| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:FGFR1-ZNF703 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: FGFR1-ZNF703 | FusionPDB ID: 30270 | FusionGDB2.0 ID: 30270 | Hgene | Tgene | Gene symbol | FGFR1 | ZNF703 | Gene ID | 2260 | 80139 |
| Gene name | fibroblast growth factor receptor 1 | zinc finger protein 703 | |
| Synonyms | BFGFR|CD331|CEK|ECCL|FGFBR|FGFR-1|FLG|FLT-2|FLT2|HBGFR|HH2|HRTFDS|KAL2|N-SAM|OGD|bFGF-R-1 | NLZ1|ZEPPO1|ZNF503L|ZPO1 | |
| Cytomap | 8p11.23 | 8p11.23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | fibroblast growth factor receptor 1FGFR1/PLAG1 fusionFMS-like tyrosine kinase 2basic fibroblast growth factor receptor 1fms-related tyrosine kinase 2heparin-binding growth factor receptorhydroxyaryl-protein kinaseproto-oncogene c-Fgr | zinc finger protein 703zinc finger elbow-related proline domain protein 1 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | Q9NVK5 | Q9H7S9 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000326324, ENST00000335922, ENST00000341462, ENST00000356207, ENST00000397091, ENST00000397103, ENST00000397108, ENST00000397113, ENST00000425967, ENST00000447712, ENST00000532791, ENST00000496629, | ENST00000331569, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 17 X 24 X 9=3672 | 1 X 1 X 1=1 |
| # samples | 20 | 1 | |
| ** MAII score | log2(20/3672*10)=-4.19849415363908 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(1/1*10)=3.32192809488736 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: FGFR1 [Title/Abstract] AND ZNF703 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | FGFR1-ZNF703 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. FGFR1-ZNF703 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. FGFR1-ZNF703 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. FGFR1-ZNF703 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | FGFR1 | GO:0008284 | positive regulation of cell proliferation | 8663044 |
| Hgene | FGFR1 | GO:0008543 | fibroblast growth factor receptor signaling pathway | 8663044 |
| Hgene | FGFR1 | GO:0010863 | positive regulation of phospholipase C activity | 18480409 |
| Hgene | FGFR1 | GO:0018108 | peptidyl-tyrosine phosphorylation | 8622701|18480409 |
| Hgene | FGFR1 | GO:0043406 | positive regulation of MAP kinase activity | 8622701|18480409 |
| Hgene | FGFR1 | GO:0046777 | protein autophosphorylation | 8622701 |
| Hgene | FGFR1 | GO:2000546 | positive regulation of endothelial cell chemotaxis to fibroblast growth factor | 21885851 |
| Tgene | ZNF703 | GO:0006355 | regulation of transcription, DNA-templated | 21328542 |
| Tgene | ZNF703 | GO:0008284 | positive regulation of cell proliferation | 21328542 |
| Tgene | ZNF703 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway | 21337521 |
| Tgene | ZNF703 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation | 21337521 |
| Tgene | ZNF703 | GO:0051726 | regulation of cell cycle | 21328542 |
| Tgene | ZNF703 | GO:0060644 | mammary gland epithelial cell differentiation | 21337521 |
| Tgene | ZNF703 | GO:0071392 | cellular response to estradiol stimulus | 21328542 |
 Fusion gene breakpoints across FGFR1 (5'-gene) Fusion gene breakpoints across FGFR1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across ZNF703 (3'-gene) Fusion gene breakpoints across ZNF703 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | FGFR1 | chr8 | 38272076 | - | ZNF703 | chr8 | 37554662 | + |
| ChimerKB3 | . | . | FGFR1 | chr8 | 38273387 | - | ZNF703 | chr8 | 37554662 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000397091 | FGFR1 | chr8 | 38272076 | - | ENST00000331569 | ZNF703 | chr8 | 37554662 | + | 5666 | 2791 | 368 | 2968 | 866 |
| ENST00000447712 | FGFR1 | chr8 | 38272076 | - | ENST00000331569 | ZNF703 | chr8 | 37554662 | + | 5865 | 2990 | 561 | 3167 | 868 |
| ENST00000341462 | FGFR1 | chr8 | 38272076 | - | ENST00000331569 | ZNF703 | chr8 | 37554662 | + | 5865 | 2990 | 561 | 3167 | 868 |
| ENST00000532791 | FGFR1 | chr8 | 38272076 | - | ENST00000331569 | ZNF703 | chr8 | 37554662 | + | 5647 | 2772 | 349 | 2949 | 866 |
| ENST00000397113 | FGFR1 | chr8 | 38272076 | - | ENST00000331569 | ZNF703 | chr8 | 37554662 | + | 5234 | 2359 | 131 | 2536 | 801 |
