| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:HACL1-RAF1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: HACL1-RAF1 | FusionPDB ID: 35563 | FusionGDB2.0 ID: 35563 | Hgene | Tgene | Gene symbol | HACL1 | RAF1 | Gene ID | 337880 | 6037 |
| Gene name | keratin associated protein 11-1 | ribonuclease A family member 3 | |
| Synonyms | HACL-1|HACL1|KAP11.1 | ECP|RAF1|RNS3 | |
| Cytomap | 21q22.11 | 14q11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | keratin-associated protein 11-1high sulfur keratin-associated protein 11.1 | eosinophil cationic proteinRNase 3cytotoxic ribonucleaseribonuclease 3ribonuclease, RNase A family, 3 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9UJ83 | P04049 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000321169, ENST00000435217, ENST00000451445, ENST00000456194, ENST00000457447, | ENST00000251849, ENST00000442415, ENST00000534997, ENST00000542177, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 7 X 6 X 4=168 | 19 X 16 X 7=2128 |
| # samples | 7 | 18 | |
| ** MAII score | log2(7/168*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(18/2128*10)=-3.56342933917152 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: HACL1 [Title/Abstract] AND RAF1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | RAF1 | GO:0002227 | innate immune response in mucosa | 12860195 |
| Tgene | RAF1 | GO:0019731 | antibacterial humoral response | 12860195 |
| Tgene | RAF1 | GO:0043152 | induction of bacterial agglutination | 23992292 |
| Tgene | RAF1 | GO:0045087 | innate immune response | 23992292 |
| Tgene | RAF1 | GO:0050829 | defense response to Gram-negative bacterium | 23992292 |
| Tgene | RAF1 | GO:0050830 | defense response to Gram-positive bacterium | 12860195|23992292 |
| Tgene | RAF1 | GO:0061844 | antimicrobial humoral immune response mediated by antimicrobial peptide | 12860195 |
 Fusion gene breakpoints across HACL1 (5'-gene) Fusion gene breakpoints across HACL1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across RAF1 (3'-gene) Fusion gene breakpoints across RAF1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | HACL1 | chr3 | 15604864 | - | RAF1 | chr3 | 12641914 | - |
| ChimerKB3 | . | . | HACL1 | chr3 | 15604864 | - | RAF1 | chr3 | 46425057 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000321169 | HACL1 | chr3 | 15604864 | - | ENST00000251849 | RAF1 | chr3 | 12641914 | - | 4098 | 2072 | 368 | 3184 | 938 |
| ENST00000321169 | HACL1 | chr3 | 15604864 | - | ENST00000442415 | RAF1 | chr3 | 12641914 | - | 3901 | 2072 | 368 | 3184 | 938 |
| ENST00000321169 | HACL1 | chr3 | 15604864 | - | ENST00000534997 | RAF1 | chr3 | 12641914 | - | 3432 | 2072 | 368 | 3184 | 938 |
| ENST00000321169 | HACL1 | chr3 | 15604864 | - | ENST00000542177 | RAF1 | chr3 | 12641914 | - | 3366 | 2072 | 368 | 3184 | 938 |
| ENST00000435217 | HACL1 | chr3 | 15604864 | - | ENST00000251849 | RAF1 | chr3 | 12641914 | - | 3113 | 1087 | 106 | 2199 | 697 |
| ENST00000435217 | HACL1 | chr3 | 15604864 | - | ENST00000442415 | RAF1 | chr3 | 12641914 | - | 2916 | 1087 | 106 | 2199 | 697 |
| ENST00000435217 | HACL1 | chr3 | 15604864 | - | ENST00000534997 | RAF1 | chr3 | 12641914 | - | 2447 | 1087 | 106 | 2199 | 697 |
| ENST00000435217 | HACL1 | chr3 | 15604864 | - | ENST00000542177 | RAF1 | chr3 | 12641914 | - | 2381 | 1087 | 106 | 2199 | 697 |
| ENST00000451445 | HACL1 | chr3 | 15604864 | - | ENST00000251849 | RAF1 | chr3 | 12641914 | - | 3608 | 1582 | 124 | 2694 | 856 |
| ENST00000451445 | HACL1 | chr3 | 15604864 | - | ENST00000442415 | RAF1 | chr3 | 12641914 | - | 3411 | 1582 | 124 | 2694 | 856 |
| ENST00000451445 | HACL1 | chr3 | 15604864 | - | ENST00000534997 | RAF1 | chr3 | 12641914 | - | 2942 | 1582 | 124 | 2694 | 856 |
| ENST00000451445 | HACL1 | chr3 | 15604864 | - | ENST00000542177 | RAF1 | chr3 | 12641914 | - | 2876 | 1582 | 124 | 2694 | 856 |
| ENST00000456194 | HACL1 | chr3 | 15604864 | - | ENST00000251849 | RAF1 | chr3 | 12641914 | - | 3755 | 1729 | 106 | 2841 | 911 |
| ENST00000456194 | HACL1 | chr3 | 15604864 | - | ENST00000442415 | RAF1 | chr3 | 12641914 | - | 3558 | 1729 | 106 | 2841 | 911 |
| ENST00000456194 | HACL1 | chr3 | 15604864 | - | ENST00000534997 | RAF1 | chr3 | 12641914 | - | 3089 | 1729 | 106 | 2841 | 911 |
| ENST00000456194 | HACL1 | chr3 | 15604864 | - | ENST00000542177 | RAF1 | chr3 | 12641914 | - | 3023 | 1729 | 106 | 2841 | 911 |
| ENST00000457447 | HACL1 | chr3 | 15604864 | - | ENST00000251849 | RAF1 | chr3 | 12641914 | - | 3656 | 1630 | 106 | 2742 | 878 |
| ENST00000457447 | HACL1 | chr3 | 15604864 | - | ENST00000442415 | RAF1 | chr3 | 12641914 | - | 3459 | 1630 | 106 | 2742 | 878 |
| ENST00000457447 | HACL1 | chr3 | 15604864 | - | ENST00000534997 | RAF1 | chr3 | 12641914 | - | 2990 | 1630 | 106 | 2742 | 878 |
| ENST00000457447 | HACL1 | chr3 | 15604864 | - | ENST00000542177 | RAF1 | chr3 | 12641914 | - | 2924 | 1630 | 106 | 2742 | 878 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >35563_35563_1_HACL1-RAF1_HACL1_chr3_15604864_ENST00000321169_RAF1_chr3_12641914_ENST00000251849_length(amino acids)=938AA_BP=567 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWPLLVIGGSSERNQETMGAFQEFPQVEACRLYTKFSARPSSIEAIPFVIEKAVRSSIYGRPGACYVDIPADFVNLQVN VNSIKYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRAL QFADVIVLFGARLNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKTLREKMKSN EAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKDRSPGQWII CVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQK SLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSY YWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLY KHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQ DNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQ -------------------------------------------------------------- >35563_35563_2_HACL1-RAF1_HACL1_chr3_15604864_ENST00000321169_RAF1_chr3_12641914_ENST00000442415_length(amino