| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:GC-DSP |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: GC-DSP | FusionPDB ID: 32749 | FusionGDB2.0 ID: 32749 | Hgene | Tgene | Gene symbol | GC | DSP | Gene ID | 2638 | 1834 |
| Gene name | GC vitamin D binding protein | dentin sialophosphoprotein | |
| Synonyms | DBP|DBP-maf|DBP/GC|GRD3|Gc-MAF|GcMAF|HEL-S-51|VDB|VDBG|VDBP | DFNA39|DGI1|DMP3|DPP|DSP | |
| Cytomap | 4q13.3 | 4q22.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | vitamin D-binding proteinepididymis secretory protein Li 51gc protein-derived macrophage activating factorgc-globulingroup-specific component (vitamin D binding protein)vitamin D-binding alpha-globulinvitamin D-binding protein-macrophage activating | dentin sialophosphoproteindeafness, autosomal dominant 39dentin phosphophoryndentin phosphoproteindentin phosphoryndentin sialoprotein | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | P23434 | P15924 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000273951, ENST00000504199, ENST00000503472, ENST00000513476, | ENST00000379802, ENST00000418664, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 3 X 3 X 3=27 | 22 X 19 X 10=4180 |
| # samples | 3 | 26 | |
| ** MAII score | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(26/4180*10)=-4.00691941393979 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: GC [Title/Abstract] AND DSP [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | GC(72607413)-DSP(7575528), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | GC-DSP seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. GC-DSP seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. GC-DSP seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. GC-DSP seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across GC (5'-gene) Fusion gene breakpoints across GC (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
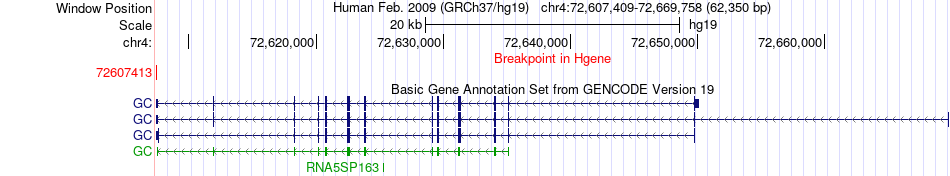 |
 Fusion gene breakpoints across DSP (3'-gene) Fusion gene breakpoints across DSP (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
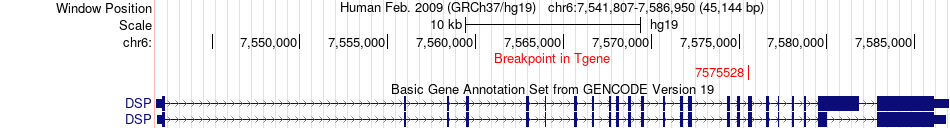 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-DD-A3A8-01A | GC | chr4 | 72607413 | - | DSP | chr6 | 7575528 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000273951 | GC | chr4 | 72607413 | - | ENST00000379802 | DSP | chr6 | 7575528 | + | 8991 | 1972 | 1915 | 8151 | 2078 |
| ENST00000273951 | GC | chr4 | 72607413 | - | ENST00000418664 | DSP | chr6 | 7575528 | + | 7049 | 1972 | 1915 | 6354 | 1479 |
| ENST00000504199 | GC | chr4 | 72607413 | - | ENST00000379802 | DSP | chr6 | 7575528 | + | 8796 | 1777 | 1723 | 7956 | 2077 |
| ENST00000504199 | GC | chr4 | 72607413 | - | ENST00000418664 | DSP | chr6 | 7575528 | + | 6854 | 1777 | 1723 | 6159 | 1478 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000273951 | ENST00000379802 | GC | chr4 | 72607413 | - | DSP | chr6 | 7575528 | + | 0.00769224 | 0.9923078 |