| ENST00000356207 | FGFR1 | chr8 | 38272076 | - | ENST00000331569 | ZNF703 | chr8 | 37554662 | + | 5380 | 2505 | 343 | 2682 | 779 |
| ENST00000326324 | FGFR1 | chr8 | 38272076 | - | ENST00000331569 | ZNF703 | chr8 | 37554662 | + | 5374 | 2499 | 343 | 2676 | 777 |
| ENST00000397103 | FGFR1 | chr8 | 38272076 | - | ENST00000331569 | ZNF703 | chr8 | 37554662 | + | 4713 | 1838 | 27 | 2015 | 662 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >30270_30270_1_FGFR1-ZNF703_FGFR1_chr8_38272076_ENST00000326324_ZNF703_chr8_37554662_ENST00000331569_length(amino acids)=777AA_BP=719 MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSRAPAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPE AEPPRRARVSLKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARPSPTLPEQDALPSSEDDDDDDDSSSEEKETD NTKPNPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPSDKGNYTCIVEN EYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKTAGVNTTDKEME VLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEERPAVMTSPLYLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMAV HKLAKSIPLRRQVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPRDRLVLGKPLGEGCFGQVVLAEAIGLDKDK PNRVTKVAVKMLKSDATEKDLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQARRPPGLEYCYNPSHNPEEQ LSSKDLVSCAYQVARGMEYLASKKCIHRDLAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDRIYTHQSDVW -------------------------------------------------------------- >30270_30270_2_FGFR1-ZNF703_FGFR1_chr8_38272076_ENST00000341462_ZNF703_chr8_37554662_ENST00000331569_length(amino acids)=868AA_BP=810 MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSRAPAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPE AEPPRRARVSLKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARPSPTLPEQAQPWGAPVEVESFLVHPGDLLQL RCRLRDDVQSINWLRDGVQLAESNRTRITGEEVEVQDSVPADSGLYACVTSSPSGSDTTYFSVNVSVPIDALPSSEDDDDDDDSSSEEKE TDNTKPNPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPSDKGNYTCIV ENEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKHSGINSSDAE VLTLFNVTEAQSGEYVCKVSNYIGEANQSAWLTVTRPVLEERPAVMTSPLYLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMA VHKLAKSIPLRRQVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPRDRLVLGKPLGEGCFGQVVLAEAIGLDKD KPNRVTKVAVKMLKSDATEKDLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQARRPPGLEYCYNPSHNPEE QLSSKDLVSCAYQVARGMEYLASKKCIHRDLAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDRIYTHQSDV -------------------------------------------------------------- >30270_30270_3_FGFR1-ZNF703_FGFR1_chr8_38272076_ENST00000356207_ZNF703_chr8_37554662_ENST00000331569_length(amino acids)=779AA_BP=721 MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSRAPAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPE AEPPRRARVSLKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARPSPTLPEQDALPSSEDDDDDDDSSSEEKETD NTKPNRMPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPSDKGNYTCIV ENEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKTAGVNTTDKE MEVLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEERPAVMTSPLYLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQM AVHKLAKSIPLRRQVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPRDRLVLGKPLGEGCFGQVVLAEAIGLDK DKPNRVTKVAVKMLKSDATEKDLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQARRPPGLEYCYNPSHNPE EQLSSKDLVSCAYQVARGMEYLASKKCIHRDLAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDRIYTHQSD -------------------------------------------------------------- >30270_30270_4_FGFR1-ZNF703_FGFR1_chr8_38272076_ENST00000397091_ZNF703_chr8_37554662_ENST00000331569_length(amino acids)=866AA_BP=808 MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSRAPAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPE AEPPRRARVSLKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARPSPTLPEQAQPWGAPVEVESFLVHPGDLLQL RCRLRDDVQSINWLRDGVQLAESNRTRITGEEVEVQDSVPADSGLYACVTSSPSGSDTTYFSVNVSDALPSSEDDDDDDDSSSEEKETDN TKPNPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPSDKGNYTCIVENE YGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKTAGVNTTDKEMEV LHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEERPAVMTSPLYLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMAVH KLAKSIPLRRQVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPRDRLVLGKPLGEGCFGQVVLAEAIGLDKDKP NRVTKVAVKMLKSDATEKDLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQARRPPGLEYCYNPSHNPEEQL SSKDLVSCAYQVARGMEYLASKKCIHRDLAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDRIYTHQSDVWT -------------------------------------------------------------- >30270_30270_5_FGFR1-ZNF703_FGFR1_chr8_38272076_ENST00000397103_ZNF703_chr8_37554662_ENST00000331569_length(amino acids)=662AA_BP=604 MSPTSNCRTGMWSWKCLLFWAVLVTATLCTARPSPTLPEQDALPSSEDDDDDDDSSSEEKETDNTKPNPVAPYWTSPEKMEKKLHAVPAA KTVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPSDKGNYTCIVENEYGSINHTYQLDVVERSPHRPILQAGL PANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKHSGINSSDAEVLTLFNVTEAQSGEYVCKVSNYIGEANQS AWLTVTRPVAKALEERPAVMTSPLYLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMAVHKLAKSIPLRRQVTVSADSSASMNS GVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPRDRLVLGKPLGEGCFGQVVLAEAIGLDKDKPNRVTKVAVKMLKSDATEKDLSDLI SEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQARRPPGLEYCYNPSHNPEEQLSSKDLVSCAYQVARGMEYLASKKC IHRDLAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDRIYTHQSDVWTPRRAPWRCWLRPARRSASRTRRPP -------------------------------------------------------------- >30270_30270_6_FGFR1-ZNF703_FGFR1_chr8_38272076_ENST00000397113_ZNF703_chr8_37554662_ENST00000331569_length(amino acids)=801AA_BP=743 MRPLPALVPPLFPPPNFSSNSRSGAGEAAAAPQVSLKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARPSPTLP EQAQPWGAPVEVESFLVHPGDLLQLRCRLRDDVQSINWLRDGVQLAESNRTRITGEEVEVQDSVPADSGLYACVTSSPSGSDTTYFSVNV SDALPSSEDDDDDDDSSSEEKETDNTKPNPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRY ATWSIIMDSVVPSDKGNYTCIVENEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKI GPDNLPYVQILKTAGVNTTDKEMEVLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEERPAVMTSPLYLEIIIYCTGAFLISCM VGSVIVYKMKSGTKKSDFHSQMAVHKLAKSIPLRRQVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPRDRLVL GKPLGEGCFGQVVLAEAIGLDKDKPNRVTKVAVKMLKSDATEKDLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLR EYLQARRPPGLEYCYNPSHNPEEQLSSKDLVSCAYQVARGMEYLASKKCIHRDLAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNG -------------------------------------------------------------- >30270_30270_7_FGFR1-ZNF703_FGFR1_chr8_38272076_ENST00000447712_ZNF703_chr8_37554662_ENST00000331569_length(amino acids)=868AA_BP=810 MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSRAPAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPE AEPPRRARVSLKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARPSPTLPEQAQPWGAPVEVESFLVHPGDLLQL RCRLRDDVQSINWLRDGVQLAESNRTRITGEEVEVQDSVPADSGLYACVTSSPSGSDTTYFSVNVSDALPSSEDDDDDDDSSSEEKETDN TKPNRMPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPSDKGNYTCIVE NEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKTAGVNTTDKEM EVLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEERPAVMTSPLYLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMA VHKLAKSIPLRRQVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPRDRLVLGKPLGEGCFGQVVLAEAIGLDKD KPNRVTKVAVKMLKSDATEKDLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQARRPPGLEYCYNPSHNPEE QLSSKDLVSCAYQVARGMEYLASKKCIHRDLAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDRIYTHQSDV -------------------------------------------------------------- >30270_30270_8_FGFR1-ZNF703_FGFR1_chr8_38272076_ENST00000532791_ZNF703_chr8_37554662_ENST00000331569_length(amino acids)=866AA_BP=808 MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSRAPAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPE AEPPRRARVSLKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARPSPTLPEQAQPWGAPVEVESFLVHPGDLLQL RCRLRDDVQSINWLRDGVQLAESNRTRITGEEVEVQDSVPADSGLYACVTSSPSGSDTTYFSVNVSDALPSSEDDDDDDDSSSEEKETDN TKPNRMPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPSDKGNYTCIVE NEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKTAGVNTTDKEM EVLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEERPAVMTSPLYLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMA VHKLAKSIPLRRQVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPRDRLVLGKPLGEGCFGQVVLAEAIGLDKDKP NRVTKVAVKMLKSDATEKDLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQARRPPGLEYCYNPSHNPEEQL SSKDLVSCAYQVARGMEYLASKKCIHRDLAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDRIYTHQSDVWT -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:/chr8:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| FGFR1 | ZNF703 |