acids)=938AA_BP=567 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWPLLVIGGSSERNQETMGAFQEFPQVEACRLYTKFSARPSSIEAIPFVIEKAVRSSIYGRPGACYVDIPADFVNLQVN VNSIKYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRAL QFADVIVLFGARLNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKTLREKMKSN EAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKDRSPGQWII CVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQK SLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSY YWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLY KHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQ DNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQ -------------------------------------------------------------- >35563_35563_3_HACL1-RAF1_HACL1_chr3_15604864_ENST00000321169_RAF1_chr3_12641914_ENST00000534997_length(amino acids)=938AA_BP=567 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWPLLVIGGSSERNQETMGAFQEFPQVEACRLYTKFSARPSSIEAIPFVIEKAVRSSIYGRPGACYVDIPADFVNLQVN VNSIKYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRAL QFADVIVLFGARLNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKTLREKMKSN EAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKDRSPGQWII CVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQK SLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSY YWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLY KHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQ DNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQ -------------------------------------------------------------- >35563_35563_4_HACL1-RAF1_HACL1_chr3_15604864_ENST00000321169_RAF1_chr3_12641914_ENST00000542177_length(amino acids)=938AA_BP=567 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWPLLVIGGSSERNQETMGAFQEFPQVEACRLYTKFSARPSSIEAIPFVIEKAVRSSIYGRPGACYVDIPADFVNLQVN VNSIKYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRAL QFADVIVLFGARLNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKTLREKMKSN EAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKDRSPGQWII CVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQK SLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSY YWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLY KHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQ DNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQ -------------------------------------------------------------- >35563_35563_5_HACL1-RAF1_HACL1_chr3_15604864_ENST00000435217_RAF1_chr3_12641914_ENST00000251849_length(amino acids)=697AA_BP=326 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAVDICAEELGNNVKPAVTLLGNIHAVTKQ LLEELDKTPWQYPPESKWWKTLREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAG TFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMC LLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTP VPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTR HVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLAT VKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAM -------------------------------------------------------------- >35563_35563_6_HACL1-RAF1_HACL1_chr3_15604864_ENST00000435217_RAF1_chr3_12641914_ENST00000442415_length(amino acids)=697AA_BP=326 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAVDICAEELGNNVKPAVTLLGNIHAVTKQ LLEELDKTPWQYPPESKWWKTLREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAG TFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMC LLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTP VPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTR HVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLAT VKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAM -------------------------------------------------------------- >35563_35563_7_HACL1-RAF1_HACL1_chr3_15604864_ENST00000435217_RAF1_chr3_12641914_ENST00000534997_length(amino acids)=697AA_BP=326 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAVDICAEELGNNVKPAVTLLGNIHAVTKQ LLEELDKTPWQYPPESKWWKTLREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAG TFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMC LLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTP VPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTR HVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLAT VKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAM -------------------------------------------------------------- >35563_35563_8_HACL1-RAF1_HACL1_chr3_15604864_ENST00000435217_RAF1_chr3_12641914_ENST00000542177_length(amino acids)=697AA_BP=326 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAVDICAEELGNNVKPAVTLLGNIHAVTKQ LLEELDKTPWQYPPESKWWKTLREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAG TFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMC LLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTP VPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTR HVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLAT VKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAM -------------------------------------------------------------- >35563_35563_9_HACL1-RAF1_HACL1_chr3_15604864_ENST00000451445_RAF1_chr3_12641914_ENST00000251849_length(amino acids)=856AA_BP=485 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCV GAARSRALQFADVIVLFGARLNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKT LREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKD RSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFV QTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRP RGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQ WCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWM APEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQI -------------------------------------------------------------- >35563_35563_10_HACL1-RAF1_HACL1_chr3_15604864_ENST00000451445_RAF1_chr3_12641914_ENST00000442415_length(amino acids)=856AA_BP=485 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCV GAARSRALQFADVIVLFGARLNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKT LREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKD RSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFV QTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRP RGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQ WCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWM APEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQI -------------------------------------------------------------- >35563_35563_11_HACL1-RAF1_HACL1_chr3_15604864_ENST00000451445_RAF1_chr3_12641914_ENST00000534997_length(amino acids)=856AA_BP=485 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCV GAARSRALQFADVIVLFGARLNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKT LREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKD RSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFV QTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRP RGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQ WCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWM APEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQI -------------------------------------------------------------- >35563_35563_12_HACL1-RAF1_HACL1_chr3_15604864_ENST00000451445_RAF1_chr3_12641914_ENST00000542177_length(amino acids)=856AA_BP=485 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCV GAARSRALQFADVIVLFGARLNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKT LREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKD RSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFV QTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRP RGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQ WCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWM APEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQI -------------------------------------------------------------- >35563_35563_13_HACL1-RAF1_HACL1_chr3_15604864_ENST00000456194_RAF1_chr3_12641914_ENST00000251849_length(amino acids)=911AA_BP=540 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPLLVIGGSSERNQE TMGAFQEFPQVEACRLYTKFSARPSSIEAIPFVIEKAVRSSIYGRPGACYVDIPADFVNLQVNVNSIKYMERCMSPPISMAETSAVCTAA SVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVIVLFGARLNWILHFGLPPRYQP DVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKTLREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQ LPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIIL LVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKA QDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGK WHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMD YLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELP YSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQHSLPKINRSASEPSLHRAAHTEDINAC -------------------------------------------------------------- >35563_35563_14_HACL1-RAF1_HACL1_chr3_15604864_ENST00000456194_RAF1_chr3_12641914_ENST00000442415_length(amino acids)=911AA_BP=540 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPLLVIGGSSERNQE TMGAFQEFPQVEACRLYTKFSARPSSIEAIPFVIEKAVRSSIYGRPGACYVDIPADFVNLQVNVNSIKYMERCMSPPISMAETSAVCTAA SVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVIVLFGARLNWILHFGLPPRYQP DVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKTLREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQ LPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIIL LVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKA QDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGK WHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMD YLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELP YSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQHSLPKINRSASEPSLHRAAHTEDINAC -------------------------------------------------------------- >35563_35563_15_HACL1-RAF1_HACL1_chr3_15604864_ENST00000456194_RAF1_chr3_12641914_ENST00000534997_length(amino acids)=911AA_BP=540 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPLLVIGGSSERNQE TMGAFQEFPQVEACRLYTKFSARPSSIEAIPFVIEKAVRSSIYGRPGACYVDIPADFVNLQVNVNSIKYMERCMSPPISMAETSAVCTAA SVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVIVLFGARLNWILHFGLPPRYQP DVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKTLREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQ LPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIIL LVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKA QDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGK WHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMD YLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELP YSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQHSLPKINRSASEPSLHRAAHTEDINAC -------------------------------------------------------------- >35563_35563_16_HACL1-RAF1_HACL1_chr3_15604864_ENST00000456194_RAF1_chr3_12641914_ENST00000542177_length(amino acids)=911AA_BP=540 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPLLVIGGSSERNQE TMGAFQEFPQVEACRLYTKFSARPSSIEAIPFVIEKAVRSSIYGRPGACYVDIPADFVNLQVNVNSIKYMERCMSPPISMAETSAVCTAA SVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVIVLFGARLNWILHFGLPPRYQP DVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWWKTLREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQ LPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIIL LVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKA QDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGK WHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMD YLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELP YSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQHSLPKINRSASEPSLHRAAHTEDINAC -------------------------------------------------------------- >35563_35563_17_HACL1-RAF1_HACL1_chr3_15604864_ENST00000457447_RAF1_chr3_12641914_ENST00000251849_length(amino acids)=878AA_BP=507 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWPLLVIGGSSERNQETMGAFQEFPQAVRSSIYGRPGACYVDIPADFVNLQVNVNSIKYMERCMSPPISMAETSAVCTA ASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVIVLFGARLNWILHFGLPPRYQ PDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDA GTFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPM CLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKT PVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKT RHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLA TVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKA -------------------------------------------------------------- >35563_35563_18_HACL1-RAF1_HACL1_chr3_15604864_ENST00000457447_RAF1_chr3_12641914_ENST00000442415_length(amino acids)=878AA_BP=507 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWPLLVIGGSSERNQETMGAFQEFPQAVRSSIYGRPGACYVDIPADFVNLQVNVNSIKYMERCMSPPISMAETSAVCTA ASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVIVLFGARLNWILHFGLPPRYQ PDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDA GTFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPM CLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKT PVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKT RHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLA TVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKA -------------------------------------------------------------- >35563_35563_19_HACL1-RAF1_HACL1_chr3_15604864_ENST00000457447_RAF1_chr3_12641914_ENST00000534997_length(amino acids)=878AA_BP=507 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWPLLVIGGSSERNQETMGAFQEFPQAVRSSIYGRPGACYVDIPADFVNLQVNVNSIKYMERCMSPPISMAETSAVCTA ASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVIVLFGARLNWILHFGLPPRYQ PDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDA GTFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPM CLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKT PVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKT RHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLA TVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKA -------------------------------------------------------------- >35563_35563_20_HACL1-RAF1_HACL1_chr3_15604864_ENST00000457447_RAF1_chr3_12641914_ENST00000542177_length(amino acids)=878AA_BP=507 MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQLGIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIH ALGGMANANMNCWPLLVIGGSSERNQETMGAFQEFPQAVRSSIYGRPGACYVDIPADFVNLQVNVNSIKYMERCMSPPISMAETSAVCTA ASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVIVLFGARLNWILHFGLPPRYQ PDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDA GTFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPM CLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKT PVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKT RHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLA TVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKA -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:/chr3:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HACL1 | RAF1 |
| FUNCTION: Peroxisomal 2-OH acyl-CoA lyase involved in the cleavage (C1 removal) reaction in the fatty acid alpha-oxydation in a thiamine pyrophosphate (TPP)-dependent manner (PubMed:28289220, PubMed:21708296, PubMed:10468558). Involved in the degradation of 3-methyl-branched fatty acids like phytanic acid and the shortening of 2-hydroxy long-chain fatty acids (PubMed:28289220, PubMed:21708296, PubMed:10468558). Plays a significant role in the biosynthesis of heptadecanal in the liver (By similarity). {ECO:0000250|UniProtKB:Q9QXE0, ECO:0000269|PubMed:10468558, ECO:0000269|PubMed:21708296, ECO:0000269|PubMed:28289220}. | FUNCTION: Serine/threonine-protein kinase that acts as a regulatory link between the membrane-associated Ras GTPases and the MAPK/ERK cascade, and this critical regulatory link functions as a switch determining cell fate decisions including proliferation, differentiation, apoptosis, survival and oncogenic transformation. RAF1 activation initiates a mitogen-activated protein kinase (MAPK) cascade that comprises a sequential phosphorylation of the dual-specific MAPK kinases (MAP2K1/MEK1 and MAP2K2/MEK2) and the extracellular signal-regulated kinases (MAPK3/ERK1 and MAPK1/ERK2). The phosphorylated form of RAF1 (on residues Ser-338 and Ser-339, by PAK1) phosphorylates BAD/Bcl2-antagonist of cell death at 'Ser-75'. Phosphorylates adenylyl cyclases: ADCY2, ADCY5 and ADCY6, resulting in their activation. Phosphorylates PPP1R12A resulting in inhibition of the phosphatase activity. Phosphorylates TNNT2/cardiac muscle troponin T. Can promote NF-kB activation and inhibit signal transducers involved in motility (ROCK2), apoptosis (MAP3K5/ASK1 and STK3/MST2), proliferation and angiogenesis (RB1). Can protect cells from apoptosis also by translocating to the mitochondria where it binds BCL2 and displaces BAD/Bcl2-antagonist of cell death. Regulates Rho signaling and migration, and is required for normal wound healing. Plays a role in the oncogenic transformation of epithelial cells via repression of the TJ protein, occludin (OCLN) by inducing the up-regulation of a transcriptional repressor SNAI2/SLUG, which induces down-regulation of OCLN. Restricts caspase activation in response to selected stimuli, notably Fas stimulation, pathogen-mediated macrophage apoptosis, and erythroid differentiation. {ECO:0000269|PubMed:11427728, ECO:0000269|PubMed:11719507, ECO:0000269|PubMed:15385642, ECO:0000269|PubMed:15618521, ECO:0000269|PubMed:15849194, ECO:0000269|PubMed:16892053, ECO:0000269|PubMed:16924233, ECO:0000269|PubMed:9360956}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (564) >>>564.pdbFusion protein BP residue: 326 CIF file (564) >>>564.cif | HACL1 | chr3 | 15604864 | - | RAF1 | chr3 | 12641914 | - | MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQL GIKYIGMRNEQAVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPW QYPPESKWWKTLREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQLPRD CFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAK DRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDT DTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQKS LRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLS PTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVML STRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTR HVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQ TAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQ VEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHI NNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQ | 697 |