| ENST00000273951 | ENST00000418664 | GC | chr4 | 72607413 | - | DSP | chr6 | 7575528 | + | 0.004043234 | 0.9959567 |
| ENST00000504199 | ENST00000379802 | GC | chr4 | 72607413 | - | DSP | chr6 | 7575528 | + | 0.00876331 | 0.9912367 |
| ENST00000504199 | ENST00000418664 | GC | chr4 | 72607413 | - | DSP | chr6 | 7575528 | + | 0.004509968 | 0.99549 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >32749_32749_1_GC-DSP_GC_chr4_72607413_ENST00000273951_DSP_chr6_7575528_ENST00000379802_length(amino acids)=2078AA_BP=19 MTSMMICYQNKLKYNANHKKIKNDLNLKKSLLATMKTELQKAQQIHSQTSQQYPLYDLDLGKFGEKVTQLTDRWQRIDKQIDFRLWDLEK QIKQLRNYRDNYQAFCKWLYDAKRRQDSLESMKFGDSNTVMRFLNEQKNLHSEISGKRDKSEEVQKIAELCANSIKDYELQLASYTSGLE TLLNIPIKRTMIQSPSGVILQEAADVHARYIELLTRSGDYYRFLSEMLKSLEDLKLKNTKIEVLEEELRLARDANSENCNKNKFLDQNLQ KYQAECSQFKAKLASLEELKRQAELDGKSAKQNLDKCYGQIKELNEKITRLTYEIEDEKRRRKSVEDRFDQQKNDYDQLQKARQCEKENL GWQKLESEKAIKEKEYEIERLRVLLQEEGTRKREYENELAKVRNHYNEEMSNLRNKYETEINITKTTIKEISMQKEDDSKNLRNQLDRLS RENRDLKDEIVRLNDSILQATEQRRRAEENALQQKACGSEIMQKKQHLEIELKQVMQQRSEDNARHKQSLEEAAKTIQDKNKEIERLKAE FQEEAKRRWEYENELSKVRNNYDEEIISLKNQFETEINITKTTIHQLTMQKEEDTSGYRAQIDNLTRENRSLSEEIKRLKNTLTQTTENL RRVEEDIQQQKATGSEVSQRKQQLEVELRQVTQMRTEESVRYKQSLDDAAKTIQDKNKEIERLKQLIDKETNDRKCLEDENARLQRVQYD LQKANSSATETINKLKVQEQELTRLRIDYERVSQERTVKDQDITRFQNSLKELQLQKQKVEEELNRLKRTASEDSCKRKKLEEELEGMRR SLKEQAIKITNLTQQLEQASIVKKRSEDDLRQQRDVLDGHLREKQRTQEELRRLSSEVEALRRQLLQEQESVKQAHLRNEHFQKAIEDKS RSLNESKIEIERLQSLTENLTKEHLMLEEELRNLRLEYDDLRRGRSEADSDKNATILELRSQLQISNNRTLELQGLINDLQRERENLRQE IEKFQKQALEASNRIQESKNQCTQVVQERESLLVKIKVLEQDKARLQRLEDELNRAKSTLEAETRVKQRLECEKQQIQNDLNQWKTQYSR KEEAIRKIESEREKSEREKNSLRSEIERLQAEIKRIEERCRRKLEDSTRETQSQLETERSRYQREIDKLRQRPYGSHRETQTECEWTVDT SKLVFDGLRKKVTAMQLYECQLIDKTTLDKLLKGKKSVEEVASEIQPFLRGAGSIAGASASPKEKYSLVEAKRKKLISPESTVMLLEAQA ATGGIIDPHRNEKLTVDSAIARDLIDFDDRQQIYAAEKAITGFDDPFSGKTVSVSEAIKKNLIDRETGMRLLEAQIASGGVVDPVNSVFL PKDVALARGLIDRDLYRSLNDPRDSQKNFVDPVTKKKVSYVQLKERCRIEPHTGLLLLSVQKRSMSFQGIRQPVTVTELVDSGILRPSTV NELESGQISYDEVGERIKDFLQGSSCIAGIYNETTKQKLGIYEAMKIGLVRPGTALELLEAQAATGFIVDPVSNLRLPVEEAYKRGLVGI EFKEKLLSAERAVTGYNDPETGNIISLFQAMNKELIEKGHGIRLLEAQIATGGIIDPKESHRLPVDIAYKRGYFNEELSEILSDPSDDTK GFFDPNTEENLTYLQLKERCIKDEETGLCLLPLKEKKKQVQTSQKNTLRKRRVVIVDPETNKEMSVQEAYKKGLIDYETFKELCEQECEW EEITITGSDGSTRVVLVDRKTGSQYDIQDAIDKGLVDRKFFDQYRSGSLSLTQFADMISLKNGVGTSSSMGSGVSDDVFSSSRHESVSKI STISSVRNLTIRSSSFSDTLEESSPIAAIFDTENLEKISITEGIERGIVDSITGQRLLEAQACTGGIIHPTTGQKLSLQDAVSQGVIDQD MATRLKPAQKAFIGFEGVKGKKKMSAAEAVKEKWLPYEAGQRFLEFQYLTGGLVDPEVHGRISTEEAIRKGFIDGRAAQRLQDTSSYAKI LTCPKTKLKISYKDAINRSMVEDITGLRLLEAASVSSKGLPSPYNMSSAPGSRSGSRSGSRSGSRSGSRSGSRRGSFDATGNSSYSYSYS -------------------------------------------------------------- >32749_32749_2_GC-DSP_GC_chr4_72607413_ENST00000273951_DSP_chr6_7575528_ENST00000418664_length(amino acids)=1479AA_BP=19 MTSMMICYQNKLKYNANHKKIKNDLNLKKSLLATMKTELQKAQQIHSQTSQQYPLYDLDLGKFGEKVTQLTDRWQRIDKQIDFRLWDLEK