| FUNCTION: May be involved in wound healing pathway. {ECO:0000250}. | FUNCTION: Transcriptional corepressor which does not bind directly to DNA and may regulate transcription through recruitment of histone deacetylases to gene promoters. Regulates cell adhesion, migration and proliferation. May be required for segmental gene expression during hindbrain development. {ECO:0000269|PubMed:21328542, ECO:0000269|PubMed:21337521}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
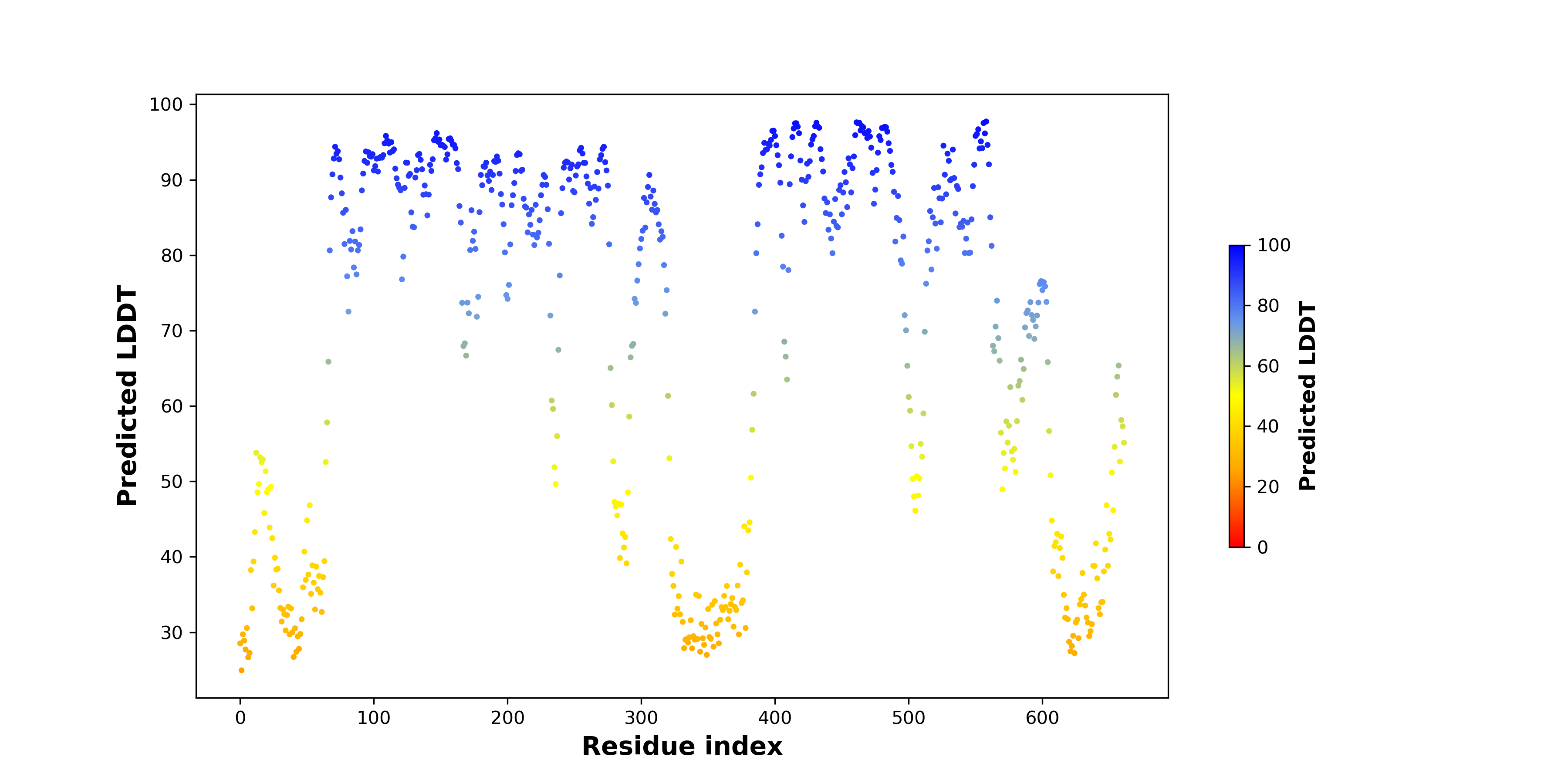
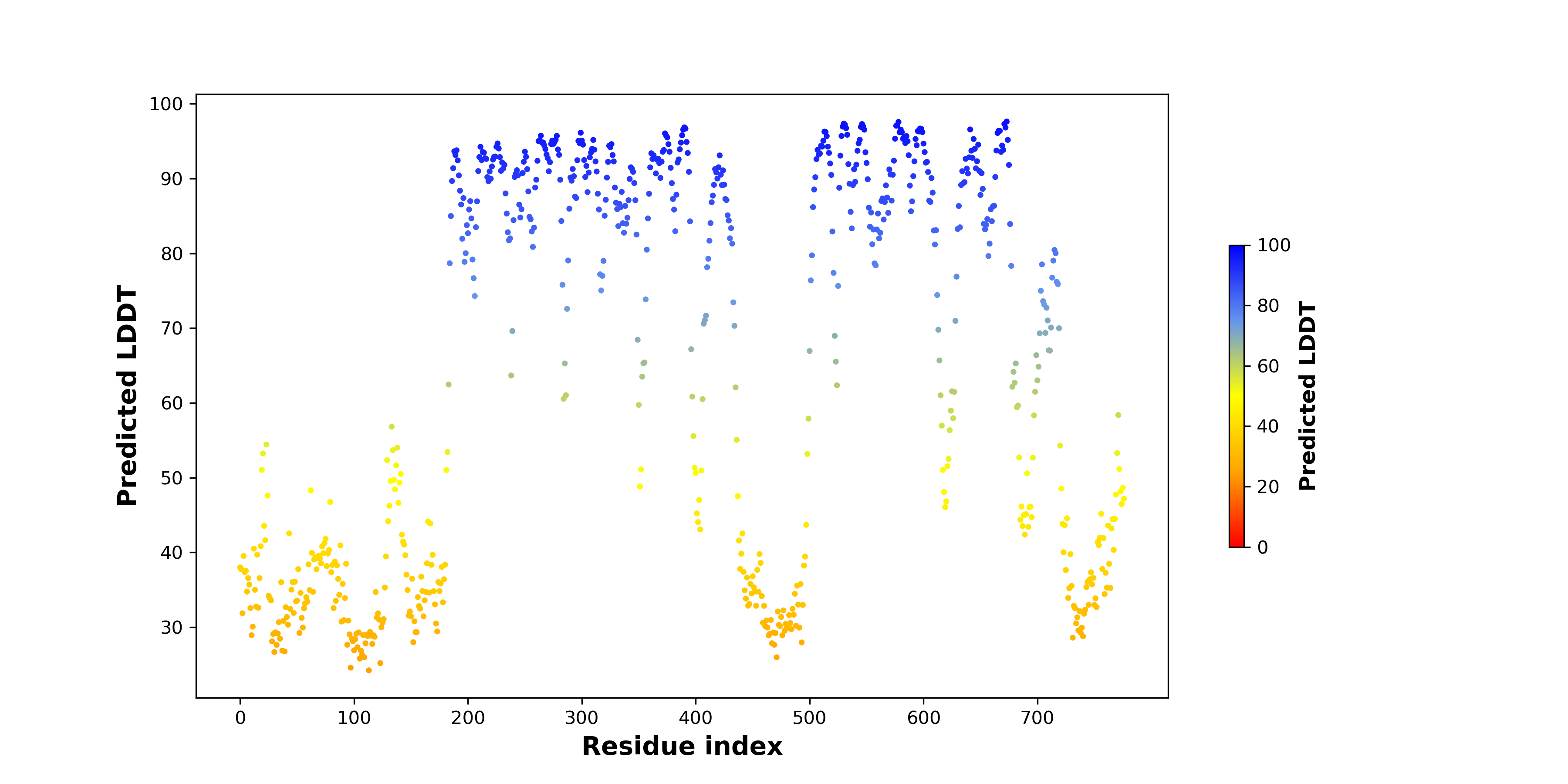
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (540) >>>540.pdbFusion protein BP residue: 604 CIF file (540) >>>540.cif | FGFR1 | chr8 | 38272076 | - | ZNF703 | chr8 | 37554662 | + | MSPTSNCRTGMWSWKCLLFWAVLVTATLCTARPSPTLPEQDALPSSEDDD DDDDSSSEEKETDNTKPNPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSS GTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPSDKGNYTCI VENEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYS DPQPHIQWLKHIEVNGSKIGPDNLPYVQILKHSGINSSDAEVLTLFNVTE AQSGEYVCKVSNYIGEANQSAWLTVTRPVAKALEERPAVMTSPLYLEIII YCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMAVHKLAKSIPLRRQVTV SADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPRDRLV LGKPLGEGCFGQVVLAEAIGLDKDKPNRVTKVAVKMLKSDATEKDLSDLI SEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQARRPP GLEYCYNPSHNPEEQLSSKDLVSCAYQVARGMEYLASKKCIHRDLAARNV LVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDRIYTH QSDVWTPRRAPWRCWLRPARRSASRTRRPPPNSTRWRRRPTGWERRRTPA | 662 |
| 3D view using mol* of 540 (AA BP:604) | ||||||||||
 | ||||||||||
| PDB file (635) >>>635.pdbFusion protein BP residue: 719 CIF file (635) >>>635.cif | FGFR1 | chr8 | 38272076 | - | ZNF703 | chr8 | 37554662 | + | MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSR APAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPEAEPPRRARVS LKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARP SPTLPEQDALPSSEDDDDDDDSSSEEKETDNTKPNPVAPYWTSPEKMEKK LHAVPAAKTVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWS IIMDSVVPSDKGNYTCIVENEYGSINHTYQLDVVERSPHRPILQAGLPAN KTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKTA GVNTTDKEMEVLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEE RPAVMTSPLYLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMAV HKLAKSIPLRRQVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYE LPEDPRWELPRDRLVLGKPLGEGCFGQVVLAEAIGLDKDKPNRVTKVAVK MLKSDATEKDLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYA SKGNLREYLQARRPPGLEYCYNPSHNPEEQLSSKDLVSCAYQVARGMEYL ASKKCIHRDLAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPV KWMAPEALFDRIYTHQSDVWTPRRAPWRCWLRPARRSASRTRRPPPNSTR | 777 |
| 3D view using mol* of 635 (AA BP:719) | ||||||||||
 | ||||||||||
| PDB file (637) >>>637.pdbFusion protein BP residue: 721 CIF file (637) >>>637.cif | FGFR1 | chr8 | 38272076 | - | ZNF703 | chr8 | 37554662 | + | MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSR APAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPEAEPPRRARVS LKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARP SPTLPEQDALPSSEDDDDDDDSSSEEKETDNTKPNRMPVAPYWTSPEKME KKLHAVPAAKTVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYAT WSIIMDSVVPSDKGNYTCIVENEYGSINHTYQLDVVERSPHRPILQAGLP ANKTVALGSNVEFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILK TAGVNTTDKEMEVLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEAL EERPAVMTSPLYLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQM AVHKLAKSIPLRRQVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSE YELPEDPRWELPRDRLVLGKPLGEGCFGQVVLAEAIGLDKDKPNRVTKVA VKMLKSDATEKDLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVE YASKGNLREYLQARRPPGLEYCYNPSHNPEEQLSSKDLVSCAYQVARGME YLASKKCIHRDLAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRL PVKWMAPEALFDRIYTHQSDVWTPRRAPWRCWLRPARRSASRTRRPPPNS | 779 |
| 3D view using mol* of 637 (AA BP:721) | ||||||||||
 | ||||||||||