| 3D view using mol* of 564 (AA BP:326) | ||||||||||
 | ||||||||||
| PDB file (681) >>>681.pdbFusion protein BP residue: 485 CIF file (681) >>>681.cif | HACL1 | chr3 | 15604864 | - | RAF1 | chr3 | 12641914 | - | MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQL GIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIHALGGMANANM NCWYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAYAHAEES IKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVIVLFGAR LNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIHAVTKQL LEELDKTPWQYPPESKWWKTLREKMKSNEAASKELASKKSLPMNYYTVFY HVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGF AIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIILLVVNN NGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGGKGYFV QTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSESASPSA LSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRDSSYYW EIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQFQAFRN EVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQETKFQM FQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDFGLATV KSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIVLYELM TGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVADCVKKV KEERPLFPQILSSIELLQHSLPKINRSASEPSLHRAAHTEDINACTLTTS | 856 |
| 3D view using mol* of 681 (AA BP:485) | ||||||||||
 | ||||||||||
| PDB file (694) >>>694.pdbFusion protein BP residue: 507 CIF file (694) >>>694.cif | HACL1 | chr3 | 15604864 | - | RAF1 | chr3 | 12641914 | - | MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQL GIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIHALGGMANANM NCWPLLVIGGSSERNQETMGAFQEFPQAVRSSIYGRPGACYVDIPADFVN LQVNVNSIKYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAY AHAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVI VLFGARLNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIH AVTKQELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQ NYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGF SGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPM CLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTTKPSLINIMIE PQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKTPVPAQRERAP VSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHG DVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIV TQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMK SNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQ DNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFMVGRGYASPDL SKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQHSLPKINRSA | 878 |
| 3D view using mol* of 694 (AA BP:507) | ||||||||||
 | ||||||||||
| PDB file (707) >>>707.pdbFusion protein BP residue: 540 CIF file (707) >>>707.cif | HACL1 | chr3 | 15604864 | - | RAF1 | chr3 | 12641914 | - | MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQL GIKYIGMRNEQAACYAASAIGYLTSRPLLVIGGSSERNQETMGAFQEFPQ VEACRLYTKFSARPSSIEAIPFVIEKAVRSSIYGRPGACYVDIPADFVNL QVNVNSIKYMERCMSPPISMAETSAVCTAASVIRNAKQPLLIIGKGAAYA HAEESIKKLVEQYKLPFLPTPMGKGVVPDNHPYCVGAARSRALQFADVIV LFGARLNWILHFGLPPRYQPDVKFIQVDICAEELGNNVKPAVTLLGNIHA VTKQLLEELDKTPWQYPPESKWWKTLREKMKSNEAASKELASKKSLPMNY YTVFYHVQEQLPRDCFVVSEGANTMDIGRTVLQNYLPRHRLDAGTFGTMG VGLGFAIAAAVVAKDRSPGQWIICVEGDSAFGFSGMEVETICRYNLPIIL LVVNNNGIYQGFDTDTWKEMLKFQDATAVVPPMCLLPNSHYEQVMTAFGG KGYFVQTPEELQKSLRQSLADTTKPSLINIMIEPQATRKAQDAIRSHSES ASPSALSSSPNNLSPTGWSQPKTPVPAQRERAPVSGTQEKNKIRPRGQRD SSYYWEIEASEVMLSTRIGSGSFGTVYKGKWHGDVAVKILKVVDPTPEQF QAFRNEVAVLRKTRHVNILLFMGYMTKDNLAIVTQWCEGSSLYKHLHVQE TKFQMFQLIDIARQTAQGMDYLHAKNIIHRDMKSNNIFLHEGLTVKIGDF GLATVKSRWSGSQQVEQPTGSVLWMAPEVIRMQDNNPFSFQSDVYSYGIV LYELMTGELPYSHINNRDQIIFMVGRGYASPDLSKLYKNCPKAMKRLVAD CVKKVKEERPLFPQILSSIELLQHSLPKINRSASEPSLHRAAHTEDINAC | 911 |
| 3D view using mol* of 707 (AA BP:540) | ||||||||||
 | ||||||||||
| PDB file (720) >>>720.pdbFusion protein BP residue: 567 CIF file (720) >>>720.cif | HACL1 | chr3 | 15604864 | - | RAF1 | chr3 | 12641914 | - | MPDSNFAERSEEQVSGAKVIAQALKTQDVEYIFGIVGIPVTEIAIAAQQL GIKYIGMRNEQAACYAASAIGYLTSRPGVCLVVSGPGLIHALGGMANANM NCWPLLVIGGSSERNQETMGAFQEFPQVEACRLYTKFSARPSSIEAIPFV IEKAVRSSIYGRPGACYVDIPADFVNLQVNVNSIKYMERCMSPPISMAET SAVCTAASVIRNAKQPLLIIGKGAAYAHAEESIKKLVEQYKLPFLPTPMG KGVVPDNHPYCVGAARSRALQFADVIVLFGARLNWILHFGLPPRYQPDVK FIQVDICAEELGNNVKPAVTLLGNIHAVTKQLLEELDKTPWQYPPESKWW KTLREKMKSNEAASKELASKKSLPMNYYTVFYHVQEQLPRDCFVVSEGAN TMDIGRTVLQNYLPRHRLDAGTFGTMGVGLGFAIAAAVVAKDRSPGQWII CVEGDSAFGFSGMEVETICRYNLPIILLVVNNNGIYQGFDTDTWKEMLKF QDATAVVPPMCLLPNSHYEQVMTAFGGKGYFVQTPEELQKSLRQSLADTT KPSLINIMIEPQATRKAQDAIRSHSESASPSALSSSPNNLSPTGWSQPKT PVPAQRERAPVSGTQEKNKIRPRGQRDSSYYWEIEASEVMLSTRIGSGSF GTVYKGKWHGDVAVKILKVVDPTPEQFQAFRNEVAVLRKTRHVNILLFMG YMTKDNLAIVTQWCEGSSLYKHLHVQETKFQMFQLIDIARQTAQGMDYLH AKNIIHRDMKSNNIFLHEGLTVKIGDFGLATVKSRWSGSQQVEQPTGSVL WMAPEVIRMQDNNPFSFQSDVYSYGIVLYELMTGELPYSHINNRDQIIFM VGRGYASPDLSKLYKNCPKAMKRLVADCVKKVKEERPLFPQILSSIELLQ | 938 |
| 3D view using mol* of 720 (AA BP:567) | ||||||||||
 | ||||||||||
Top |
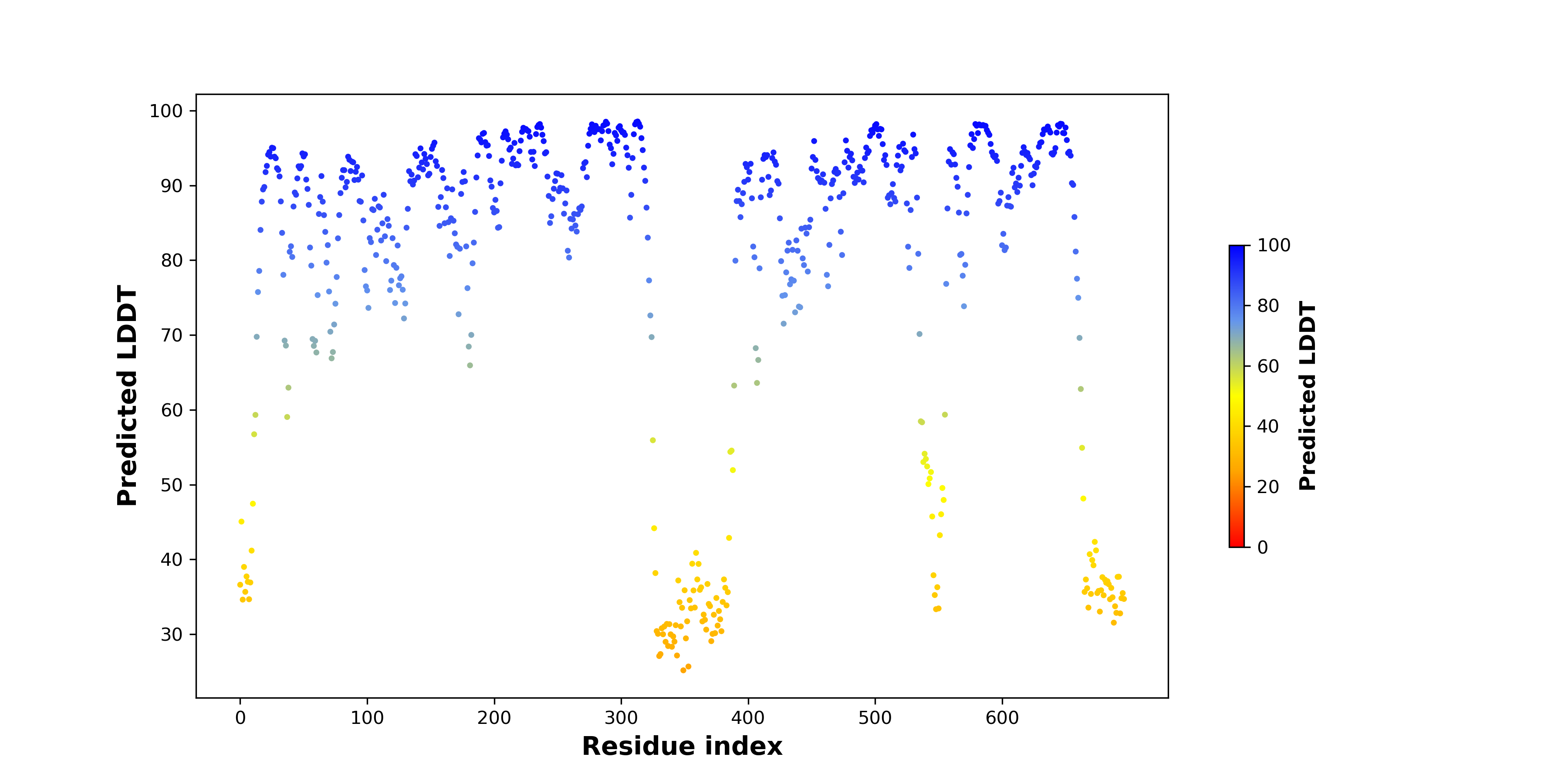
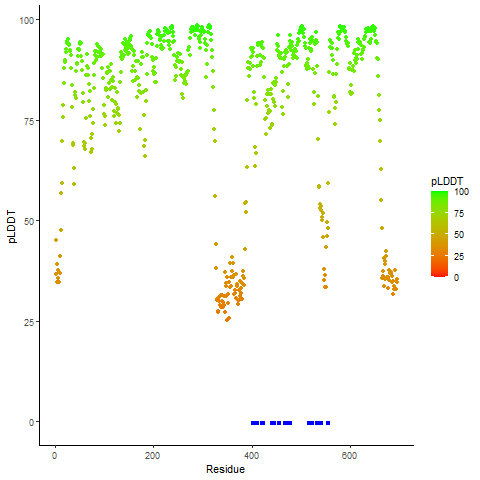
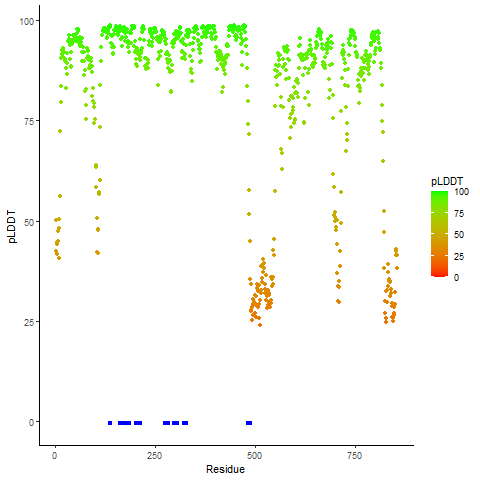
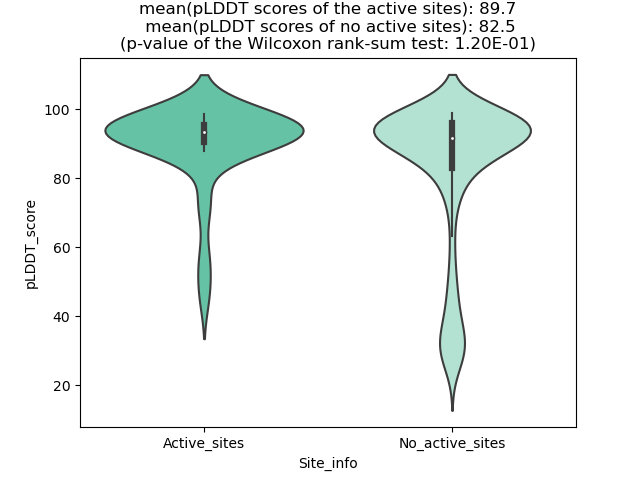
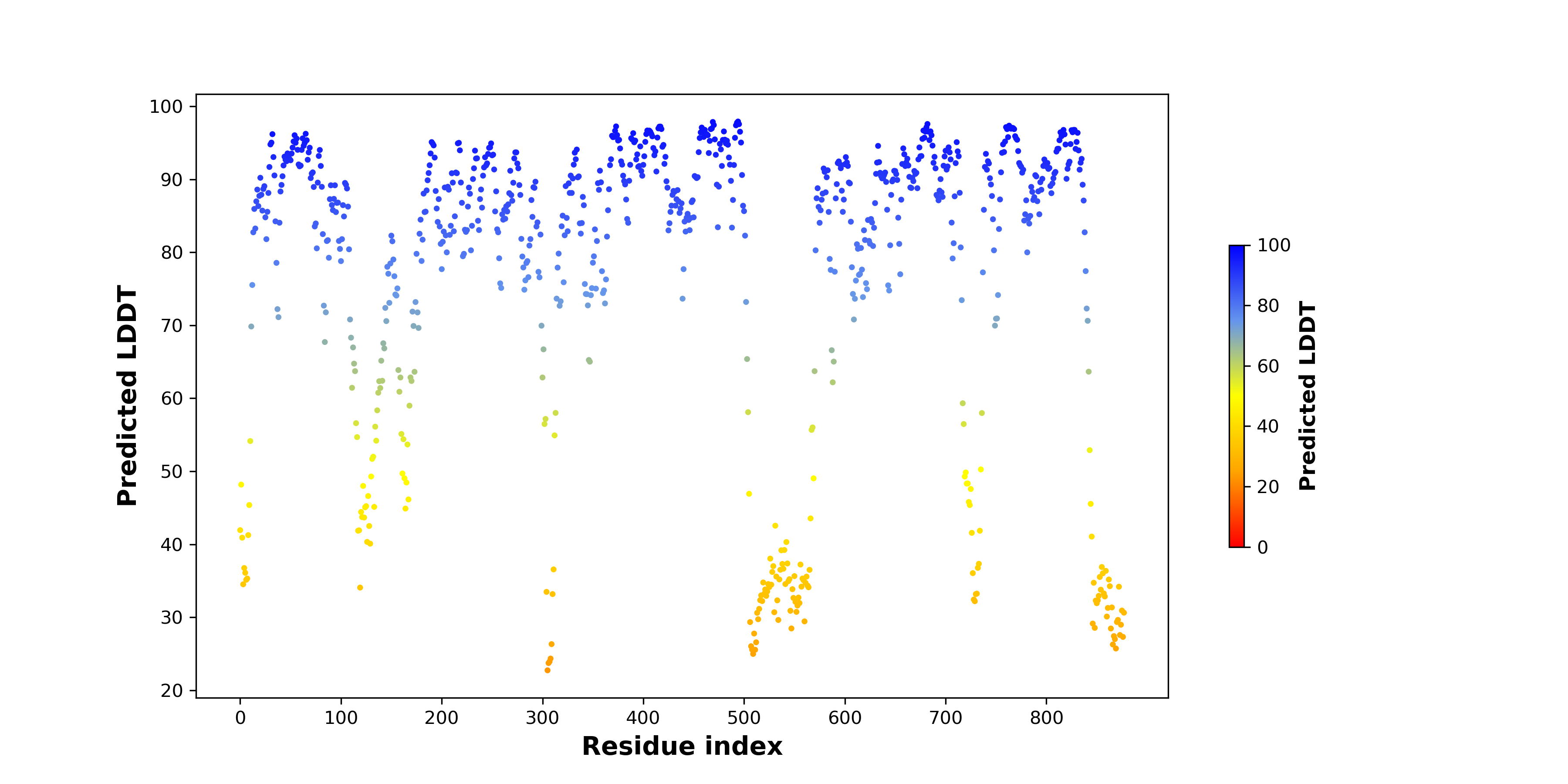
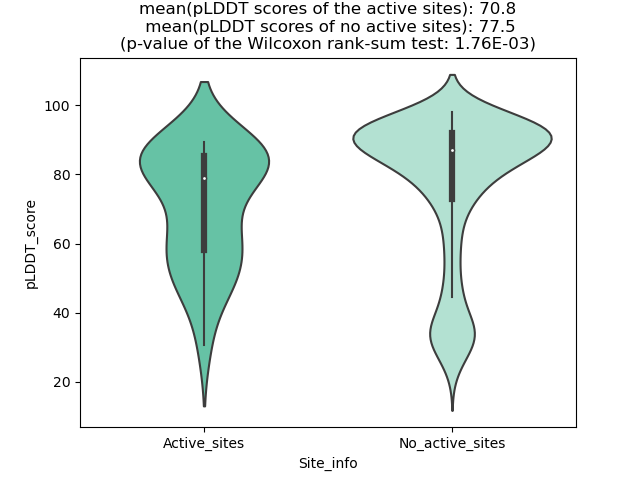
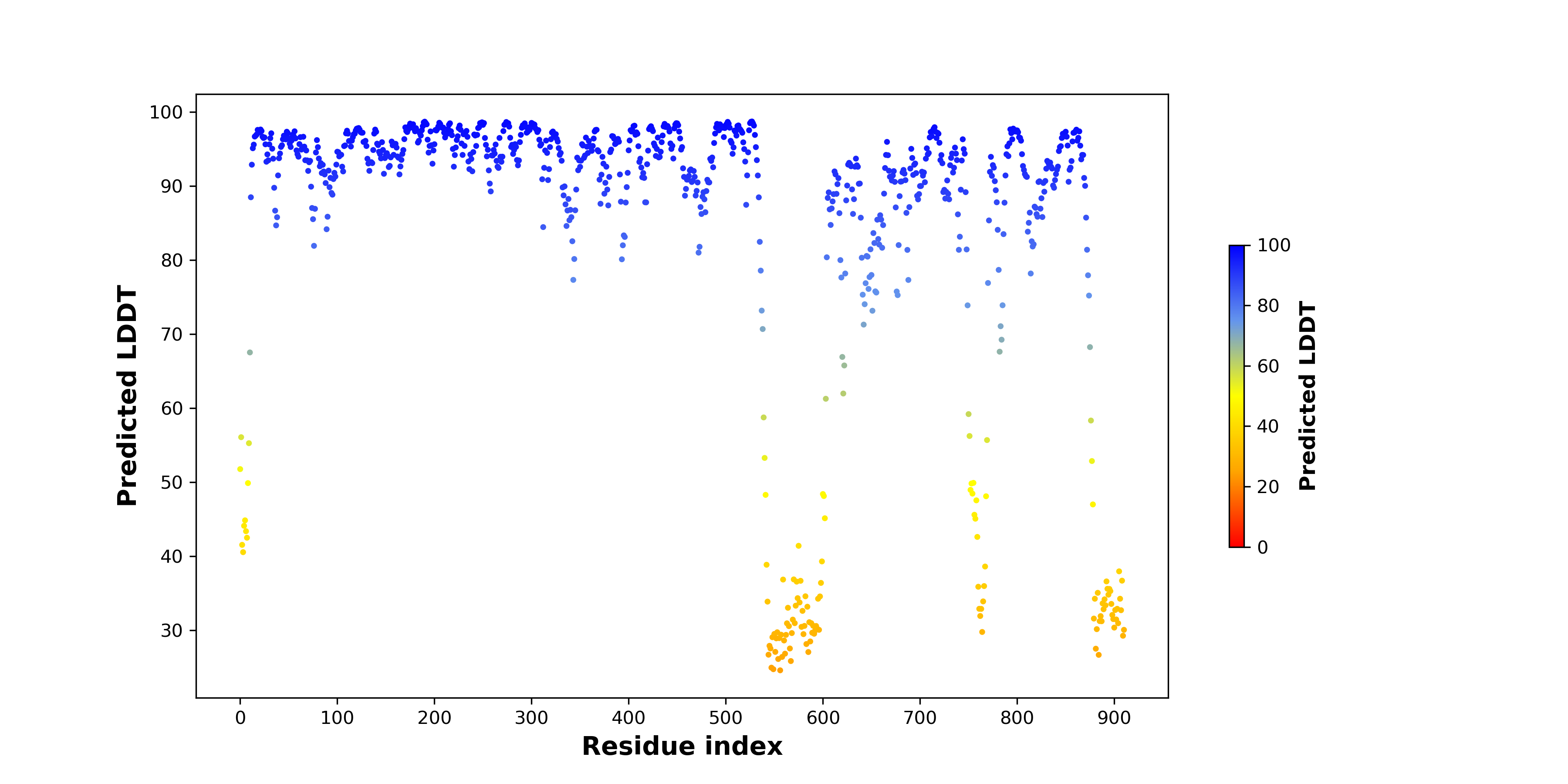
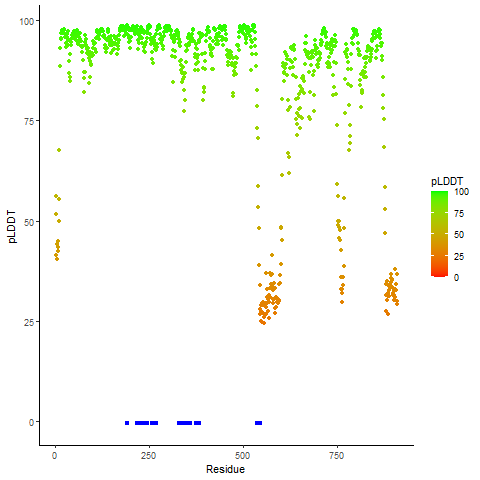
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
HACL1_pLDDT.png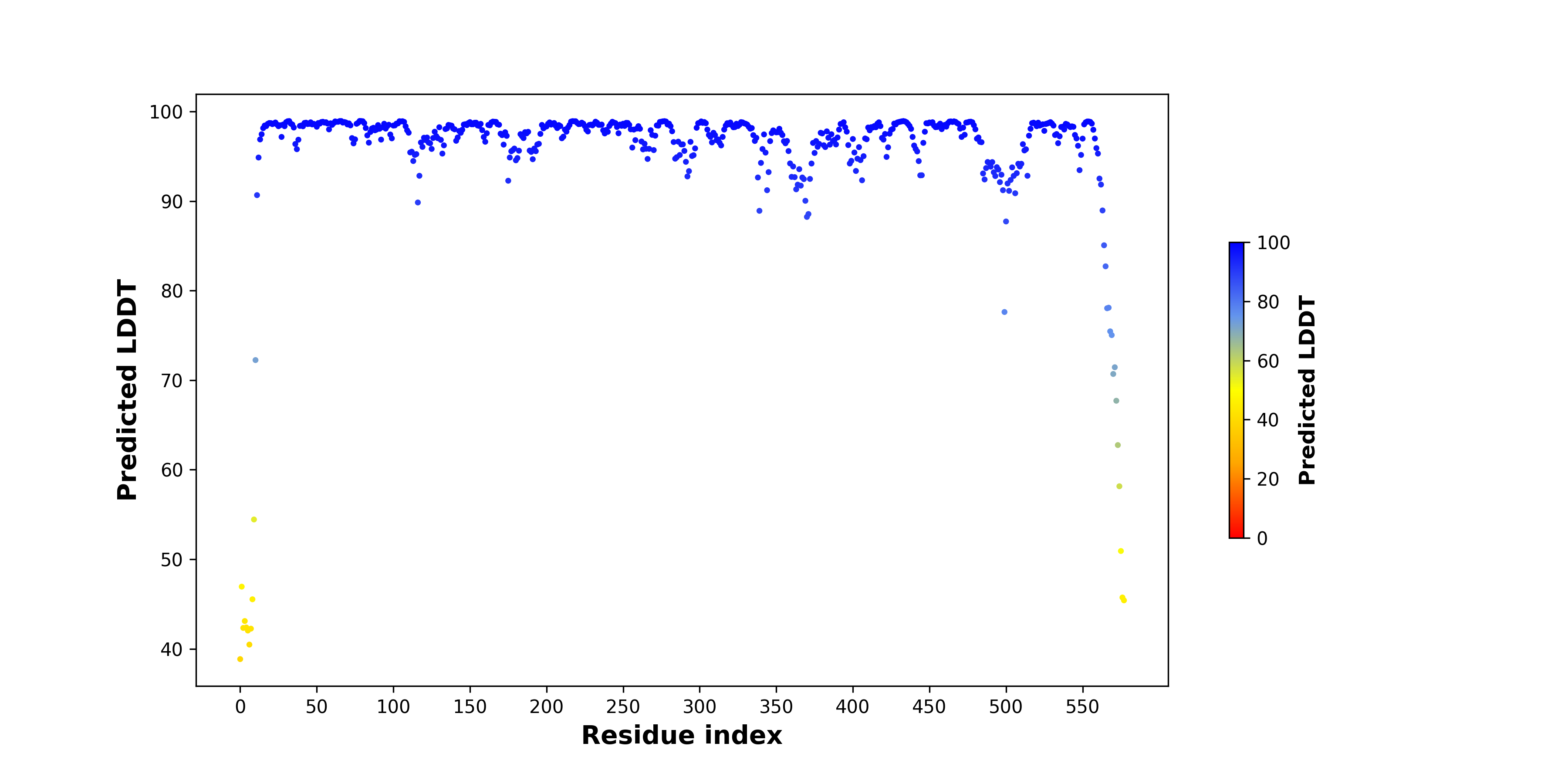 |
RAF1_pLDDT.png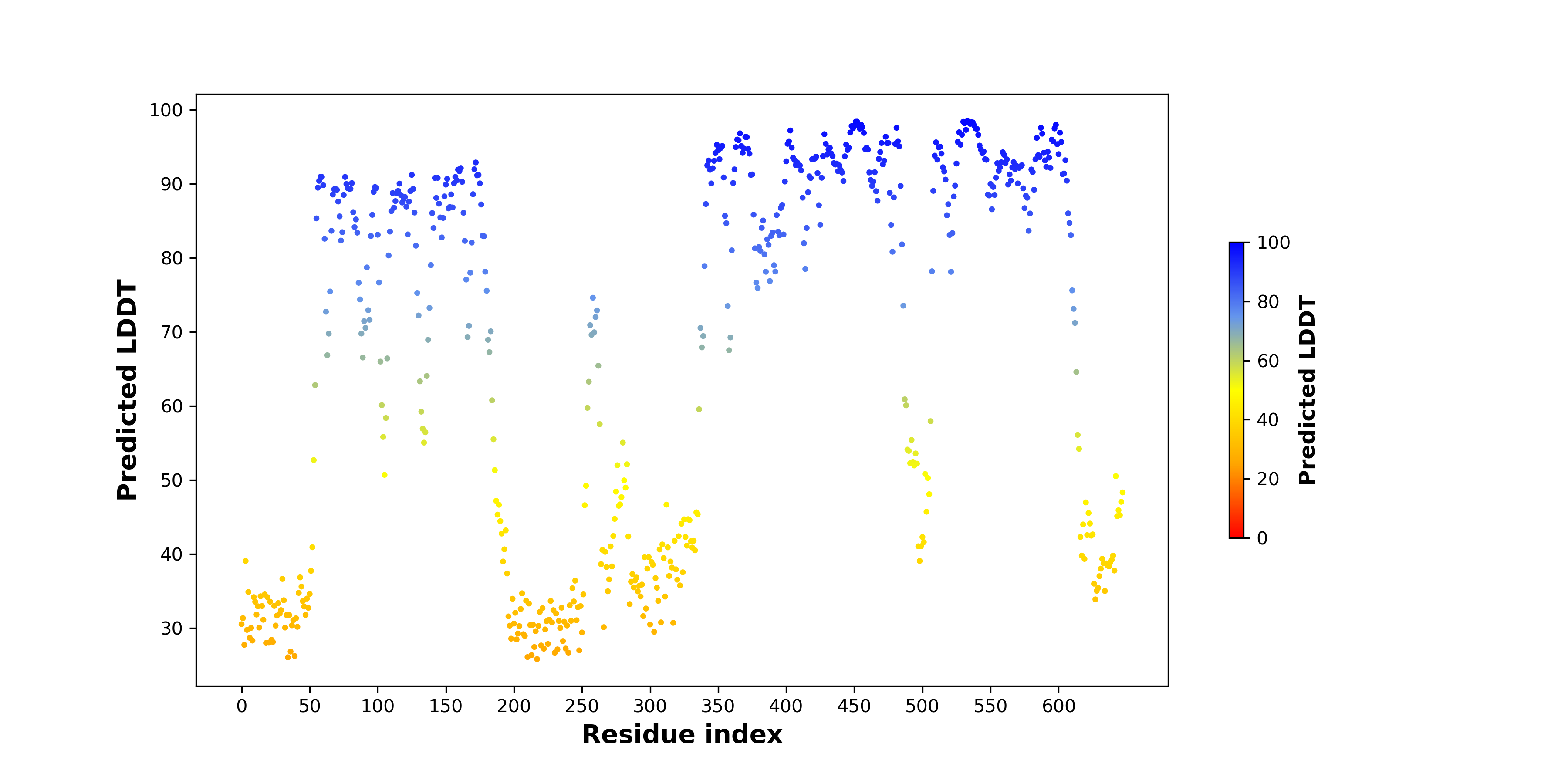 |
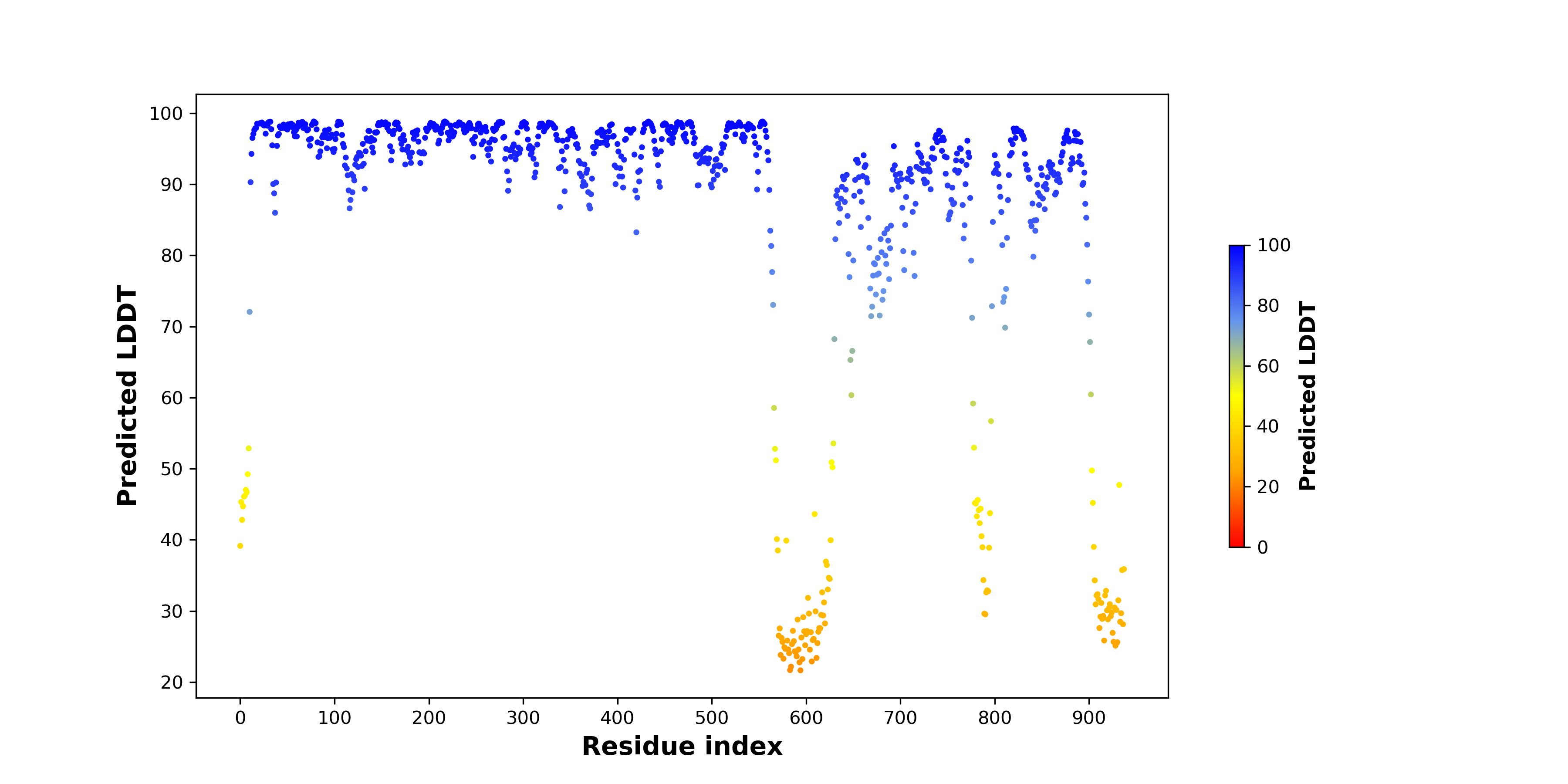
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
