729 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(19/1530*10)=-3.00946032924907 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: AGRN [Title/Abstract] AND MXI1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | AGRN(970704)-MXI1(111987946), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | AGRN-MXI1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AGRN-MXI1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. AGRN-MXI1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. AGRN-MXI1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | AGRN | GO:0043113 | receptor clustering | 15340048 |
| Tgene | MXI1 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 11875718 |
 Fusion gene breakpoints across AGRN (5'-gene) Fusion gene breakpoints across AGRN (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
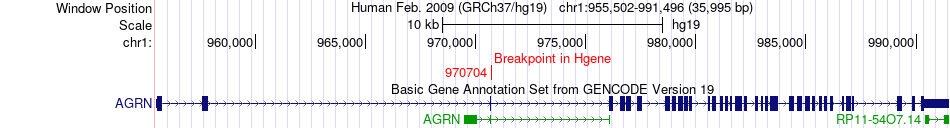 |
 Fusion gene breakpoints across MXI1 (3'-gene) Fusion gene breakpoints across MXI1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
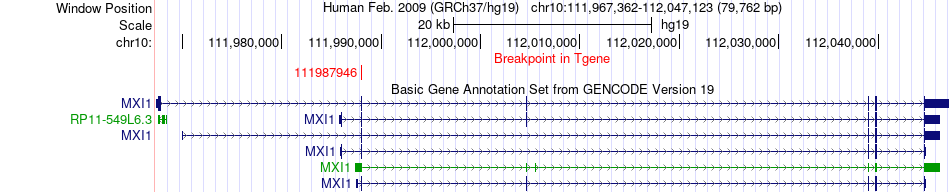 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-FP-8631 | AGRN | chr1 | 970704 | + | MXI1 | chr10 | 111987946 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000379370 | AGRN | chr1 | 970704 | + | ENST00000332674 | MXI1 | chr10 | 111987946 | + | 3553 | 561 | 50 | 1174 | 374 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000379370 | ENST00000332674 | AGRN | chr1 | 970704 | + | MXI1 | chr10 | 111987946 | + | 0.001221085 | 0.9987789 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >3001_3001_1_AGRN-MXI1_AGRN_chr1_970704_ENST00000379370_MXI1_chr10_111987946_ENST00000332674_length(amino acids)=374AA_BP=170 MAGRSHPGPLRPLLPLLVVAACVLPGAGGTCPERALERREEEANVVLTGTVEEILNVDPVQHTYSCKVRVWRYLKGKDLVARESLLDGGN KVVISGFGDPLICDNQVSTGDTRIFFVNPAPPYLWPAHKNELMLNSSLMRITLRNLEEVEFCVEDKPGTHFTPVPPTPPDECEHGYASSF PSMPSPRLQHSKPPRRLSRAQKHSSGSSNTSTANRSTHNELEKNRRAHLRLCLERLKVLIPLGPDCTRHTTLGLLNKAKAHIKKLEEAER KSQHQLENLEREQRFLKWRLEQLQGPQEMERIRMDSIGSTISSDRSDSEREEIEVDVESTEFSHGEVDNISTTSISDIDDHSSLPSIGSD -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:970704/chr10:111987946) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| AGRN | MXI1 |
| FUNCTION: [Isoform 1]: heparan sulfate basal lamina glycoprotein that plays a central role in the formation and the maintenance of the neuromuscular junction (NMJ) and directs key events in postsynaptic differentiation. Component of the AGRN-LRP4 receptor complex that induces the phosphorylation and activation of MUSK. The activation of MUSK in myotubes induces the formation of NMJ by regulating different processes including the transcription of specific genes and the clustering of AChR in the postsynaptic membrane. Calcium ions are required for maximal AChR clustering. AGRN function in neurons is highly regulated by alternative splicing, glycan binding and proteolytic processing. Modulates calcium ion homeostasis in neurons, specifically by inducing an increase in cytoplasmic calcium ions. Functions differentially in the central nervous system (CNS) by inhibiting the alpha(3)-subtype of Na+/K+-ATPase and evoking depolarization at CNS synapses. This secreted isoform forms a bridge, after release from motor neurons, to basal lamina through binding laminin via the NtA domain.; FUNCTION: [Isoform 2]: transmembrane form that is the predominate form in neurons of the brain, induces dendritic filopodia and synapse formation in mature hippocampal neurons in large part due to the attached glycosaminoglycan chains and the action of Rho-family GTPases.; FUNCTION: Isoform 1, isoform 4 and isoform 5: neuron-specific (z+) isoforms that contain C-terminal insertions of 8-19 AA are potent activators of AChR clustering. Isoform 5, agrin (z+8), containing the 8-AA insert, forms a receptor complex in myotubules containing the neuronal AGRN, the muscle-specific kinase MUSK and LRP4, a member of the LDL receptor family. The splicing factors, NOVA1 and NOVA2, regulate AGRN splicing and production of the 'z' isoforms.; FUNCTION: Isoform 3 and isoform 6: lack any 'z' insert, are muscle-specific and may be involved in endothelial cell differentiation.; FUNCTION: [Agrin N-terminal 110 kDa subunit]: is involved in regulation of neurite outgrowth probably due to the presence of the glycosaminoglcan (GAG) side chains of heparan and chondroitin