| PDB file (652) >>>652.pdbFusion protein BP residue: 743 CIF file (652) >>>652.cif | FGFR1 | chr8 | 38272076 | - | ZNF703 | chr8 | 37554662 | + | MRPLPALVPPLFPPPNFSSNSRSGAGEAAAAPQVSLKRRIELTVEYPWRC GALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARPSPTLPEQAQPWGAPV EVESFLVHPGDLLQLRCRLRDDVQSINWLRDGVQLAESNRTRITGEEVEV QDSVPADSGLYACVTSSPSGSDTTYFSVNVSDALPSSEDDDDDDDSSSEE KETDNTKPNPVAPYWTSPEKMEKKLHAVPAAKTVKFKCPSSGTPNPTLRW LKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPSDKGNYTCIVENEYGSIN HTYQLDVVERSPHRPILQAGLPANKTVALGSNVEFMCKVYSDPQPHIQWL KHIEVNGSKIGPDNLPYVQILKTAGVNTTDKEMEVLHLRNVSFEDAGEYT CLAGNSIGLSHHSAWLTVLEALEERPAVMTSPLYLEIIIYCTGAFLISCM VGSVIVYKMKSGTKKSDFHSQMAVHKLAKSIPLRRQVTVSADSSASMNSG VLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPRDRLVLGKPLGEGCFG QVVLAEAIGLDKDKPNRVTKVAVKMLKSDATEKDLSDLISEMEMMKMIGK HKNIINLLGACTQDGPLYVIVEYASKGNLREYLQARRPPGLEYCYNPSHN PEEQLSSKDLVSCAYQVARGMEYLASKKCIHRDLAARNVLVTEDNVMKIA DFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDRIYTHQSDVWTPRRAP WRCWLRPARRSASRTRRPPPNSTRWRRRPTGWERRRTPAAQPRAPPPQPR | 801 |
| 3D view using mol* of 652 (AA BP:743) | ||||||||||
 | ||||||||||
| PDB file (688) >>>688.pdbFusion protein BP residue: 808 CIF file (688) >>>688.cif | FGFR1 | chr8 | 38272076 | - | ZNF703 | chr8 | 37554662 | + | MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSR APAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPEAEPPRRARVS LKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARP SPTLPEQAQPWGAPVEVESFLVHPGDLLQLRCRLRDDVQSINWLRDGVQL AESNRTRITGEEVEVQDSVPADSGLYACVTSSPSGSDTTYFSVNVSDALP SSEDDDDDDDSSSEEKETDNTKPNPVAPYWTSPEKMEKKLHAVPAAKTVK FKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPSDK GNYTCIVENEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNVEF MCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKTAGVNTTDKEMEV LHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEERPAVMTSPLYL EIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMAVHKLAKSIPLRR QVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPR DRLVLGKPLGEGCFGQVVLAEAIGLDKDKPNRVTKVAVKMLKSDATEKDL SDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQA RRPPGLEYCYNPSHNPEEQLSSKDLVSCAYQVARGMEYLASKKCIHRDLA ARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDR IYTHQSDVWTPRRAPWRCWLRPARRSASRTRRPPPNSTRWRRRPTGWERR | 866 |
| 3D view using mol* of 688 (AA BP:808) | ||||||||||
 | ||||||||||
| PDB file (689) >>>689.pdbFusion protein BP residue: 808 CIF file (689) >>>689.cif | FGFR1 | chr8 | 38272076 | - | ZNF703 | chr8 | 37554662 | + | MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSR APAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPEAEPPRRARVS LKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARP SPTLPEQAQPWGAPVEVESFLVHPGDLLQLRCRLRDDVQSINWLRDGVQL AESNRTRITGEEVEVQDSVPADSGLYACVTSSPSGSDTTYFSVNVSDALP SSEDDDDDDDSSSEEKETDNTKPNRMPVAPYWTSPEKMEKKLHAVPAAKT VKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPS DKGNYTCIVENEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNV EFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKTAGVNTTDKEM EVLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEERPAVMTSPL YLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMAVHKLAKSIPL RRQVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWELPR DRLVLGKPLGEGCFGQVVLAEAIGLDKDKPNRVTKVAVKMLKSDATEKDL SDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLQA RRPPGLEYCYNPSHNPEEQLSSKDLVSCAYQVARGMEYLASKKCIHRDLA ARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALFDR IYTHQSDVWTPRRAPWRCWLRPARRSASRTRRPPPNSTRWRRRPTGWERR | 866 |
| 3D view using mol* of 689 (AA BP:808) | ||||||||||
 | ||||||||||
| PDB file (690) >>>690.pdbFusion protein BP residue: 810 CIF file (690) >>>690.cif | FGFR1 | chr8 | 38272076 | - | ZNF703 | chr8 | 37554662 | + | MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSR APAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPEAEPPRRARVS LKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARP SPTLPEQAQPWGAPVEVESFLVHPGDLLQLRCRLRDDVQSINWLRDGVQL AESNRTRITGEEVEVQDSVPADSGLYACVTSSPSGSDTTYFSVNVSDALP SSEDDDDDDDSSSEEKETDNTKPNRMPVAPYWTSPEKMEKKLHAVPAAKT VKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVPS DKGNYTCIVENEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSNV EFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKTAGVNTTDKEM EVLHLRNVSFEDAGEYTCLAGNSIGLSHHSAWLTVLEALEERPAVMTSPL YLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMAVHKLAKSIPL RRQVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWEL PRDRLVLGKPLGEGCFGQVVLAEAIGLDKDKPNRVTKVAVKMLKSDATEK DLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYL QARRPPGLEYCYNPSHNPEEQLSSKDLVSCAYQVARGMEYLASKKCIHRD LAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALF DRIYTHQSDVWTPRRAPWRCWLRPARRSASRTRRPPPNSTRWRRRPTGWE | 868 |