QIKQLRNYRDNYQAFCKWLYDAKRRQDSLESMKFGDSNTVMRFLNEQKNLHSEISGKRDKSEEVQKIAELCANSIKDYELQLASYTSGLE TLLNIPIKRTMIQSPSGVILQEAADVHARYIELLTRSGDYYRFLSEMLKSLEDLKLKNTKIEVLEEELRLARDANSENCNKNKFLDQNLQ KYQAECSQFKAKLASLEELKRQAELDGKSAKQNLDKCYGQIKELNEKITRLTYEIEDEKRRRKSVEDRFDQQKNDYDQLQKARQCEKENL GWQKLESEKAIKEKEYEIERLRVLLQEEGTRKREYENELAKASNRIQESKNQCTQVVQERESLLVKIKVLEQDKARLQRLEDELNRAKST LEAETRVKQRLECEKQQIQNDLNQWKTQYSRKEEAIRKIESEREKSEREKNSLRSEIERLQAEIKRIEERCRRKLEDSTRETQSQLETER SRYQREIDKLRQRPYGSHRETQTECEWTVDTSKLVFDGLRKKVTAMQLYECQLIDKTTLDKLLKGKKSVEEVASEIQPFLRGAGSIAGAS ASPKEKYSLVEAKRKKLISPESTVMLLEAQAATGGIIDPHRNEKLTVDSAIARDLIDFDDRQQIYAAEKAITGFDDPFSGKTVSVSEAIK KNLIDRETGMRLLEAQIASGGVVDPVNSVFLPKDVALARGLIDRDLYRSLNDPRDSQKNFVDPVTKKKVSYVQLKERCRIEPHTGLLLLS VQKRSMSFQGIRQPVTVTELVDSGILRPSTVNELESGQISYDEVGERIKDFLQGSSCIAGIYNETTKQKLGIYEAMKIGLVRPGTALELL EAQAATGFIVDPVSNLRLPVEEAYKRGLVGIEFKEKLLSAERAVTGYNDPETGNIISLFQAMNKELIEKGHGIRLLEAQIATGGIIDPKE SHRLPVDIAYKRGYFNEELSEILSDPSDDTKGFFDPNTEENLTYLQLKERCIKDEETGLCLLPLKEKKKQVQTSQKNTLRKRRVVIVDPE TNKEMSVQEAYKKGLIDYETFKELCEQECEWEEITITGSDGSTRVVLVDRKTGSQYDIQDAIDKGLVDRKFFDQYRSGSLSLTQFADMIS LKNGVGTSSSMGSGVSDDVFSSSRHESVSKISTISSVRNLTIRSSSFSDTLEESSPIAAIFDTENLEKISITEGIERGIVDSITGQRLLE AQACTGGIIHPTTGQKLSLQDAVSQGVIDQDMATRLKPAQKAFIGFEGVKGKKKMSAAEAVKEKWLPYEAGQRFLEFQYLTGGLVDPEVH GRISTEEAIRKGFIDGRAAQRLQDTSSYAKILTCPKTKLKISYKDAINRSMVEDITGLRLLEAASVSSKGLPSPYNMSSAPGSRSGSRSG -------------------------------------------------------------- >32749_32749_3_GC-DSP_GC_chr4_72607413_ENST00000504199_DSP_chr6_7575528_ENST00000379802_length(amino acids)=2077AA_BP=18 MTSMMICYQNKLKYNANHKIKNDLNLKKSLLATMKTELQKAQQIHSQTSQQYPLYDLDLGKFGEKVTQLTDRWQRIDKQIDFRLWDLEKQ IKQLRNYRDNYQAFCKWLYDAKRRQDSLESMKFGDSNTVMRFLNEQKNLHSEISGKRDKSEEVQKIAELCANSIKDYELQLASYTSGLET LLNIPIKRTMIQSPSGVILQEAADVHARYIELLTRSGDYYRFLSEMLKSLEDLKLKNTKIEVLEEELRLARDANSENCNKNKFLDQNLQK YQAECSQFKAKLASLEELKRQAELDGKSAKQNLDKCYGQIKELNEKITRLTYEIEDEKRRRKSVEDRFDQQKNDYDQLQKARQCEKENLG WQKLESEKAIKEKEYEIERLRVLLQEEGTRKREYENELAKVRNHYNEEMSNLRNKYETEINITKTTIKEISMQKEDDSKNLRNQLDRLSR ENRDLKDEIVRLNDSILQATEQRRRAEENALQQKACGSEIMQKKQHLEIELKQVMQQRSEDNARHKQSLEEAAKTIQDKNKEIERLKAEF QEEAKRRWEYENELSKVRNNYDEEIISLKNQFETEINITKTTIHQLTMQKEEDTSGYRAQIDNLTRENRSLSEEIKRLKNTLTQTTENLR RVEEDIQQQKATGSEVSQRKQQLEVELRQVTQMRTEESVRYKQSLDDAAKTIQDKNKEIERLKQLIDKETNDRKCLEDENARLQRVQYDL QKANSSATETINKLKVQEQELTRLRIDYERVSQERTVKDQDITRFQNSLKELQLQKQKVEEELNRLKRTASEDSCKRKKLEEELEGMRRS LKEQAIKITNLTQQLEQASIVKKRSEDDLRQQRDVLDGHLREKQRTQEELRRLSSEVEALRRQLLQEQESVKQAHLRNEHFQKAIEDKSR SLNESKIEIERLQSLTENLTKEHLMLEEELRNLRLEYDDLRRGRSEADSDKNATILELRSQLQISNNRTLELQGLINDLQRERENLRQEI EKFQKQALEASNRIQESKNQCTQVVQERESLLVKIKVLEQDKARLQRLEDELNRAKSTLEAETRVKQRLECEKQQIQNDLNQWKTQYSRK EEAIRKIESEREKSEREKNSLRSEIERLQAEIKRIEERCRRKLEDSTRETQSQLETERSRYQREIDKLRQRPYGSHRETQTECEWTVDTS KLVFDGLRKKVTAMQLYECQLIDKTTLDKLLKGKKSVEEVASEIQPFLRGAGSIAGASASPKEKYSLVEAKRKKLISPESTVMLLEAQAA TGGIIDPHRNEKLTVDSAIARDLIDFDDRQQIYAAEKAITGFDDPFSGKTVSVSEAIKKNLIDRETGMRLLEAQIASGGVVDPVNSVFLP