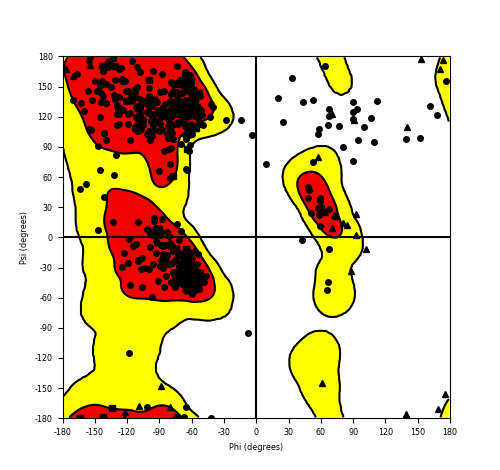
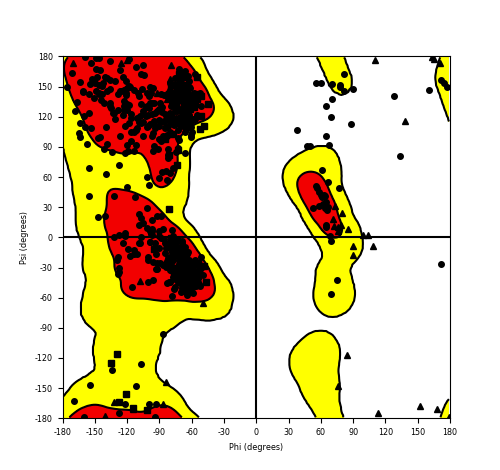
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| HACL1_RAF1_564.png |
 |
| HACL1_RAF1_681.png |
 |
| HACL1_RAF1_694.png |
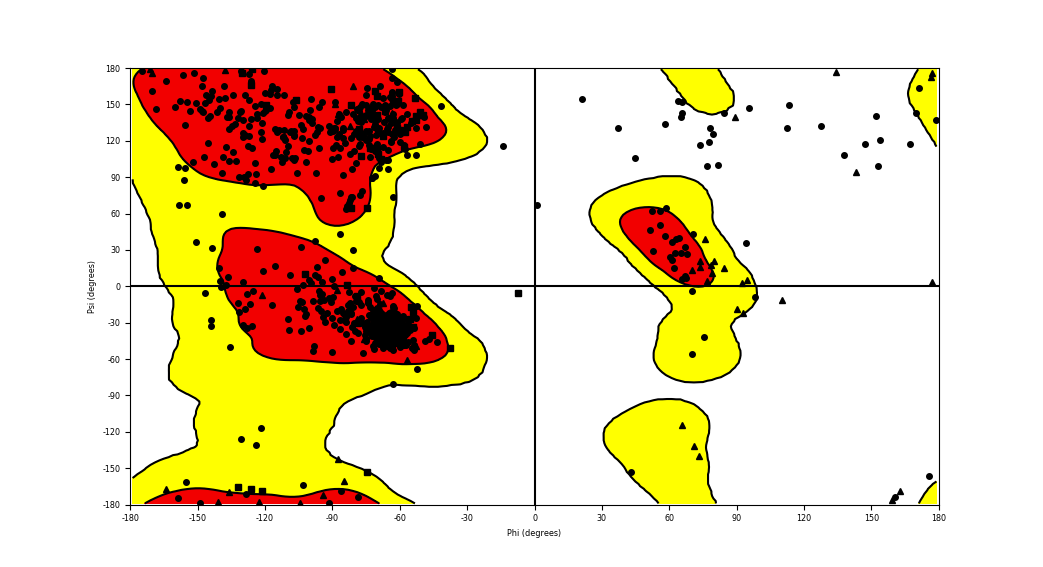 |
| HACL1_RAF1_707.png |
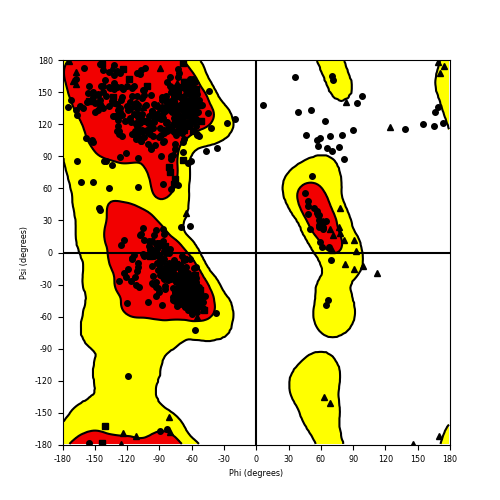 |
| HACL1_RAF1_720.png |
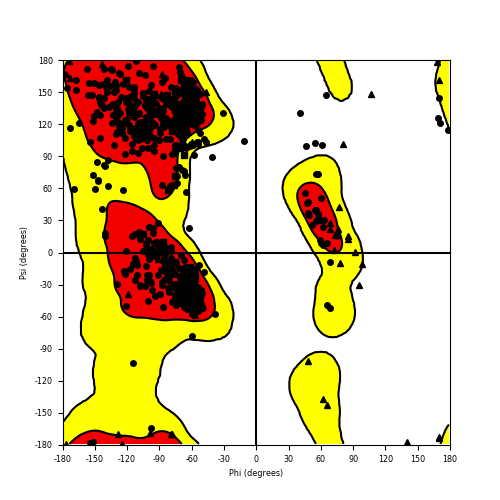 |
Top |
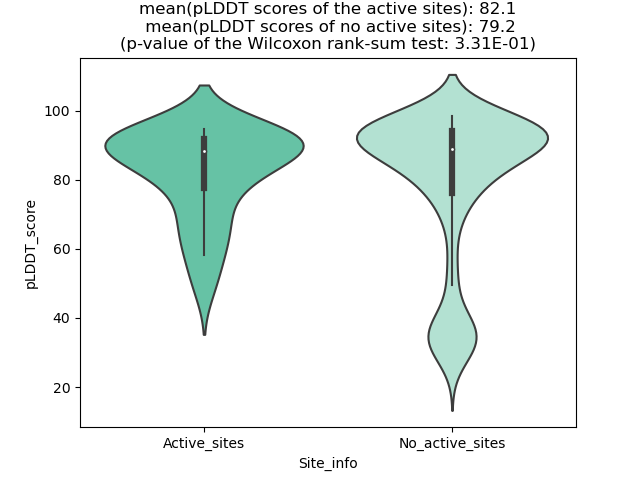
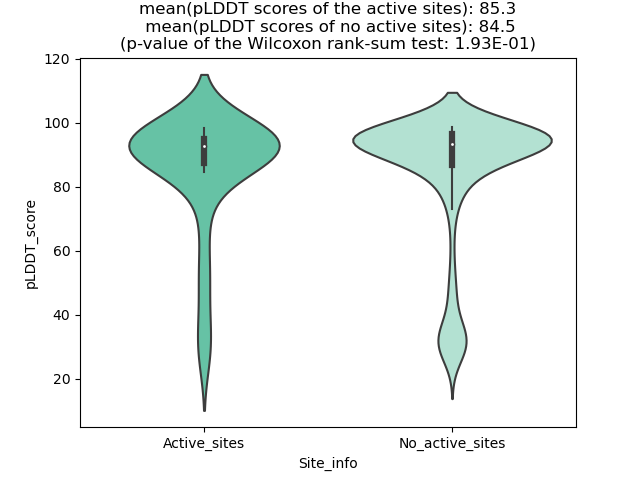
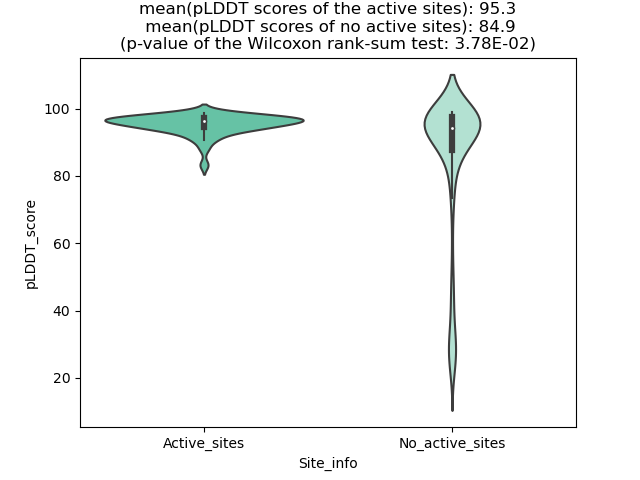
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 564 | 1.053 | 146 | 1.064 | 407.484 | 0.51 | 0.777 | 0.983 | 1.001 | 1.05 | 0.953 | 1.001 | Chain A: 404,405,406,407,408,409,410,412,422,424,4 42,446,455,457,468,470,471,472,473,474,475,476,477 ,479,517,519,521,522,524,534,535,536,538,542,555,5 56 |
| 681 | 1.076 | 310 | 1.108 | 666.449 | 0.428 | 0.775 | 0.995 | 0.857 | 0.912 | 0.939 | 0.702 | Chain A: 137,163,164,165,166,169,174,175,176,177,1 79,181,182,183,184,185,203,204,205,206,212,274,275 ,278,279,282,283,296,300,303,304,321,322,324,325,3 26,328,483,484,485,486,487 |
| 694 | 1.107498 | 102 | 1.19435 | 412.629 | 0.680251 | 0.711796 | 0.815935 | 1.660206 | 0.529841 | 3.133406 | 0.96704 | Chain A: 16,19,20,23,24,26,27,29,76,77,106,108,129 ,132,133,142,144,148,151,153,158,159,160,161,162,1 64,878 |
| 707 | 1.048 | 377 | 1.066 | 768.663 | 0.438 | 0.761 | 0.997 | 0.802 | 1.014 | 0.791 | 0.658 | Chain A: 192,218,219,220,221,224,229,230,231,232,2 34,235,236,237,238,239,240,241,244,245,258,259,260 ,261,266,267,329,330,333,334,336,337,338,341,342,3 51,355,358,359,376,377,379,380,381,383,538,539,540 ,541,542,543,544,545,546 |
| 720 | 1.0832 | 233 | 1.096 | 396.508 | 0.3995 | 0.8182 | 1.0753 | 1.1234 | 1.0273 | 1.0936 | 0.7176 | Chain A: 72,75,76,101,102,103,159,160,161,162,163, 189,192,193,194,195,221,222,223,226,227,228,247,24 8,249,250,280,281,282,284,286,287,304,305,306,324, 325,326,406,419,421,422 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
| 720 | ZINC000000121541 | DB00643 | Mebendazole | -8.63597 | -9.31867 |
| 720 | ZINC000068247389 | DB12483 | Copanlisib | -8.42822 | -8.85532 |
| 720 | ZINC000000000655 | DB01058 | Praziquantel | -8.78817 | -8.78817 |
| 720 | ZINC000029571072 | DB06636 | Isavuconazonium | -6.98077 | -8.75307 |
| 720 | ZINC000003964126 | DB06228 | Rivaroxaban | -8.69525 | -8.69525 |
| 720 | ZINC000040430143 | DB09074 | Olaparib | -8.60124 | -8.60124 |
| 720 | ZINC000004099200 | DB00213 | Pantoprazole | -8.08373 | -8.38543 |
| 720 | ZINC000003802690 | DB00442 | Entecavir | -8.3393 | -8.3393 |
| 720 | ZINC000000020220 | DB00537 | Ciprofloxacin | -7.23159 | -8.33599 |
| 720 | ZINC000002008866 | DB00601 | Linezolid | -8.28785 | -8.28785 |
| 720 | ZINC000100378061 | DB11691 | Naldemedine | -7.92656 | -8.28096 |
| 720 | ZINC000043206370 | DB11793 | Niraparib | -8.03337 | -8.03337 |
| 720 | ZINC000000000850 | DB00744 | Zileuton | -7.99829 | -8.01689 |
| 720 | ZINC000000968328 | DB00412 | Rosiglitazone | -7.44757 | -7.97177 |
| 720 | ZINC000098023177 | DB09330 | Osimertinib | -7.86467 | -7.87237 |
| 720 | ZINC000011616882 | DB06626 | Axitinib | -7.8458 | -7.849 |
| 720 | ZINC000000000973 | DB09495 | Avobenzone | -7.48205 | -7.80725 |
| 720 | Netarsudil | DB09495 | -7.79041 | -7.80151 | |
| 720 | ZINC000019796080 | DB00450 | Droperidol | -7.69718 | -7.79558 |
| 720 | ZINC000000005560 | DB01009 | Ketoprofen | -7.79303 | -7.79353 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
| ZINC000000121541 | DB00643 | Mebendazole | Small molecule | COC(=O)NC1=NC2=C(N1)C=C(C=C2)C(=O)C1=CC=CC=C1 | Approved|Vet_approved |
| ZINC000068247389 | DB12483 | Copanlisib | Small molecule | COC1=C(OCCCN2CCOCC2)C=CC2=C1N=C(NC(=O)C1=CN=C(N)N=C1)N1CCN=C21 | Approved|Investigational |
| ZINC000003964126 | DB06228 | Rivaroxaban | Small molecule | ClC1=CC=C(S1)C(=O)NC[C@H]1CN(C(=O)O1)C1=CC=C(C=C1)N1CCOCC1=O | Approved |
| ZINC000040430143 | DB09074 | Olaparib | Small molecule | FC1=CC=C(CC2=NNC(=O)C3=CC=CC=C23)C=C1C(=O)N1CCN(CC1)C(=O)C1CC1 | Approved |
| ZINC000003802690 | DB00442 | Entecavir | Small molecule | NC1=NC(=O)C2=C(N1)N(C=N2)[C@H]1C[C@H](O)[C@@H](CO)C1=C | Approved|Investigational |
| ZINC000000020220 | DB00537 | Ciprofloxacin | Small molecule | OC(=O)C1=CN(C2CC2)C2=CC(N3CCNCC3)=C(F)C=C2C1=O | Approved|Investigational |
| ZINC000002008866 | DB00601 | Linezolid | Small molecule | CC(=O)NC[C@H]1CN(C(=O)O1)C1=CC(F)=C(C=C1)N1CCOCC1 | Approved|Investigational |
| ZINC000100378061 | DB11691 | Naldemedine | Small molecule | [H][C@@]12OC3=C4C(C[C@@]5([H])N(CC6CC6)CC[C@@]14[C@@]5(O)CC(C(=O)NC(C)(C)C1=NC(=NO1)C1=CC=CC=C1)=C2O)=CC=C3O | Approved|Investigational |
| ZINC000043206370 | DB11793 | Niraparib | Small molecule | NC(=O)C1=CC=CC2=CN(N=C12)C1=CC=C(C=C1)[C@@H]1CCCNC1 | Approved|Investigational |
| ZINC000098023177 | DB09330 | Osimertinib | Small molecule | COC1=C(NC2=NC=CC(=N2)C2=CN(C)C3=C2C=CC=C3)C=C(NC(=O)C=C)C(=C1)N(C)CCN(C)C | Approved |
| ZINC000011616882 | DB06626 | Axitinib | Small molecule | CNC(=O)C1=C(SC2=CC=C3C(NN=C3C=CC3=CC=CC=N3)=C2)C=CC=C1 | Approved|Investigational |
| ZINC000000000973 | DB09495 | Avobenzone | Small molecule | COC1=CC=C(C=C1)C(=O)CC(=O)C1=CC=C(C=C1)C(C)(C)C | Approved|Investigational |
| ZINC000019796080 | DB00450 | Droperidol | Small molecule | FC1=CC=C(C=C1)C(=O)CCCN1CCC(=CC1)N1C(=O)NC2=CC=CC=C12 | Approved|Vet_approved |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
| ZINC000000121541 | 295.297 | 6.562 | 576.161 | 98.468 | 169.387 | 308.306 | 0 | 957.18 | 1 | 5 | 8.911 | 3 | 83.641 | 0 | 0 |
| ZINC000000121541 | 295.297 | 2.873 | 577.033 | 96.773 | 172.83 | 307.43 | 0 | 959.022 | 0 | 3 | 8.668 | 3 | 88.187 | 0 | 0 |
| ZINC000000121541 | 295.297 | 10.919 | 575.199 | 98.469 | 170.072 | 306.658 | 0 | 956.419 | 1 | 5 | 8.861 | 3 | 83.461 | 0 | 0 |