| 3D view using mol* of 690 (AA BP:810) | ||||||||||
 | ||||||||||
| PDB file (691) >>>691.pdbFusion protein BP residue: 810 CIF file (691) >>>691.cif | FGFR1 | chr8 | 38272076 | - | ZNF703 | chr8 | 37554662 | + | MRSGGGGHKVWRPRVADGSPPPAPPPGHQLRLHCSRPGWRRRAPSAAGSR APAAELLRPRQDPNRARGRRAGAGDAGTRPLAQATADSPEAEPPRRARVS LKRRIELTVEYPWRCGALSPTSNCRTGMWSWKCLLFWAVLVTATLCTARP SPTLPEQAQPWGAPVEVESFLVHPGDLLQLRCRLRDDVQSINWLRDGVQL AESNRTRITGEEVEVQDSVPADSGLYACVTSSPSGSDTTYFSVNVSVPID ALPSSEDDDDDDDSSSEEKETDNTKPNPVAPYWTSPEKMEKKLHAVPAAK TVKFKCPSSGTPNPTLRWLKNGKEFKPDHRIGGYKVRYATWSIIMDSVVP SDKGNYTCIVENEYGSINHTYQLDVVERSPHRPILQAGLPANKTVALGSN VEFMCKVYSDPQPHIQWLKHIEVNGSKIGPDNLPYVQILKHSGINSSDAE VLTLFNVTEAQSGEYVCKVSNYIGEANQSAWLTVTRPVLEERPAVMTSPL YLEIIIYCTGAFLISCMVGSVIVYKMKSGTKKSDFHSQMAVHKLAKSIPL RRQVTVSADSSASMNSGVLLVRPSRLSSSGTPMLAGVSEYELPEDPRWEL PRDRLVLGKPLGEGCFGQVVLAEAIGLDKDKPNRVTKVAVKMLKSDATEK DLSDLISEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYL QARRPPGLEYCYNPSHNPEEQLSSKDLVSCAYQVARGMEYLASKKCIHRD LAARNVLVTEDNVMKIADFGLARDIHHIDYYKKTTNGRLPVKWMAPEALF DRIYTHQSDVWTPRRAPWRCWLRPARRSASRTRRPPPNSTRWRRRPTGWE | 868 |
| 3D view using mol* of 691 (AA BP:810) | ||||||||||
 | ||||||||||
Top |
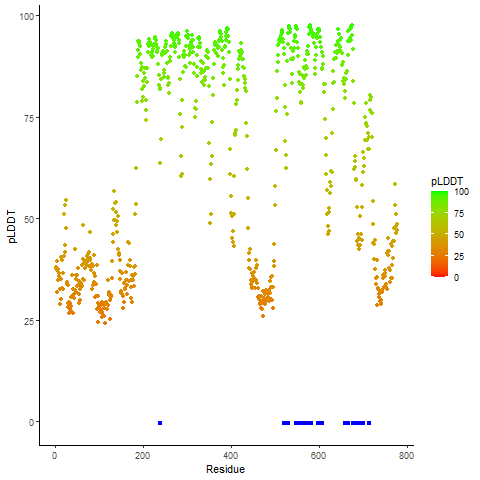
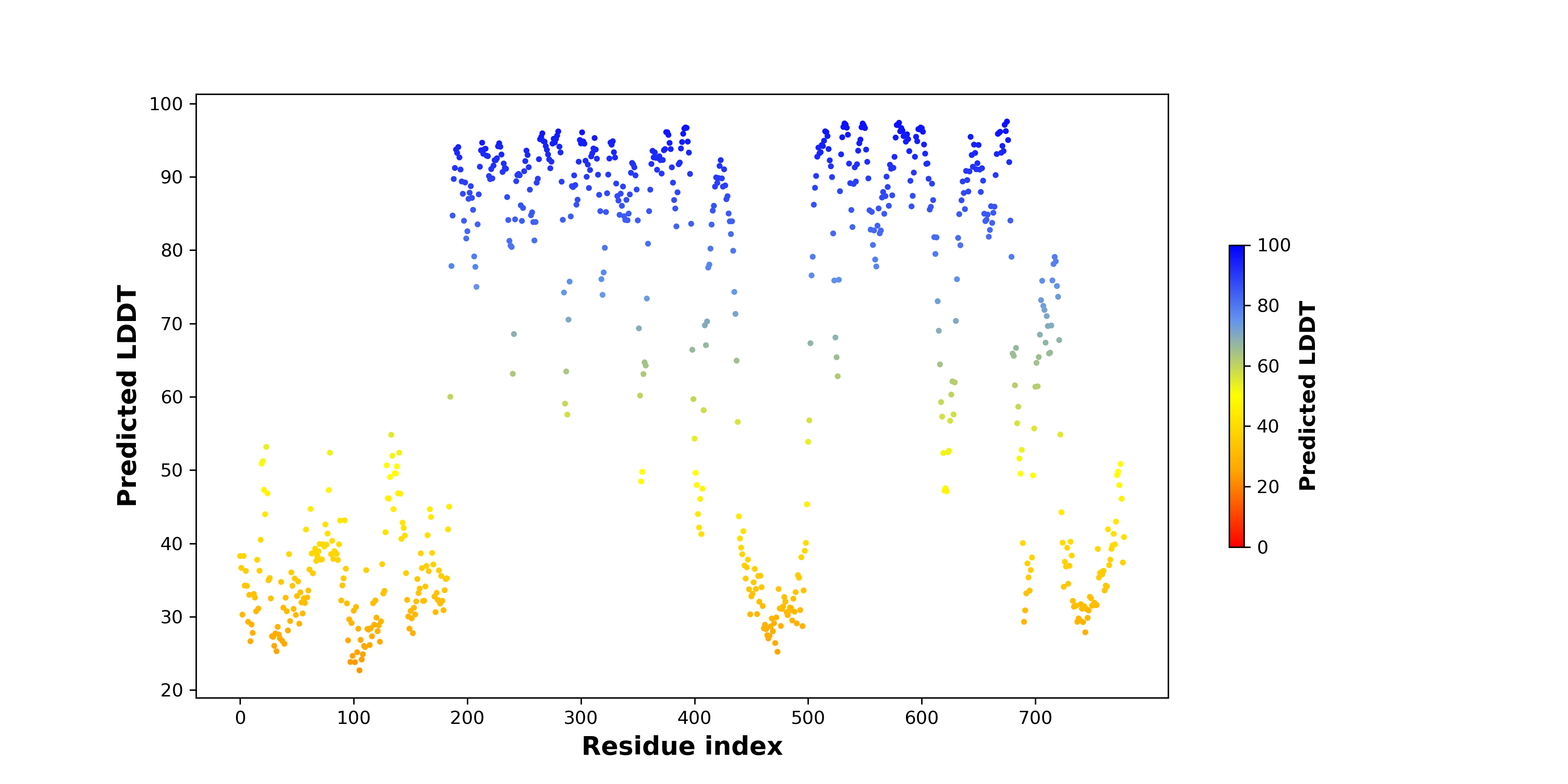
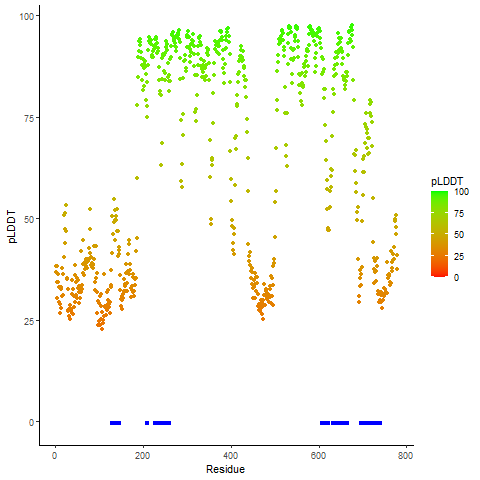
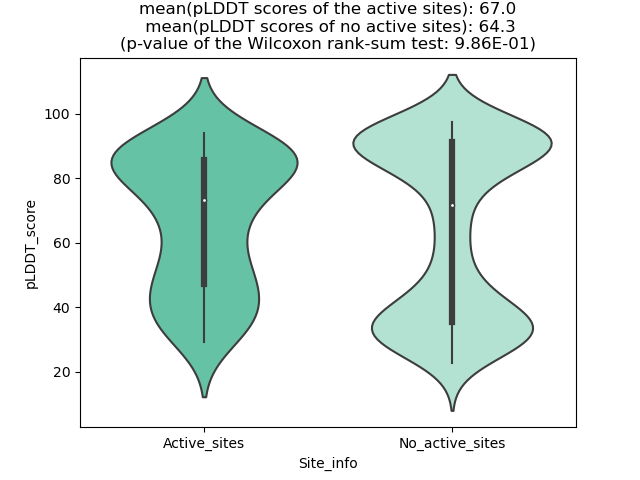
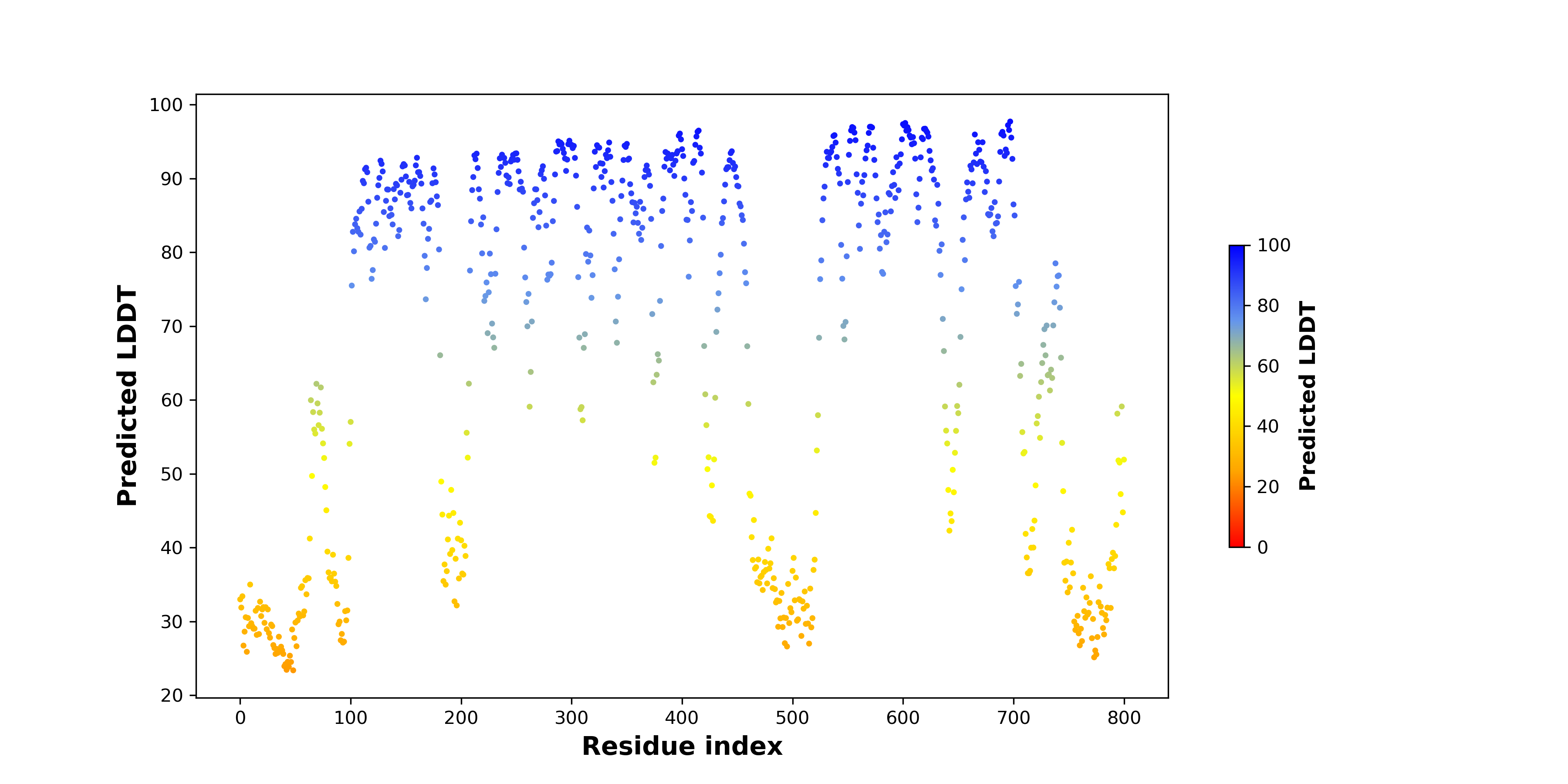
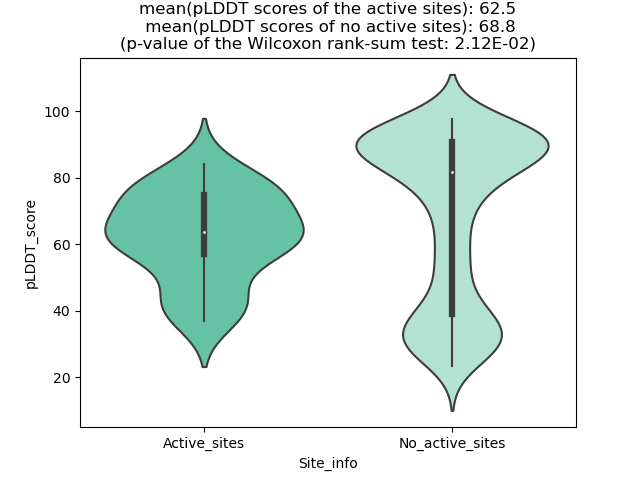
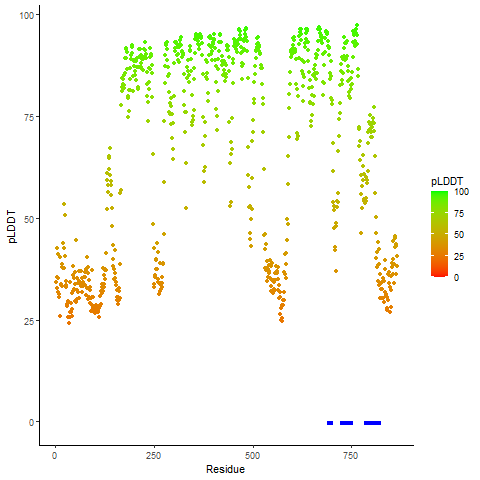
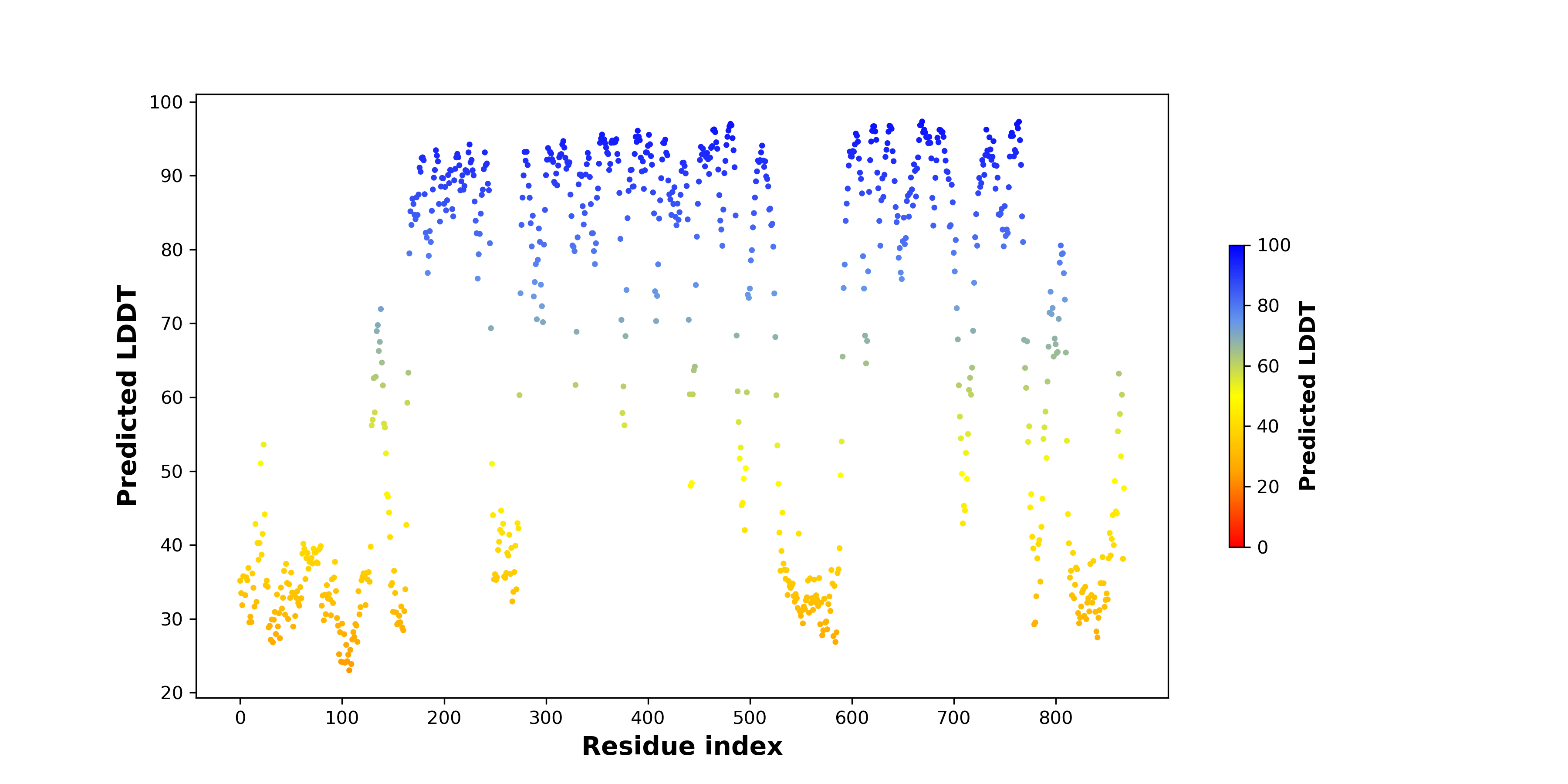
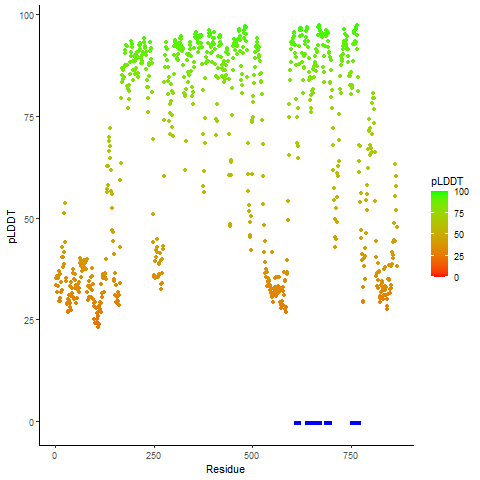
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
FGFR1_pLDDT.png |
ZNF703_pLDDT.png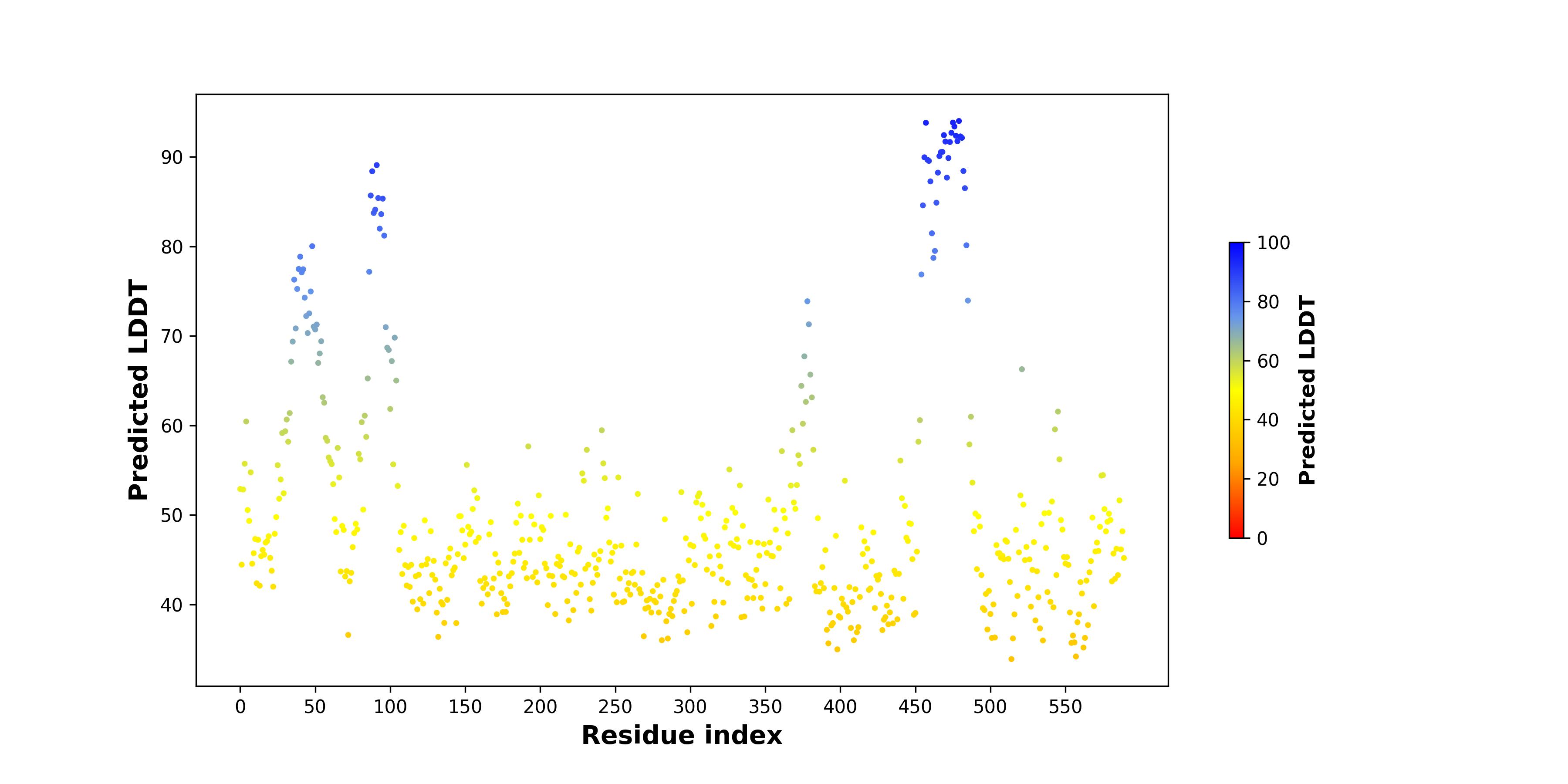 |
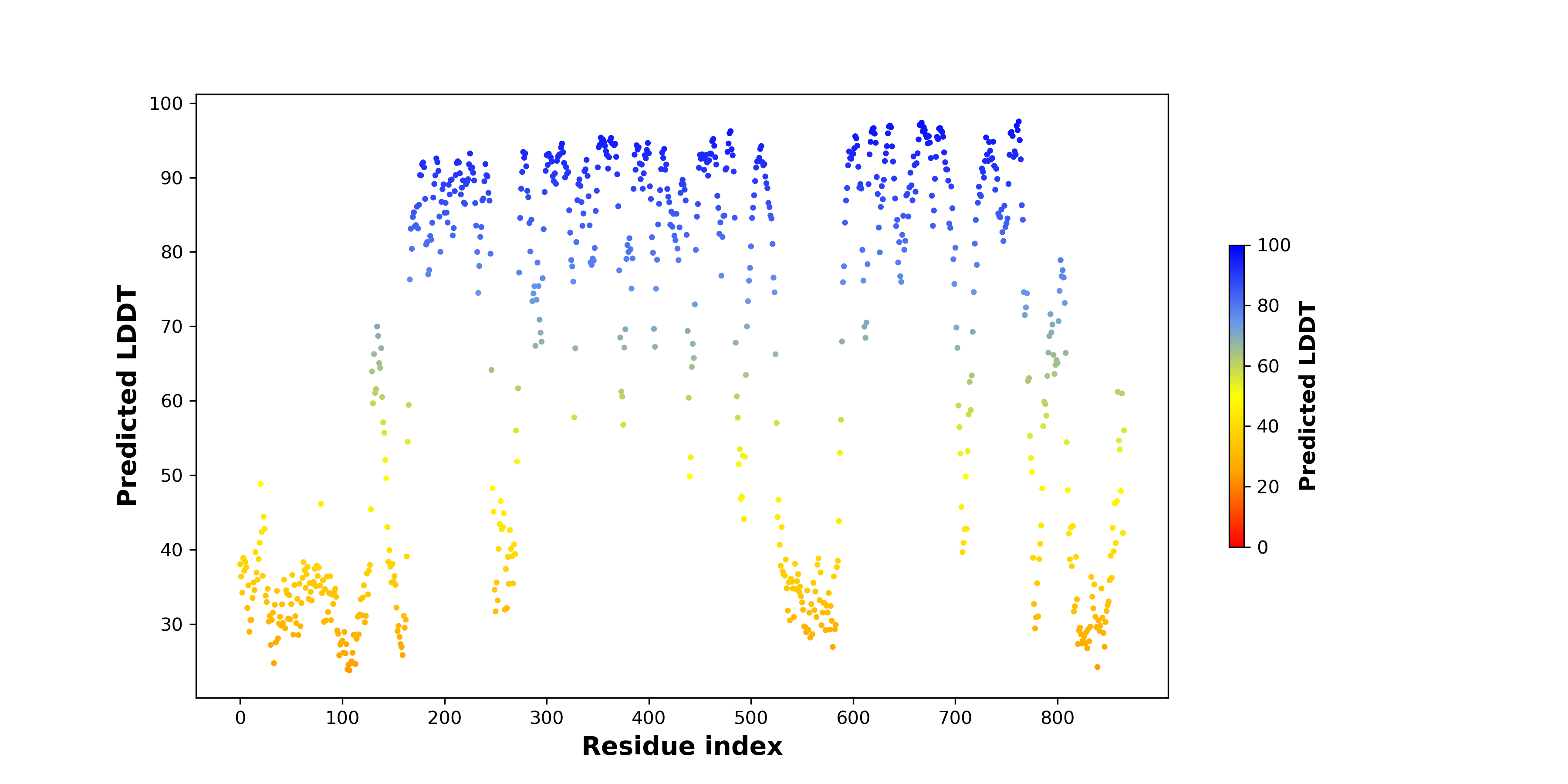
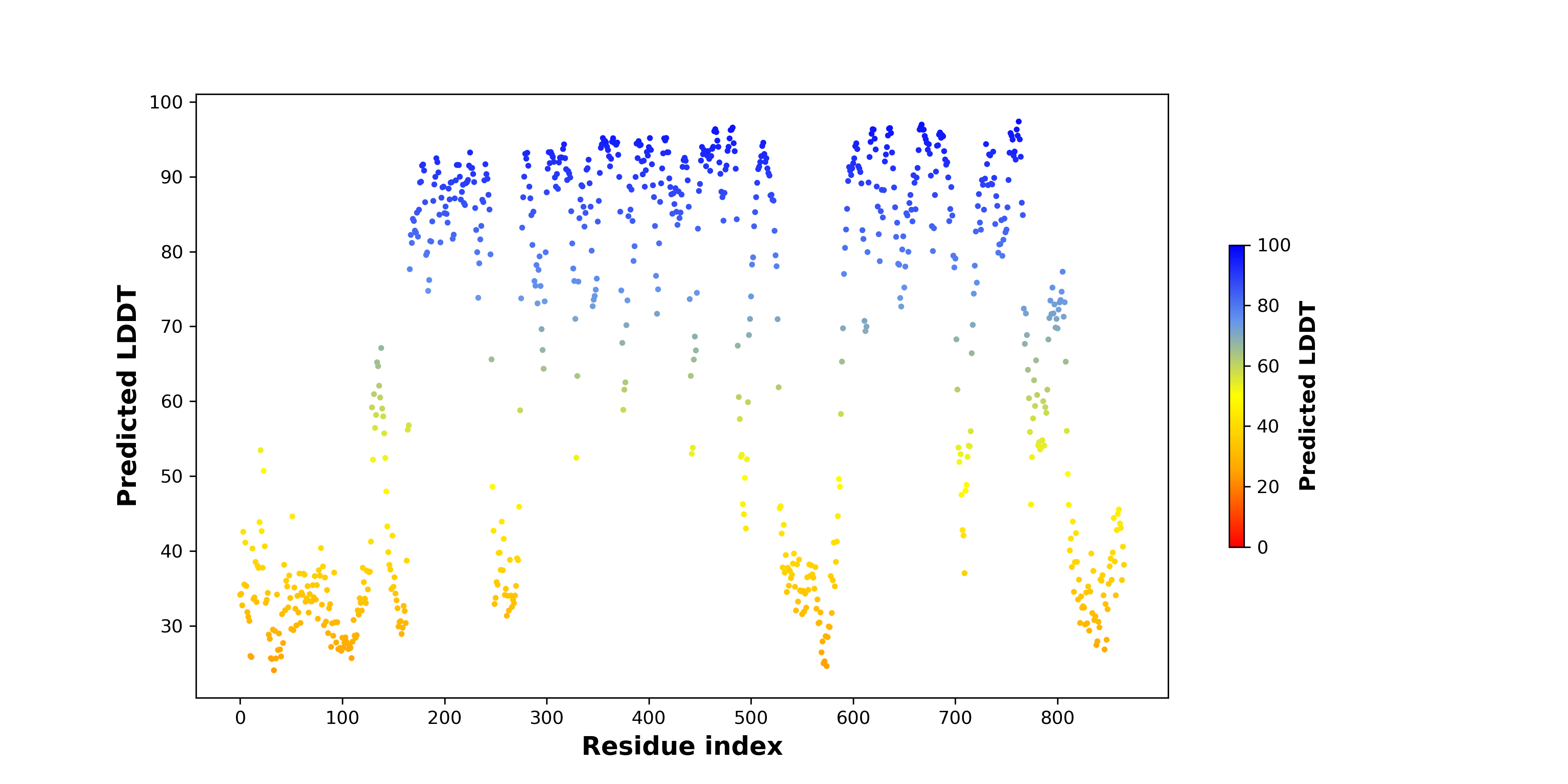
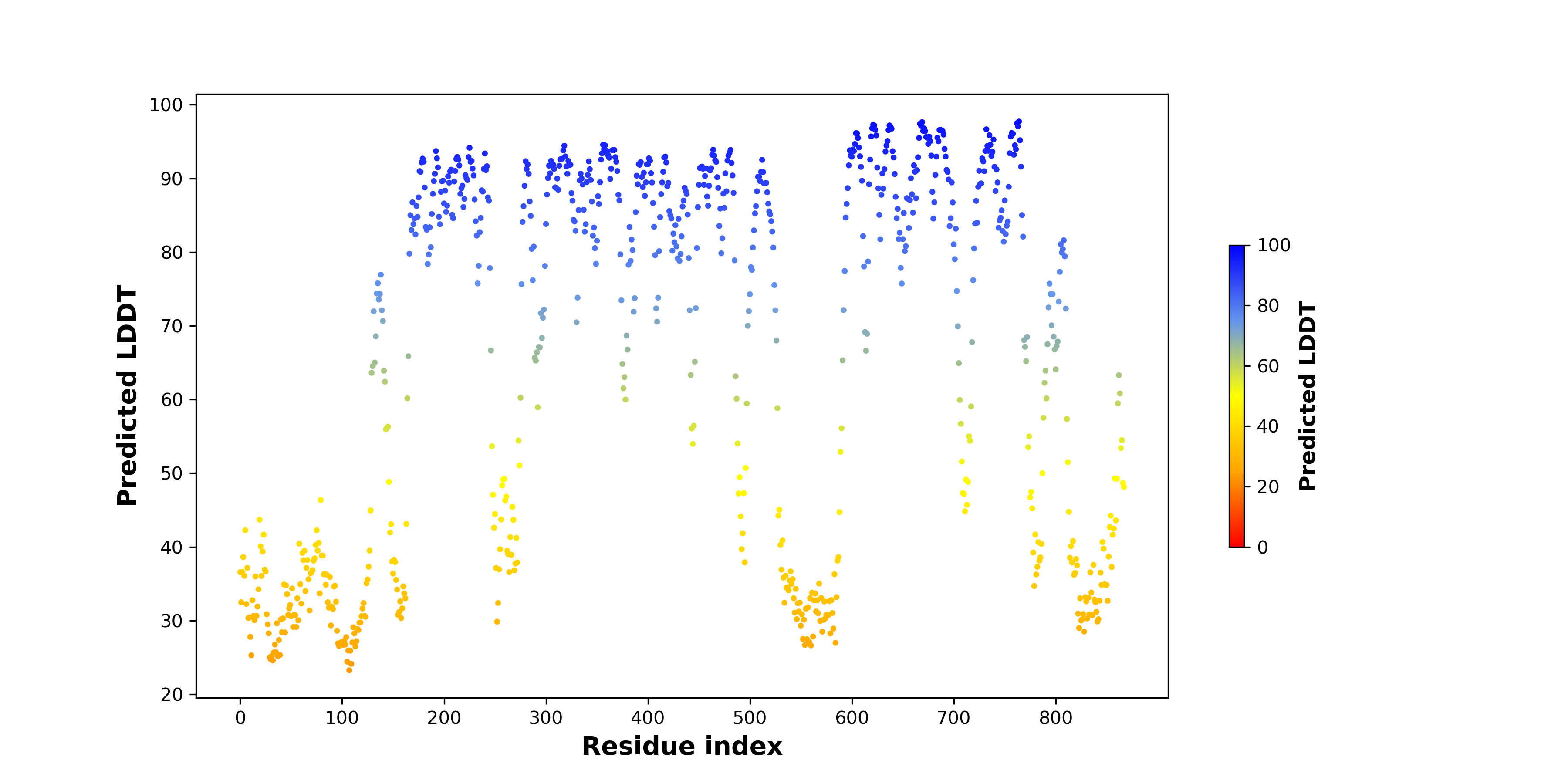
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
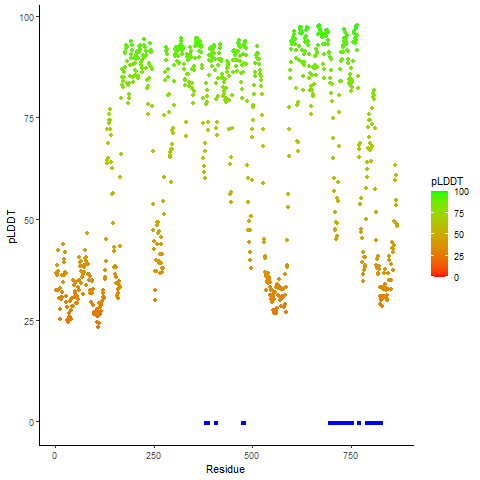
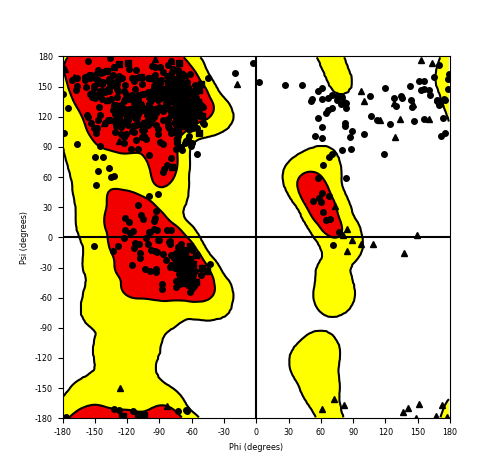
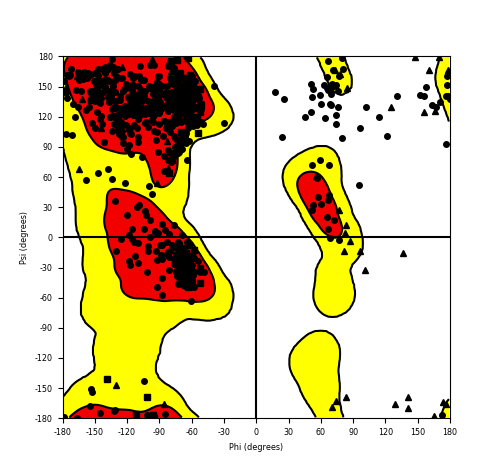
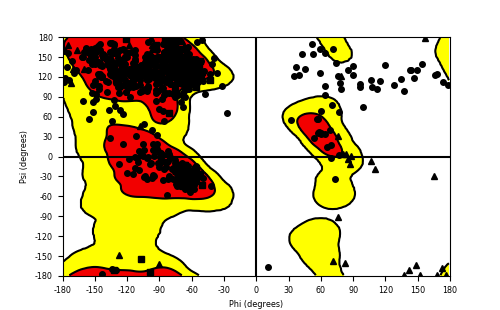
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| FGFR1_ZNF703_635.png |
 |
| FGFR1_ZNF703_637.png |
 |
| FGFR1_ZNF703_652.png |
 |
| FGFR1_ZNF703_688.png |
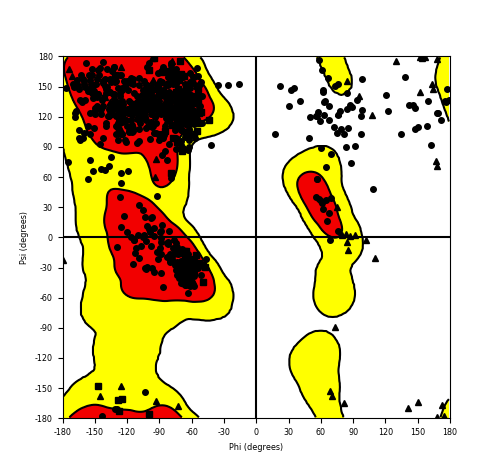 |
| FGFR1_ZNF703_689.png |
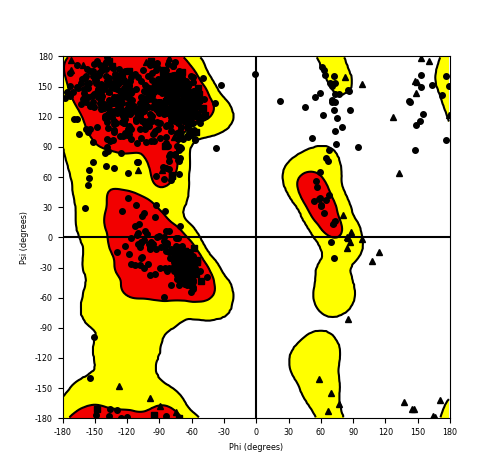 |
| FGFR1_ZNF703_690.png |
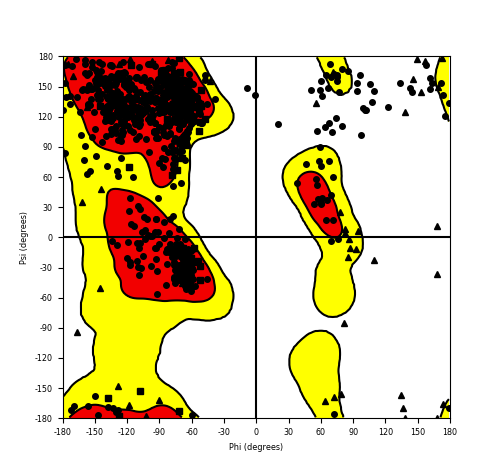 |
| FGFR1_ZNF703_691.png |
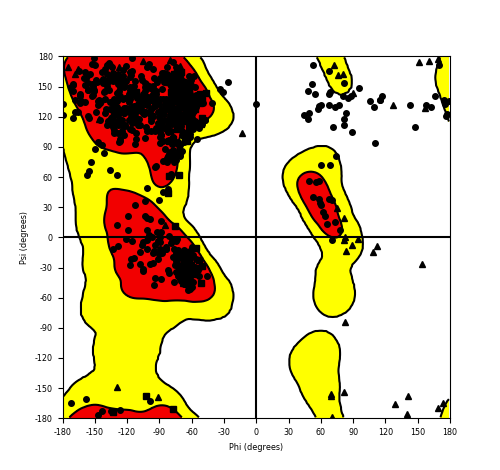 |
Top |
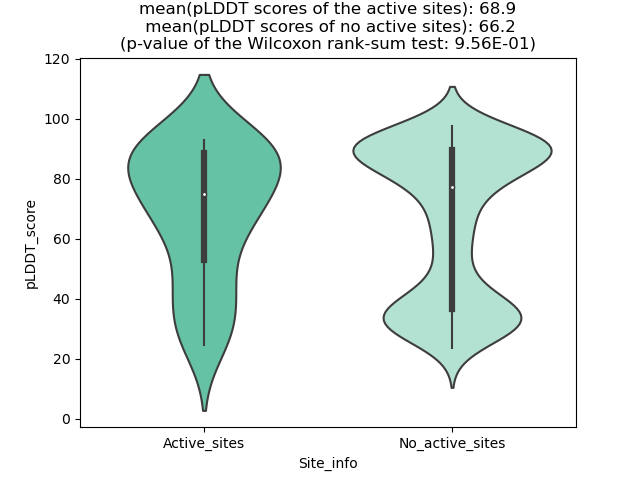
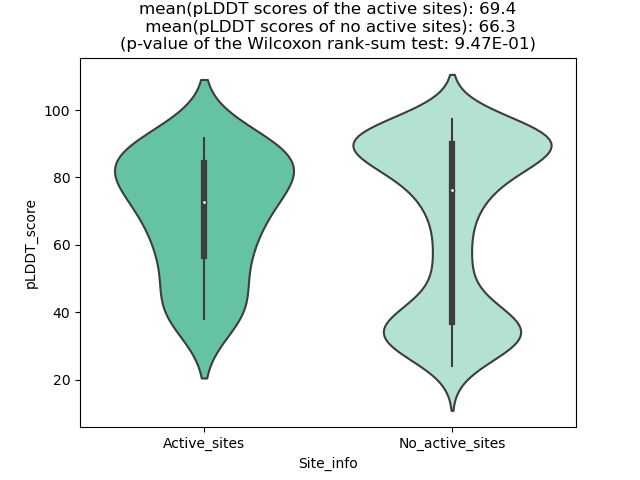
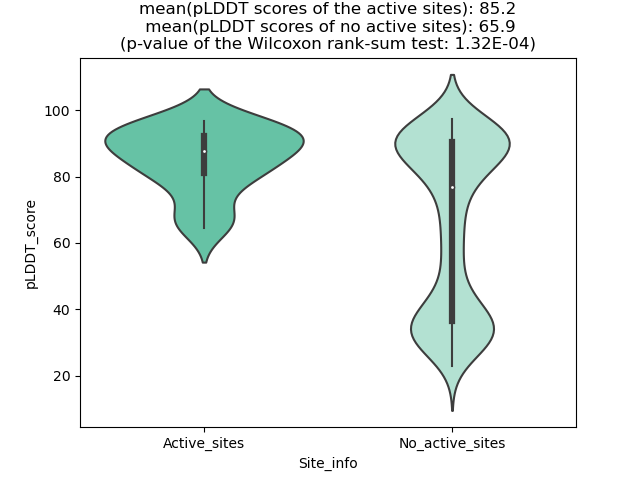
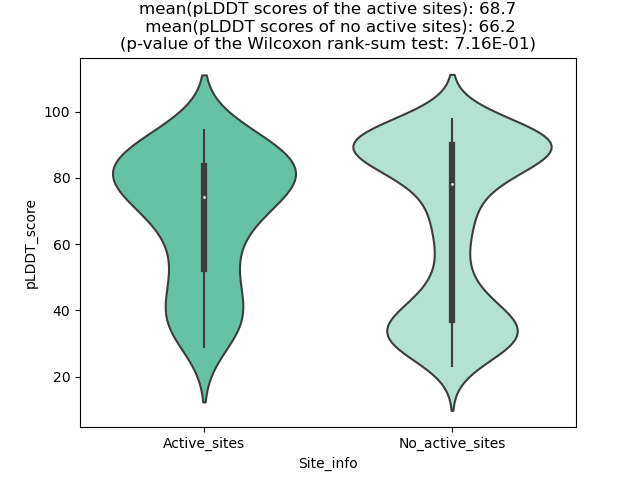
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 540 | 1.07 | 153 | 1.099 | 526.162 | 0.517 | 0.771 | 0.943 | 0.984 | 0.93 | 1.059 | 0.845 | Chain A: 405,406,408,409,410,411,412,413,433,435,4 36,437,444,445,448,449,452,456,466,482,483,484,485 ,488,489,548,549,551,561,562,563,564,565,582 |
| 635 | 1.062 | 171 | 1.09 | 709.667 | 0.557 | 0.763 | 0.974 | 1.119 | 0.946 | 1.182 | 0.685 | Chain A: 239,520,521,523,524,525,526,527,528,548,5 50,551,552,559,560,563,564,566,567,571,581,597,598 ,599,600,601,603,604,607,658,663,664,666,676,677,6 78,679,680,682,690,692,697,698,699,713 |
| 637 | 1.085 | 595 | 1.127 | 1892.674 | 0.496 | 0.766 | 1.022 | 1.34 | 0.836 | 1.602 | 1.117 | Chain A: 129,131,134,135,137,138,139,141,142,145,1 46,209,226,227,228,234,235,236,237,238,240,241,242 ,243,244,245,246,248,256,257,259,607,608,611,612,6 14,615,616,619,621,622,633,635,638,639,641,642,643 ,645,646,649,653,660,661,662,663,664,665,696,698,6 99,700,701,702,703,704,705,706,715,717,718,719,720 ,721,722,723,724,725,727,728,729,730,731,732,733,7 34,735,736,737,738,739 |
| 652 | 1.081 | 150 | 1.11 | 314.188 | 0.466 | 0.785 | 1.021 | 1.199 | 0.923 | 1.299 | 0.871 | Chain A: 229,230,231,232,262,263,264,279,280,281,2 82,283,284,309,310,311,716,717,718,719,720,721,722 ,724,727,732,733,735 |