KDVALARGLIDRDLYRSLNDPRDSQKNFVDPVTKKKVSYVQLKERCRIEPHTGLLLLSVQKRSMSFQGIRQPVTVTELVDSGILRPSTVN ELESGQISYDEVGERIKDFLQGSSCIAGIYNETTKQKLGIYEAMKIGLVRPGTALELLEAQAATGFIVDPVSNLRLPVEEAYKRGLVGIE FKEKLLSAERAVTGYNDPETGNIISLFQAMNKELIEKGHGIRLLEAQIATGGIIDPKESHRLPVDIAYKRGYFNEELSEILSDPSDDTKG FFDPNTEENLTYLQLKERCIKDEETGLCLLPLKEKKKQVQTSQKNTLRKRRVVIVDPETNKEMSVQEAYKKGLIDYETFKELCEQECEWE EITITGSDGSTRVVLVDRKTGSQYDIQDAIDKGLVDRKFFDQYRSGSLSLTQFADMISLKNGVGTSSSMGSGVSDDVFSSSRHESVSKIS TISSVRNLTIRSSSFSDTLEESSPIAAIFDTENLEKISITEGIERGIVDSITGQRLLEAQACTGGIIHPTTGQKLSLQDAVSQGVIDQDM ATRLKPAQKAFIGFEGVKGKKKMSAAEAVKEKWLPYEAGQRFLEFQYLTGGLVDPEVHGRISTEEAIRKGFIDGRAAQRLQDTSSYAKIL TCPKTKLKISYKDAINRSMVEDITGLRLLEAASVSSKGLPSPYNMSSAPGSRSGSRSGSRSGSRSGSRSGSRRGSFDATGNSSYSYSYSF -------------------------------------------------------------- >32749_32749_4_GC-DSP_GC_chr4_72607413_ENST00000504199_DSP_chr6_7575528_ENST00000418664_length(amino acids)=1478AA_BP=18 MTSMMICYQNKLKYNANHKIKNDLNLKKSLLATMKTELQKAQQIHSQTSQQYPLYDLDLGKFGEKVTQLTDRWQRIDKQIDFRLWDLEKQ IKQLRNYRDNYQAFCKWLYDAKRRQDSLESMKFGDSNTVMRFLNEQKNLHSEISGKRDKSEEVQKIAELCANSIKDYELQLASYTSGLET LLNIPIKRTMIQSPSGVILQEAADVHARYIELLTRSGDYYRFLSEMLKSLEDLKLKNTKIEVLEEELRLARDANSENCNKNKFLDQNLQK YQAECSQFKAKLASLEELKRQAELDGKSAKQNLDKCYGQIKELNEKITRLTYEIEDEKRRRKSVEDRFDQQKNDYDQLQKARQCEKENLG WQKLESEKAIKEKEYEIERLRVLLQEEGTRKREYENELAKASNRIQESKNQCTQVVQERESLLVKIKVLEQDKARLQRLEDELNRAKSTL EAETRVKQRLECEKQQIQNDLNQWKTQYSRKEEAIRKIESEREKSEREKNSLRSEIERLQAEIKRIEERCRRKLEDSTRETQSQLETERS RYQREIDKLRQRPYGSHRETQTECEWTVDTSKLVFDGLRKKVTAMQLYECQLIDKTTLDKLLKGKKSVEEVASEIQPFLRGAGSIAGASA SPKEKYSLVEAKRKKLISPESTVMLLEAQAATGGIIDPHRNEKLTVDSAIARDLIDFDDRQQIYAAEKAITGFDDPFSGKTVSVSEAIKK NLIDRETGMRLLEAQIASGGVVDPVNSVFLPKDVALARGLIDRDLYRSLNDPRDSQKNFVDPVTKKKVSYVQLKERCRIEPHTGLLLLSV QKRSMSFQGIRQPVTVTELVDSGILRPSTVNELESGQISYDEVGERIKDFLQGSSCIAGIYNETTKQKLGIYEAMKIGLVRPGTALELLE AQAATGFIVDPVSNLRLPVEEAYKRGLVGIEFKEKLLSAERAVTGYNDPETGNIISLFQAMNKELIEKGHGIRLLEAQIATGGIIDPKES HRLPVDIAYKRGYFNEELSEILSDPSDDTKGFFDPNTEENLTYLQLKERCIKDEETGLCLLPLKEKKKQVQTSQKNTLRKRRVVIVDPET NKEMSVQEAYKKGLIDYETFKELCEQECEWEEITITGSDGSTRVVLVDRKTGSQYDIQDAIDKGLVDRKFFDQYRSGSLSLTQFADMISL KNGVGTSSSMGSGVSDDVFSSSRHESVSKISTISSVRNLTIRSSSFSDTLEESSPIAAIFDTENLEKISITEGIERGIVDSITGQRLLEA QACTGGIIHPTTGQKLSLQDAVSQGVIDQDMATRLKPAQKAFIGFEGVKGKKKMSAAEAVKEKWLPYEAGQRFLEFQYLTGGLVDPEVHG RISTEEAIRKGFIDGRAAQRLQDTSSYAKILTCPKTKLKISYKDAINRSMVEDITGLRLLEAASVSSKGLPSPYNMSSAPGSRSGSRSGS -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:72607413/chr6:7575528) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| GC | DSP |
| FUNCTION: The glycine cleavage system catalyzes the degradation of glycine. The H protein (GCSH) shuttles the methylamine group of glycine from the P protein (GLDC) to the T protein (GCST). {ECO:0000269|PubMed:1671321}. | FUNCTION: Major high molecular weight protein of desmosomes. Involved in the organization of the desmosomal cadherin-plakoglobin complexes into discrete plasma membrane domains and in the anchoring of intermediate filaments to the desmosomes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | GC | chr4:72607413 | chr6:7575528 | ENST00000273951 | - | 13 | 13 | 17_208 | 542.6666666666666 | 395.3333333333333 | Domain | Albumin 1 |