| ZINC000000000655 | 312.411 | 8.136 | 587.283 | 357.505 | 79.355 | 150.424 | 0 | 1051.38 | 0 | 6 | 9.514 | 3 | 85.555 | 0 | 0 |
| ZINC000003964126 | 435.881 | 14.098 | 691.698 | 245.278 | 129.622 | 200.433 | 116.364 | 1226.142 | 1 | 10.7 | 8.72 | 3 | 89.646 | 0 | 0 |
| ZINC000040430143 | 434.469 | 9.89 | 732.917 | 311 | 167.143 | 218.809 | 35.965 | 1326.981 | 1 | 8 | 9.389 | 3 | 83.359 | 0 | 0 |
| ZINC000004099200 | 383.369 | 8.318 | 623.128 | 213.244 | 77.412 | 222.302 | 110.17 | 1084.47 | 1 | 8 | 9.124 | 3 | 76.692 | 0 | 0 |
| ZINC000004099200 | 383.369 | 8.299 | 620.121 | 215.65 | 78.983 | 216.916 | 108.573 | 1082.558 | 1 | 8 | 9.183 | 3 | 76.221 | 0 | 0 |
| ZINC000004099200 | 383.369 | 8.355 | 619.5 | 212.861 | 77.163 | 219.295 | 110.182 | 1080.981 | 1 | 8 | 9.019 | 3 | 76.603 | 0 | 0 |
| ZINC000004099200 | 383.369 | 8.428 | 625.396 | 215.335 | 78.49 | 221.055 | 110.516 | 1084.724 | 1 | 8 | 9.007 | 3 | 76.459 | 0 | 0 |
| ZINC000003802690 | 277.282 | 6.742 | 504.225 | 143.129 | 289.374 | 71.721 | 0 | 851.662 | 5 | 8.9 | 8.512 | 2 | 42.613 | 0 | 1 |
| ZINC000000020220 | 331.346 | 8.67 | 578.412 | 299.833 | 178.539 | 68.487 | 31.553 | 1017.371 | 1 | 6 | 8.925 | 2 | 48.33 | 0 | 1 |
| ZINC000000020220 | 331.346 | 8.111 | 575.511 | 298.04 | 176.667 | 69.246 | 31.558 | 1012.813 | 1 | 6 | 8.79 | 2 | 48.648 | 0 | 1 |
| ZINC000002008866 | 337.35 | 11.384 | 587.905 | 370.553 | 99.012 | 86.525 | 31.815 | 1034.058 | 1 | 8.7 | 8.411 | 3 | 80.732 | 0 | 0 |
| ZINC000100378061 | 570.644 | 7.56 | 854.021 | 394.296 | 164.643 | 295.081 | 0 | 1642.452 | 2 | 10.75 | 8.853 | 2 | 62.781 | 1 | 0 |
| ZINC000100378061 | 570.644 | 8.331 | 882.482 | 435.335 | 156.89 | 290.257 | 0 | 1665.202 | 2 | 10.75 | 8.834 | 2 | 64.48 | 1 | 0 |
| ZINC000100378061 | 570.644 | 10.805 | 763.051 | 379.112 | 156.9 | 227.039 | 0 | 1550.783 | 2 | 10.75 | 8.848 | 3 | 60.844 | 1 | 0 |
| ZINC000100378061 | 570.644 | 6.025 | 879.032 | 420.86 | 169.846 | 288.326 | 0 | 1665.947 | 2 | 10.75 | 8.837 | 2 | 62.171 | 1 | 0 |
| ZINC000100378061 | 570.644 | 5.057 | 763.557 | 369.607 | 150.289 | 243.661 | 0 | 1542.607 | 2 | 7.75 | 8.754 | 3 | 74.615 | 1 | 0 |
| ZINC000100378061 | 570.644 | 10.054 | 868.556 | 406.179 | 167.203 | 295.174 | 0 | 1648.407 | 2 | 10.75 | 8.918 | 2 | 62.166 | 1 | 0 |
| ZINC000100378061 | 570.644 | 5.965 | 873.468 | 425.996 | 156.952 | 290.52 | 0 | 1657.709 | 2 | 10.75 | 8.842 | 2 | 64.594 | 1 | 0 |
| ZINC000100378061 | 570.644 | 8.012 | 750.848 | 378.176 | 172.899 | 199.772 | 0 | 1535.752 | 2 | 7.75 | 8.801 | 3 | 69.361 | 1 | 0 |
| ZINC000100378061 | 570.644 | 7.635 | 772.058 | 387.087 | 130.302 | 254.669 | 0 | 1568.531 | 2 | 10.75 | 8.606 | 3 | 67.334 | 1 | 0 |
| ZINC000100378061 | 570.644 | 5.989 | 867.147 | 425.966 | 151.587 | 289.595 | 0 | 1651.992 | 2 | 10.75 | 8.574 | 2 | 65.463 | 1 | 0 |
| ZINC000100378061 | 570.644 | 8.675 | 845.172 | 419.952 | 137.137 | 288.083 | 0 | 1633.122 | 2 | 10.75 | 8.655 | 3 | 68.333 | 1 | 0 |
| ZINC000100378061 | 570.644 | 5.076 | 883.549 | 427.334 | 168.007 | 288.209 | 0 | 1671.805 | 2 | 10.75 | 8.637 | 2 | 62.812 | 1 | 0 |
| ZINC000100378061 | 570.644 | 6.815 | 864.11 | 391.791 | 178.38 | 293.94 | 0 | 1641.252 | 2 | 7.75 | 8.82 | 1 | 72.388 | 1 | 1 |
| ZINC000043206370 | 320.393 | 3.754 | 607.688 | 185.794 | 143.417 | 278.476 | 0 | 1055.805 | 3 | 5 | 8.546 | 3 | 76.72 | 0 | 0 |
| ZINC000000000850 | 236.288 | 5.297 | 457.642 | 85.783 | 137.035 | 198.523 | 36.3 | 755.015 | 3 | 3.7 | 8.91 | 3 | 75.351 | 0 | 0 |
| ZINC000000000850 | 236.288 | 5.297 | 450.837 | 85.532 | 135.419 | 201.893 | 27.994 | 746.839 | 3 | 3.7 | 8.57 | 3 | 76.555 | 0 | 0 |
| ZINC000000968328 | 357.426 | 1.444 | 634.074 | 175.676 | 137.918 | 287.735 | 32.745 | 1113.731 | 1 | 5.25 | 8.373 | 3 | 96.012 | 0 | 0 |
| ZINC000000968328 | 357.426 | 2.563 | 632.369 | 175.708 | 136.258 | 287.719 | 32.684 | 1111.853 | 1 | 5.25 | 8.348 | 3 | 96.294 | 0 | 0 |
| ZINC000000968328 | 357.426 | 1.098 | 615.115 | 188.498 | 120.189 | 271.15 | 35.279 | 1101.068 | 1 | 5.25 | 8.417 | 3 | 100 | 0 | 0 |
| ZINC000000968328 | 357.426 | 4.059 | 637.846 | 177.05 | 121.495 | 296.234 | 43.067 | 1111.876 | 1 | 5.25 | 8.521 | 3 | 100 | 0 | 0 |
| ZINC000098023177 | 499.614 | 4.129 | 911.961 | 486.025 | 62.254 | 363.681 | 0 | 1655.003 | 2 | 8.75 | 7.891 | 1 | 95.185 | 1 | 1 |
| ZINC000011616882 | 386.47 | 5.573 | 701.348 | 134.589 | 97.85 | 441.453 | 27.456 | 1225.038 | 2 | 4.5 | 8.259 | 1 | 100 | 0 | 1 |
| ZINC000000000973 | 310.392 | 6.186 | 619.54 | 304.394 | 82.62 | 232.526 | 0 | 1088.974 | 0 | 4.75 | 9.463 | 3 | 100 | 0 | 0 |
| ZINC000000000973 | 310.392 | 6.564 | 618.531 | 285.769 | 72.419 | 260.343 | 0 | 1082.178 | 0 | 2.5 | 9.061 | 3 | 100 | 0 | 0 |
| ZINC000000000973 | 310.392 | 7.653 | 619.32 | 285.592 | 72.423 | 261.305 | 0 | 1082.954 | 0 | 2.5 | 9.142 | 3 | 100 | 0 | 0 |
| ZINC000000000973 | 310.392 | 3.937 | 616.628 | 300.438 | 70.725 | 245.465 | 0 | 1078.245 | 0 | 2.5 | 9.238 | 3 | 100 | 0 | 0 |
| ZINC000019796080 | 379.433 | 5.336 | 690.754 | 177.289 | 126.948 | 339.689 | 46.828 | 1214.622 | 1 | 6 | 8.72 | 3 | 86.625 | 0 | 0 |
| ZINC000019796080 | 379.433 | 4.227 | 703.161 | 188.25 | 122.749 | 345.337 | 46.825 | 1231.012 | 1 | 6 | 8.619 | 3 | 87.345 | 0 | 0 |
| ZINC000019796080 | 379.433 | 6.26 | 700.949 | 186.412 | 123.449 | 344.263 | 46.825 | 1227.254 | 1 | 6 | 8.598 | 3 | 87.215 | 0 | 0 |
| ZINC000000005560 | 254.285 | 5.998 | 511.945 | 84.096 | 135.311 | 292.538 | 0 | 867.146 | 1 | 4 | 9.894 | 3 | 83.089 | 0 | 0 |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
| ZINC000000121541 | COC(=O)/N=c1[nH]c2ccc(C(=O)c3ccccc3)cc2[nH]1 | 0.209696342 | 0.592445172 |
| ZINC000068247389 | COc1c(OCCCN2CCOCC2)ccc2c1N=C(/N=C(O)c1cnc(N)nc1)N1CCN=C21 | 0.095668162 | 0.218817189 |
| ZINC000000000655 | O=C(C1CCCCC1)N1CC(=O)N2CCc3ccccc3[C@@H]2C1 | 0.180997967 | 0.220112088 |
| ZINC000029571072 | CNCC(=O)OCc1cccnc1N(C)C(=O)O[C@H](C)[n+]1cnn(C[C@](O)(c2cc(F)ccc2F)[C@@H](C)c2nc(-c3ccc(C#N)cc3)cs2)c1 | 0.025416027 | 0.235213823 |