| 688 | 1.1 | 138 | 1.151 | 435.61 | 0.57 | 0.768 | 0.959 | 1.268 | 0.765 | 1.658 | 0.851 | Chain A: 105,107,694,728,729,730,732,733,736,737,7 40,747,748,749,750,751,754,788,791,792,793,796,802 ,804,806,807,808,809,810,811,812,814,816,817,820,8 21 |
| 689 | 1.145 | 150 | 1.166 | 249.704 | 0.356 | 0.882 | 1.287 | 1.968 | 0.929 | 2.118 | 1.028 | Chain A: 694,698,725,726,728,729,732,733,736,737,7 47,748,749,750,787,788,790,791,792,793,796,802,806 ,807,808,809,810,811,814,815,816,818,819,820 |
| 690 | 1.115 | 151 | 1.169 | 545.37 | 0.548 | 0.782 | 0.903 | 1.299 | 0.736 | 1.766 | 0.8 | Chain A: 611,612,613,614,615,616,617,618,619,639,6 41,642,643,651,654,655,658,662,672,688,689,690,691 ,694,695,698,754,755,757,767,768,769,770,771 |
| 691 | 1.061 | 472 | 1.08 | 973.434 | 0.405 | 0.777 | 1.065 | 1.125 | 1.001 | 1.124 | 0.883 | Chain A: 382,384,385,386,387,388,408,476,477,478,6 96,697,700,701,703,704,705,708,709,710,711,713,716 ,722,727,730,731,734,735,738,739,742,749,750,751,7 52,753,754,771,790,791,792,793,794,795,796,798,799 ,800,804,806,808,809,810,811,812,813,814,815,816,8 17,818,819,820,821,822,823,824,825,826 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| FGFR1 | FDPS, FRS3, NCAM1, FGF1, FGFR1, FRS2, PLCG1, FGF2, SHB, GRB14, BNIP2, KPNB1, CREBBP, STAT3, JAK2, SRC, NEDD4, PPP2R1B, FGF23, PTK6, ITK, NCK1, VAV1, HSP90AA1, ERBB3, CTPS2, CDK9, RPS6KA3, CDC20, TSPAN3, CEACAM21, IL20RB, LGALS8, TGFBR2, GINM1, DLK1, LTBR, TNFRSF10B, EVI2A, MREG, HEPACAM2, GOLGA7B, LDLRAD4, CRK, CTNNB1, CDH1, FGF8, SOS1, PIK3R1, PKM, FGF3, FGF5, FGF6, FGF9, FGF10, FGF17, CD44, RPS6KA1, SLA, PIK3R2, SH3BP2, NCK2, TENC1, RTN1, RTN3, PVRL1, EPHA4, YES1, L1CAM, TNK2, AKT1, PDP1, FGFR2, FAM8A1, TMEM30B, LRRN2, LITAF, C16orf58, ZDHHC11, IFNGR1, GAD1, FCGRT, IL27RA, RAET1E, SLC1A1, C14orf37, MANSC1, TPCN2, CD79B, SCN3B, C19orf45, HTR1A, FLRT1, KAL1, RASA1, CDH2, ZMYM2, FGF4, OPCML, SHC1, S100B, AGER, KLB, CDH11, TGFBR3, PTPN1, IL17RD, FBXO7, CRKL, RPN1, STOML2, DDOST, CANX, LMAN2, TMED9, RCN2, HSPA6, CDC37, HSPA2, ACTG1, ACTA1, PARD3, PGRMC1, PHGDH, ATP5B, VDAC1, VDAC2, SLC25A6, RUVBL2, RPA1, PLCG2, RAP1A, PHB, RAB1A, RAB5A, RAB8A, RAB4A, RAB14, RAB15, ARF4, RAB2B, RAB9A, COPA, SEC22B, TMEM109, STX12, CLPTM1, HSPA1L, DNAJC13, DNAJB1, TUBB4A, TUBA4A, AFAP1, TUBA1B, MTCH1, BAX, IMMT, GNB1, RAB7A, RAB11B, GAB1, PTPN11, RXRA, RARA, Nr4a2, H3F3A, Nr4a1, TH, UBC, SHE, Il17rd, PDGFRB, MFHAS1, PDK1, HEXIM1, LARP7, GNAI1, RIN1, ITGB3, KIAA1429, ERBB2, APEX1, ORF3a, ORF3b, CORO1C, HSP90AB3P, HSP90AB1, BAG2, MRPS7, MRPS31, MRPS14, P4HB, CDK1, HPX, ATP5O, LIMA1, TMOD3, CORO1B, ACTB, PLEC, RPL22L1, PPM1B, TBL2, DIMT1, SPATS2L, FARSB, HSD17B10, NUFIP2, TRMT10C, SND1, OBSCN, AIMP2, RARS, SRP19, POLRMT, MRPS2, MRPS26, BOLA2, ARGLU1, PFKFB3, MRPS6, CFL1, MRPS33, RPL22, KCTD5, HSPD1, EGFR, HULC, LDHA, MKRN2, DDX58, MLC1, IL3RA, IL32, AQP9, KCNE3, UGT1A7, GJA8, CLEC4A, AVPR1B, DEFB135, RNF149, C7orf34, EPB41L4A, CRLF2, DEFB109P1B, TBXA2R, NRSN1, OR1M1, IFNAR1, SYT2, CDH16, FBXO2, TSHR, MFI2, BCAN, CSNK1G2, CDHR4, CTSG, C1QTNF9B, PCDHA8, IZUMO1, CD8B, SFTPC, GREM2, DHFRL1, CD83, KCNC3, ANK1, PTPRN2, TMEM59, PCDHB7, P2RX4, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| FGFR1 |  |
| ZNF703 | 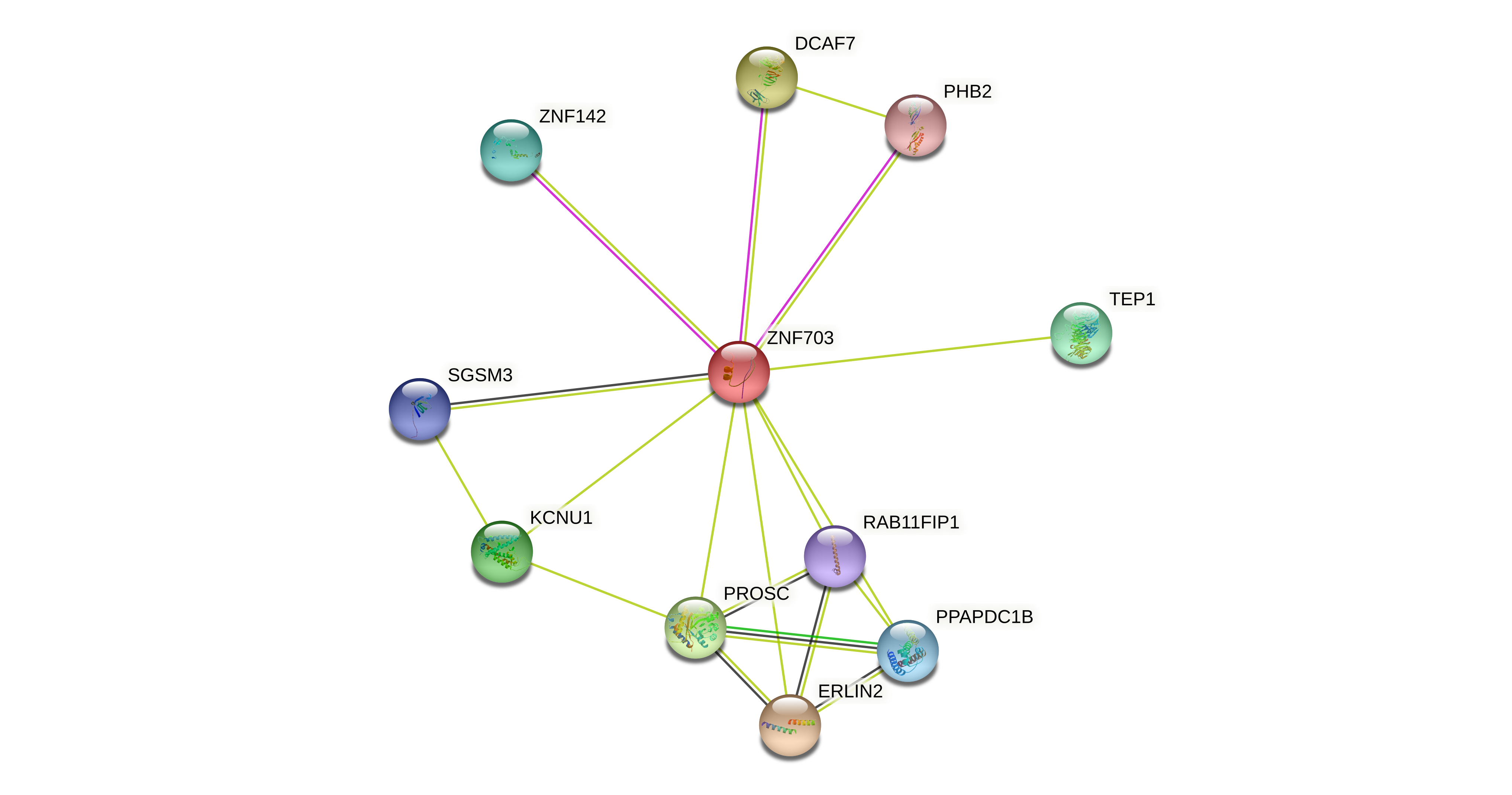 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to FGFR1-ZNF703 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to FGFR1-ZNF703 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | FGFR1 | C1563720 | Kallmann Syndrome 2 (disorder) | 18 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | FGFR1 | C1845146 | Holoprosencephaly, Ectrodactyly, and Bilateral Cleft Lip-Palate | 6 | GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | FGFR1 | C0011570 | Mental Depression | 5 | PSYGENET |
| Hgene | FGFR1 | C0011581 | Depressive disorder | 5 | CTD_human;PSYGENET |
| Hgene | FGFR1 | C0220658 | Pfeiffer Syndrome | 5 | GENOMICS_ENGLAND;UNIPROT |
| Hgene | FGFR1 | C0432283 | Osteoglophonic dwarfism | 5 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | FGFR1 | C0041696 | Unipolar Depression | 4 | CTD_human;PSYGENET |