| Hgene | GC | chr4:72607413 | chr6:7575528 | ENST00000273951 | - | 13 | 13 | 209_394 | 542.6666666666666 | 395.3333333333333 | Domain | Albumin 2 |
| Hgene | GC | chr4:72607413 | chr6:7575528 | ENST00000273951 | - | 13 | 13 | 395_474 | 542.6666666666666 | 395.3333333333333 | Domain | Albumin 3 |
| Hgene | GC | chr4:72607413 | chr6:7575528 | ENST00000504199 | - | 14 | 14 | 17_208 | 560.6666666666666 | 500.6666666666667 | Domain | Albumin 1 |
| Hgene | GC | chr4:72607413 | chr6:7575528 | ENST00000504199 | - | 14 | 14 | 209_394 | 560.6666666666666 | 500.6666666666667 | Domain | Albumin 2 |
| Hgene | GC | chr4:72607413 | chr6:7575528 | ENST00000504199 | - | 14 | 14 | 395_474 | 560.6666666666666 | 500.6666666666667 | Domain | Albumin 3 |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 1018_1945 | 812.0 | 2872.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 1018_1945 | 812.0 | 2273.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 1057_1945 | 812.0 | 2872.0 | Region | Note=Central fibrous rod domain | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 1946_2871 | 812.0 | 2872.0 | Region | Note=Globular 2 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 1960_2208 | 812.0 | 2872.0 | Region | Note=4.5 X 38 AA tandem repeats (Domain A) | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2244_2446 | 812.0 | 2872.0 | Region | Note=4.5 X 38 AA tandem repeats (Domain B) | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2609_2822 | 812.0 | 2872.0 | Region | Note=4.5 X 38 AA tandem repeats (Domain C) | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2824_2847 | 812.0 | 2872.0 | Region | Note=6 X 4 AA tandem repeats of G-S-R-[SR] | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 1057_1945 | 812.0 | 2273.0 | Region | Note=Central fibrous rod domain | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 1946_2871 | 812.0 | 2273.0 | Region | Note=Globular 2 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 1960_2208 | 812.0 | 2273.0 | Region | Note=4.5 X 38 AA tandem repeats (Domain A) | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2244_2446 | 812.0 | 2273.0 | Region | Note=4.5 X 38 AA tandem repeats (Domain B) | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2609_2822 | 812.0 | 2273.0 | Region | Note=4.5 X 38 AA tandem repeats (Domain C) | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2824_2847 | 812.0 | 2273.0 | Region | Note=6 X 4 AA tandem repeats of G-S-R-[SR] | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2009_2045 | 812.0 | 2872.0 | Repeat | Note=Plectin 1 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2046_2083 | 812.0 | 2872.0 | Repeat | Note=Plectin 2 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2084_2121 | 812.0 | 2872.0 | Repeat | Note=Plectin 3 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2122_2159 | 812.0 | 2872.0 | Repeat | Note=Plectin 4 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2163_2197 | 812.0 | 2872.0 | Repeat | Note=Plectin 5 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2198_2233 | 812.0 | 2872.0 | Repeat | Note=Plectin 6 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2251_2288 | 812.0 | 2872.0 | Repeat | Note=Plectin 7 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2289_2326 | 812.0 | 2872.0 | Repeat | Note=Plectin 8 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2327_2364 | 812.0 | 2872.0 | Repeat | Note=Plectin 9 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2365_2402 | 812.0 | 2872.0 | Repeat | Note=Plectin 10 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2406_2440 | 812.0 | 2872.0 | Repeat | Note=Plectin 11 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2456_2493 | 812.0 | 2872.0 | Repeat | Note=Plectin 12 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2507_2544 | 812.0 | 2872.0 | Repeat | Note=Plectin 13 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2610_2647 | 812.0 | 2872.0 | Repeat | Note=Plectin 14 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2648_2685 | 812.0 | 2872.0 | Repeat | Note=Plectin 15 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2724_2761 | 812.0 | 2872.0 | Repeat | Note=Plectin 16 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 2762_2799 | 812.0 | 2872.0 | Repeat | Note=Plectin 17 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2009_2045 | 812.0 | 2273.0 | Repeat | Note=Plectin 1 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2046_2083 | 812.0 | 2273.0 | Repeat | Note=Plectin 2 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2084_2121 | 812.0 | 2273.0 | Repeat | Note=Plectin 3 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2122_2159 | 812.0 | 2273.0 | Repeat | Note=Plectin 4 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2163_2197 | 812.0 | 2273.0 | Repeat | Note=Plectin 5 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2198_2233 | 812.0 | 2273.0 | Repeat | Note=Plectin 6 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2251_2288 | 812.0 | 2273.0 | Repeat | Note=Plectin 7 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2289_2326 | 812.0 | 2273.0 | Repeat | Note=Plectin 8 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2327_2364 | 812.0 | 2273.0 | Repeat | Note=Plectin 9 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2365_2402 | 812.0 | 2273.0 | Repeat | Note=Plectin 10 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2406_2440 | 812.0 | 2273.0 | Repeat | Note=Plectin 11 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2456_2493 | 812.0 | 2273.0 | Repeat | Note=Plectin 12 