sulfate attached to the Ser/Thr- and Gly/Ser-rich regions. Also involved in modulation of growth factor signaling (By similarity). {ECO:0000250, ECO:0000269|PubMed:19631309, ECO:0000269|PubMed:21969364}.; FUNCTION: [Agrin C-terminal 22 kDa fragment]: this released fragment is important for agrin signaling and to exert a maximal dendritic filopodia-inducing effect. All 'z' splice variants (z+) of this fragment also show an increase in the number of filopodia. | FUNCTION: Transcriptional repressor. MXI1 binds with MAX to form a sequence-specific DNA-binding protein complex which recognizes the core sequence 5'-CAC[GA]TG-3'. MXI1 thus antagonizes MYC transcriptional activity by competing for MAX. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 30_157 | 170.33333333333334 | 2046.0 | Domain | NtA |
| Tgene | MXI1 | chr1:970704 | chr10:111987946 | ENST00000239007 | 0 | 6 | 67_119 | 24.333333333333332 | 229.0 | Domain | bHLH | |
| Tgene | MXI1 | chr1:970704 | chr10:111987946 | ENST00000361248 | 0 | 5 | 67_119 | 0 | 183.0 | Domain | bHLH | |
| Tgene | MXI1 | chr1:970704 | chr10:111987946 | ENST00000369612 | 0 | 6 | 67_119 | 0 | 193.0 | Domain | bHLH |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1941_2009 | 170.33333333333334 | 2046.0 | Calcium binding | Ontology_term=ECO:0000250 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1058_1097 | 170.33333333333334 | 2046.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1254_1324 | 170.33333333333334 | 2046.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 671_677 | 170.33333333333334 | 2046.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 974_1099 | 170.33333333333334 | 2046.0 | Compositional bias | Note=Ser/Thr-rich |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1130_1252 | 170.33333333333334 | 2046.0 | Domain | SEA |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1329_1367 | 170.33333333333334 | 2046.0 | Domain | EGF-like 1 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1372_1548 | 170.33333333333334 | 2046.0 | Domain | Laminin G-like 1 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1549_1586 | 170.33333333333334 | 2046.0 | Domain | EGF-like 2 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1588_1625 | 170.33333333333334 | 2046.0 | Domain | EGF-like 3 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1635_1822 | 170.33333333333334 | 2046.0 | Domain | Laminin G-like 2 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1818_1857 | 170.33333333333334 | 2046.0 | Domain | EGF-like 4 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 1868_2065 | 170.33333333333334 | 2046.0 | Domain | Laminin G-like 3 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 191_244 | 170.33333333333334 | 2046.0 | Domain | Kazal-like 1 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 264_319 | 170.33333333333334 | 2046.0 | Domain | Kazal-like 2 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 337_391 | 170.33333333333334 | 2046.0 | Domain | Kazal-like 3 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 408_463 | 170.33333333333334 | 2046.0 | Domain | Kazal-like 4 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 484_536 | 170.33333333333334 | 2046.0 | Domain | Kazal-like 5 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 540_601 | 170.33333333333334 | 2046.0 | Domain | Kazal-like 6 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 607_666 | 170.33333333333334 | 2046.0 | Domain | Kazal-like 7 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 699_752 | 170.33333333333334 | 2046.0 | Domain | Kazal-like 8 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 793_846 | 170.33333333333334 | 2046.0 | Domain | Laminin EGF-like 1 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 847_893 | 170.33333333333334 | 2046.0 | Domain | Laminin EGF-like 2 |
| Hgene | AGRN | chr1:970704 | chr10:111987946 | ENST00000379370 | + | 3 | 36 | 917_971 | 170.33333333333334 | 2046.0 | Domain | Kazal-like 9 |
| Tgene | MXI1 | chr1:970704 | chr10:111987946 | ENST00000332674 | 0 | 6 | 67_119 | 91.33333333333333 | 296.0 | Domain | bHLH |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| AGRN | |
| MXI1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to AGRN-MXI1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to AGRN-MXI1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