| Hgene | FGFR1 | C0406612 | Encephalocraniocutaneous lipomatosis | 4 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | FGFR1 | C0005586 | Bipolar Disorder | 3 | PSYGENET |
| Hgene | FGFR1 | C0795998 | JACKSON-WEISS SYNDROME | 3 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | FGFR1 | C1269683 | Major Depressive Disorder | 3 | PSYGENET |
| Hgene | FGFR1 | C0006142 | Malignant neoplasm of breast | 2 | CGI;CTD_human |
| Hgene | FGFR1 | C0007131 | Non-Small Cell Lung Carcinoma | 2 | CTD_human |
| Hgene | FGFR1 | C0007137 | Squamous cell carcinoma | 2 | CTD_human |
| Hgene | FGFR1 | C0027022 | Myeloproliferative disease | 2 | CTD_human |
| Hgene | FGFR1 | C0162809 | Kallmann Syndrome | 2 | CTD_human;ORPHANET |
| Hgene | FGFR1 | C0432122 | Interfrontal craniofaciosynostosis | 2 | GENOMICS_ENGLAND;UNIPROT |
| Hgene | FGFR1 | C0678222 | Breast Carcinoma | 2 | CGI;CTD_human |
| Hgene | FGFR1 | C1257931 | Mammary Neoplasms, Human | 2 | CTD_human |
| Hgene | FGFR1 | C1458155 | Mammary Neoplasms | 2 | CTD_human |
| Hgene | FGFR1 | C4704874 | Mammary Carcinoma, Human | 2 | CTD_human |
| Hgene | FGFR1 | C0004114 | Astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0008924 | Cleft upper lip | 1 | CTD_human |
| Hgene | FGFR1 | C0008925 | Cleft Palate | 1 | CTD_human |
| Hgene | FGFR1 | C0010278 | Craniosynostosis | 1 | CTD_human;GENOMICS_ENGLAND |
| Hgene | FGFR1 | C0011573 | Endogenous depression | 1 | CTD_human |
| Hgene | FGFR1 | C0017638 | Glioma | 1 | CTD_human |
| Hgene | FGFR1 | C0018824 | Heart valve disease | 1 | CTD_human |
| Hgene | FGFR1 | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Hgene | FGFR1 | C0025193 | Melancholia | 1 | CTD_human |
| Hgene | FGFR1 | C0036341 | Schizophrenia | 1 | CTD_human |
| Hgene | FGFR1 | C0085682 | Hypophosphatemia | 1 | GENOMICS_ENGLAND |
| Hgene | FGFR1 | C0086133 | Depressive Syndrome | 1 | CTD_human |
| Hgene | FGFR1 | C0149925 | Small cell carcinoma of lung | 1 | CTD_human |
| Hgene | FGFR1 | C0205768 | Subependymal Giant Cell Astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0206726 | gliosarcoma | 1 | ORPHANET |
| Hgene | FGFR1 | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Hgene | FGFR1 | C0259783 | mixed gliomas | 1 | CTD_human |
| Hgene | FGFR1 | C0265535 | Trigonocephaly | 1 | CTD_human;ORPHANET |
| Hgene | FGFR1 | C0280783 | Juvenile Pilocytic Astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0280785 | Diffuse Astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0282126 | Depression, Neurotic | 1 | CTD_human |
| Hgene | FGFR1 | C0334579 | Anaplastic astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0334580 | Protoplasmic astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0334581 | Gemistocytic astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0334582 | Fibrillary Astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0334583 | Pilocytic Astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0334588 | Giant Cell Glioblastoma | 1 | ORPHANET |
| Hgene | FGFR1 | C0338070 | Childhood Cerebral Astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0338503 | Septo-Optic Dysplasia | 1 | ORPHANET |
| Hgene | FGFR1 | C0342384 | Idiopathic hypogonadotropic hypogonadism | 1 | GENOMICS_ENGLAND |
| Hgene | FGFR1 | C0376634 | Craniofacial Abnormalities | 1 | CTD_human |
| Hgene | FGFR1 | C0431362 | Lobar Holoprosencephaly | 1 | ORPHANET |
| Hgene | FGFR1 | C0547065 | Mixed oligoastrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0555198 | Malignant Glioma | 1 | CTD_human |
| Hgene | FGFR1 | C0750935 | Cerebral Astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0750936 | Intracranial Astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C0751617 | Semilobar Holoprosencephaly | 1 | ORPHANET |
| Hgene | FGFR1 | C1519086 | Pilomyxoid astrocytoma | 1 | ORPHANET |
| Hgene | FGFR1 | C1704230 | Grade I Astrocytoma | 1 | CTD_human |
| Hgene | FGFR1 | C1837218 | Cleft palate, isolated | 1 | CTD_human |
| Hgene | FGFR1 | C1852406 | Cutis Gyrata Syndrome of Beare And Stevenson | 1 | GENOMICS_ENGLAND |
| Hgene | FGFR1 | C2931196 | Craniofacial dysostosis type 1 | 1 | GENOMICS_ENGLAND |
| Hgene | FGFR1 | C3150773 | CHROMOSOME 8p11 MYELOPROLIFERATIVE SYNDROME | 1 | ORPHANET |