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2507_2544 | 812.0 | 2273.0 | Repeat | Note=Plectin 13 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2610_2647 | 812.0 | 2273.0 | Repeat | Note=Plectin 14 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2648_2685 | 812.0 | 2273.0 | Repeat | Note=Plectin 15 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2724_2761 | 812.0 | 2273.0 | Repeat | Note=Plectin 16 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 2762_2799 | 812.0 | 2273.0 | Repeat | Note=Plectin 17 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 458_515 | 812.0 | 2872.0 | Domain | SH3 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 458_515 | 812.0 | 2273.0 | Domain | SH3 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 1_1056 | 812.0 | 2872.0 | Region | Note=Globular 1 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 1_1056 | 812.0 | 2273.0 | Region | Note=Globular 1 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 178_271 | 812.0 | 2872.0 | Repeat | Note=Spectrin 1 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 272_375 | 812.0 | 2872.0 | Repeat | Note=Spectrin 2 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 376_446 | 812.0 | 2872.0 | Repeat | Note=Spectrin 3a | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 516_545 | 812.0 | 2872.0 | Repeat | Note=Spectrin 3b | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 546_627 | 812.0 | 2872.0 | Repeat | Note=Spectrin 4 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 654_769 | 812.0 | 2872.0 | Repeat | Note=Spectrin 5 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 770_883 | 812.0 | 2872.0 | Repeat | Note=Spectrin 6 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 178_271 | 812.0 | 2273.0 | Repeat | Note=Spectrin 1 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 272_375 | 812.0 | 2273.0 | Repeat | Note=Spectrin 2 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 376_446 | 812.0 | 2273.0 | Repeat | Note=Spectrin 3a | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 516_545 | 812.0 | 2273.0 | Repeat | Note=Spectrin 3b | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 546_627 | 812.0 | 2273.0 | Repeat | Note=Spectrin 4 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 654_769 | 812.0 | 2273.0 | Repeat | Note=Spectrin 5 | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 770_883 | 812.0 | 2273.0 | Repeat | Note=Spectrin 6 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| GC | |
| DSP |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000379802 | 16 | 24 | 1_584 | 812.0 | 2872.0 | plakophilin 1 and junction plakoglobin | |
| Tgene | DSP | chr4:72607413 | chr6:7575528 | ENST00000418664 | 16 | 24 | 1_584 | 812.0 | 2273.0 | plakophilin 1 and junction plakoglobin |
Top |
Related Drugs to GC-DSP |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to GC-DSP |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