| ZINC000003964126 | O=C(NC[C@H]1CN(c2ccc(N3CCOCC3=O)cc2)C(=O)O1)c1ccc(Cl)s1 | 0.136337489 | 0.314484681 |
| ZINC000040430143 | O=C(c1cc(Cc2n[nH]c(=O)c3ccccc23)ccc1F)N1CCN(C(=O)C2CC2)CC1 | 0.254314897 | 0.146669648 |
| ZINC000004099200 | COc1ccnc(C[S@](=O)c2nc3ccc(OC(F)F)cc3[nH]2)c1OC | 0.114667354 | 0.321842622 |
| ZINC000003802690 | C=C1[C@@H](n2cnc3c(=O)[nH]c(N)nc32)C[C@H](O)[C@H]1CO | 0.045693772 | 0.309972869 |
| ZINC000000020220 | O=C(O)c1cn(C2CC2)c2cc(N3CCNCC3)c(F)cc2c1=O | 0.231031629 | 0.323908212 |
| ZINC000002008866 | CC(=O)NC[C@H]1CN(c2ccc(N3CCOCC3)c(F)c2)C(=O)O1 | 0.172188025 | 0.409724838 |
| ZINC000100378061 | CC(C)(NC(=O)C1=C(O)[C@@H]2Oc3c(O)ccc4c3[C@@]23CCN(CC2CC2)[C@H](C4)[C@]3(O)C1)c1nc(-c2ccccc2)no1 | 0.010882818 | 0.206042928 |
| ZINC000043206370 | NC(=O)c1cccc2cn(-c3ccc([C@@H]4CCCNC4)cc3)nc12 | 0.15810306 | 0.243905197 |
| ZINC000000000850 | C[C@@H](c1cc2ccccc2s1)N(O)C(N)=O | 0.136335882 | 0.223511692 |
| ZINC000000968328 | CN(CCOc1ccc(C[C@@H]2SC(=O)NC2=O)cc1)c1ccccn1 | 0.155326871 | 0.273380926 |
| ZINC000098023177 | C=CC(=O)Nc1cc(Nc2nccc(-c3cn(C)c4ccccc34)n2)c(OC)cc1N(C)CCN(C)C | 0.14592444 | 0.194338203 |
| ZINC000011616882 | CNC(=O)c1ccccc1Sc1ccc2c(/C=Cc3ccccn3)n[nH]c2c1 | 0.19874265 | 0.230949688 |
| ZINC000000000973 | COc1ccc(C(=O)CC(=O)c2ccc(C(C)(C)C)cc2)cc1 | 0.449967764 | 0.531455882 |
| ZINC000019796080 | O=C(CCCN1CC=C(n2c(=O)[nH]c3ccccc32)CC1)c1ccc(F)cc1 | 0.219727636 | 0.259906112 |
| ZINC000000005560 | C[C@H](C(=O)O)c1cccc(C(=O)c2ccccc2)c1 | 0.317764263 | 0.285932087 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| HACL1 | ZMYND19, HACL1, MAPK6, FAM162A, PEX5, MAGEB6, GMPPB, PTPRF, WDFY1, ASPSCR1, PLXDC2, ODF2L, RNA15, CAT, APEX1, ANLN, ECT2, KIF20A, AGPS, CCNF, NBR1, |
| RAF1 | YWHAQ, YWHAG, YWHAZ, Hras, SFN, HRAS, VDAC1, PEBP4, CNKSR1, RASD2, CNKSR2, ERAS, SPRY4, SPRY2, YWHAB, OIP5, PHKA2, CCT3, MYC, AR, MRAS, KRAS, LRPAP1, RRAS2, SHOC2, TSC22D3, STK26, MAPK1, MAPK3, PRKCE, BCL2, BCL2L1, PEBP1, MAP2K1, GRB10, NRAS, MAPK8IP3, AKT1, HSP90AA1, GNG4, GNB2, VAV1, RHEB, RAP1A, SRC, FYN, RB1, RBL2, PRKG1, JAK2, YWHAH, CFLAR, CDC25A, BRAF, MAP3K1, PDGFRB, MAP3K5, MAPK7, PHB, BAG1, PAK1, PRKCZ, RRAS, PRPF6, UBE2D2, YWHAE, NR3C1, LCK, LTK, JAK1, XIAP, BIRC2, BIRC3, STUB1, Arrb2, EEF1A1, HSP82, HSPA8, HSPA5, CALM1, SGK1, NEDD4, SCNN1B, NEDD4L, STK3, BRAP, WDR83, KSR1, LATS1, MAP2K2, Rfxank, RFXANK, PPP1CC, VCP, HSPD1, BAD, BCL6, BIRC7, PIN1, MMS19, PPP2R1A, PPP2R2B, PPP2CA, HSPA4, RAF1, PPP2CB, MAPK8, RBBP8, YY1, PKM, CDK20, HIPK4, TBXA2R, SH3KBP1, ILK, GPRASP2, CDC37, IRS4, FAM96B, HUWE1, Prkab1, RANBP9, TMEM70, PACSIN3, NPLOC4, PRKAR1A, DCAF8, PURA, FYCO1, MYBPC1, COPS7A, MOV10, NXF1, EGFR, CALU, CCT8, DNAJA1, EMD, TIMM50, LOX, ARRB2, AGTR1, CASZ1, MRC2, PDLIM2, HERC2, XPO1, ACOX1, PCDH7, MAD2L1, SYNPO, TMEM63B, EEF1E1, COPS5, SS18L2, Smn1, Cep152, Aldh4a1, Ksr1, NUP133, RPGRIP1L, NCAPG2, NUP107, MSH2, LAS1L, AMER1, NUP160, EIF3D, SPATA7, NPHP4, DYNLL2, DYNLL1, IQCB1, RMDN3, TRIM25, Mapkbp1, EGLN3, RIPK4, MAPK6, CSNK1A1, HERC1, LZTR1, QKI, COPS3, TRIM28, MAEA, NANOG, ITCH, IRF7, PSMB9, MAP2K5, CDK4, CDK6, CDKN2B, FGFR4, MAP2K3, NF2, PDGFRA, TEAD2, RASSF1, ABCB5, BECN1, CBLC, GLIS2, GRM1, HGF, MAP2K6, ARNT, CCND2, CD44, CDKN2A, CDKN2C, EPHA2, ERBB2, KEAP1, LATS2, MET, NF1, TERT, DUSP2, MBP, PAK2, PLEKHA4, CRBN, ARAF, SLC25A3, TUBB4B, TUBB, HSPA6, HSP90AB3P, HSP90AB1, HSPA1A, HSPB1, BAG2, CCT7, DSG1, CCT2, CCT4, CCT6A, TCP1, AKAP8L, ATAD3A, ABCD3, SLC25A22, TMEM33, TUBA4A, HPX, MGST1, TUBB4A, TMEM161A, HSP90AB2P, CCAR2, SLC25A10, FANCD2, TUBB6, HSD17B12, SLC25A11, SLC25A1, ARL1, DNAJA2, DPM1, CCT5, AIFM1, SQSTM1, TUBG2, TUBG1, STK39, GCN1L1, ABCF2, POLD1, NUBP2, TM9SF3, NCLN, SEC61A2, SEC61A1, ALDH1A3, TUBB3, POLR2B, IARS2, TMED1, CIT, AURKB, KIF20A, BRD1, TRIM66, HAX1, CUL4A, ACTR1A, AKAP1, FLOT1, LAMTOR1, LMAN1, RAB2A, RAB4A, STX7, NPAS1, OPALIN, METTL21B, SSSCA1, FAM174A, CRKL, DDB1, DICER1, LDHB, PIK3R1, NONO, DVL2, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| HACL1 | 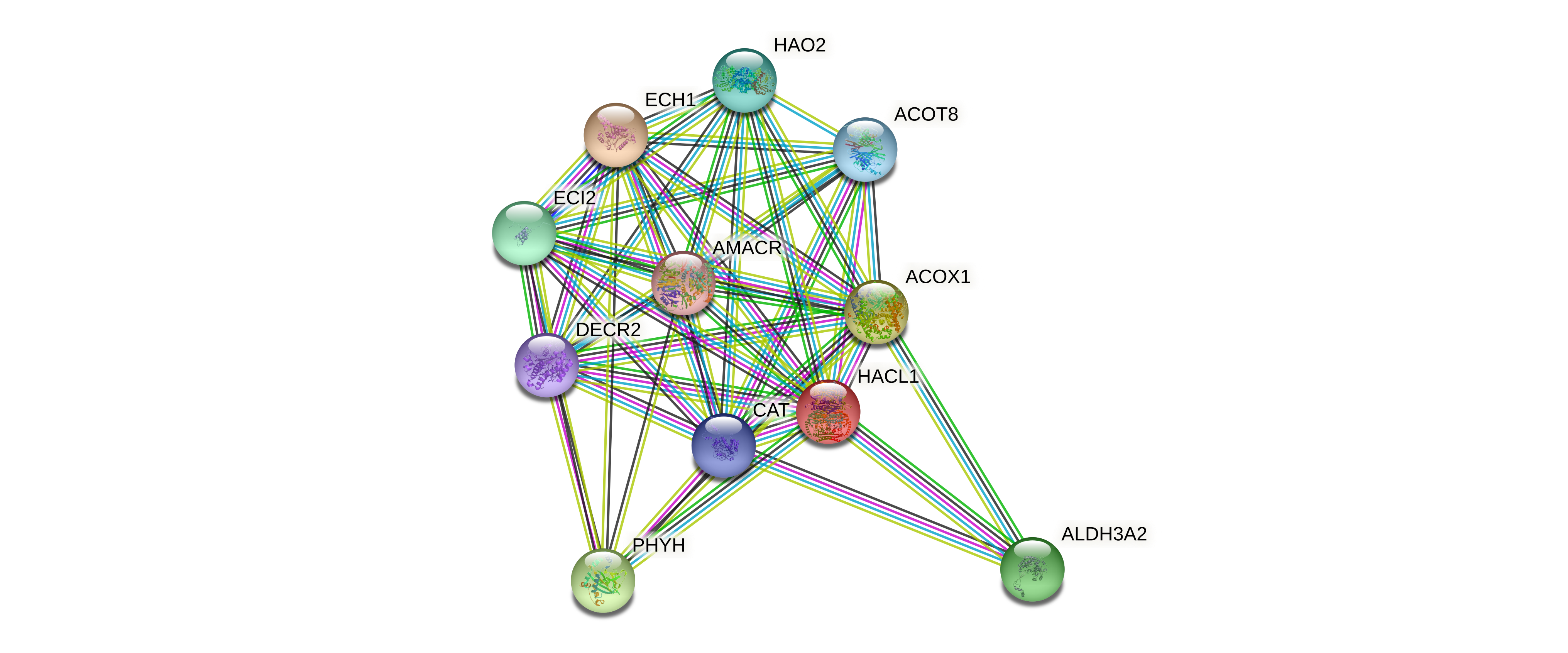 |
| RAF1 | 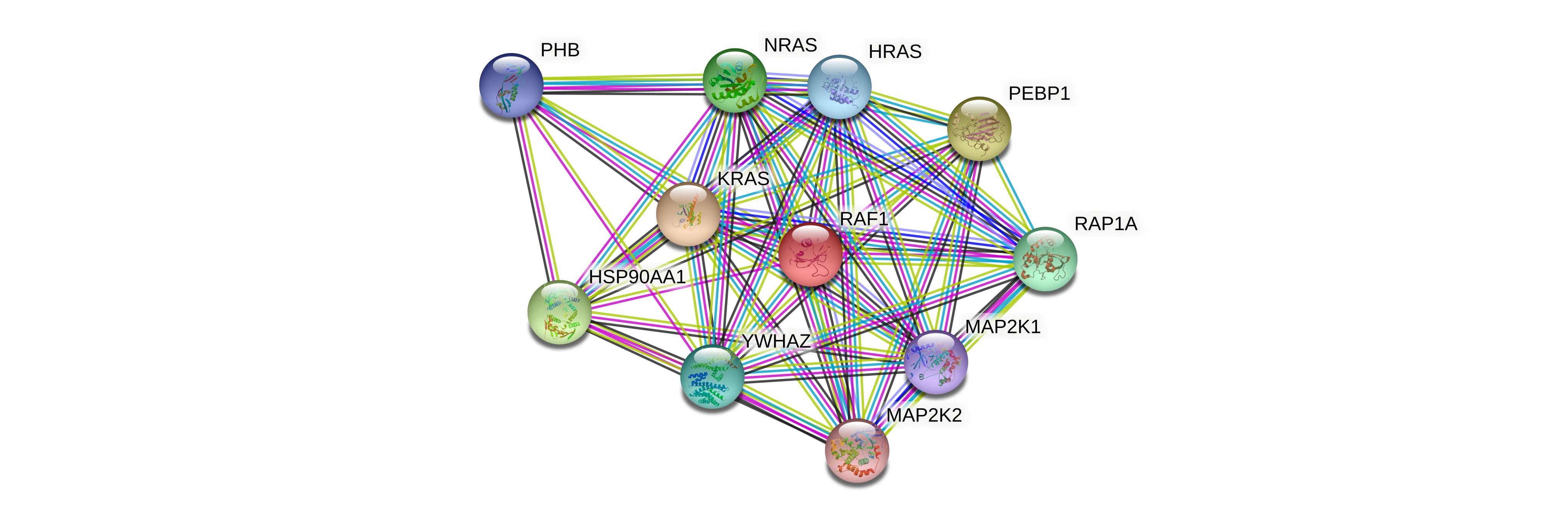 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to HACL1-RAF1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to HACL1-RAF1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| HACL1 | RAF1 | Dendritic Cell Tumor | MyCancerGenome | |
| HACL1 | RAF1 | Not Otherwise Specified | MyCancerGenome | |
| HACL1 | RAF1 | Esophageal Adenocarcinoma | MyCancerGenome | |
| HACL1 | RAF1 | Gastric Adenocarcinoma | MyCancerGenome | |
| HACL1 | RAF1 | Glioblastoma | MyCancerGenome | |
| HACL1 | RAF1 | High-Grade Glioma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | HACL1 | C0008370 | Cholestasis | 1 | CTD_human |
| Hgene | HACL1 | C0019193 | Hepatitis, Toxic | 1 | CTD_human |
| Hgene | HACL1 | C0860207 | Drug-Induced Liver Disease | 1 | CTD_human |
| Hgene | HACL1 | C1262760 | Hepatitis, Drug-Induced | 1 | CTD_human |
| Hgene | HACL1 | C3658290 | Drug-Induced Acute Liver Injury | 1 | CTD_human |
| Hgene | HACL1 | C4277682 | Chemical and Drug Induced Liver Injury | 1 | CTD_human |
| Hgene | HACL1 | C4279912 | Chemically-Induced Liver Toxicity | 1 | CTD_human |
| Tgene | RAF1 | C0028326 | Noonan Syndrome | 10 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | RAF1 | C0175704 | LEOPARD Syndrome | 7 | CLINGEN;CTD_human;GENOMICS_ENGLAND |
| Tgene | RAF1 | C1969057 | Noonan Syndrome 5 | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | RAF1 | C1969056 | LEOPARD SYNDROME 2 | 3 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | RAF1 | C0007194 | Hypertrophic Cardiomyopathy | 2 | CTD_human |
| Tgene | RAF1 | C0041409 | Turner Syndrome, Male | 2 | CTD_human |
| Tgene | RAF1 | C1519086 | Pilomyxoid astrocytoma | 2 | ORPHANET |
| Tgene | RAF1 | C1527404 | Female Pseudo-Turner Syndrome | 2 | CTD_human |
| Tgene | RAF1 | C4551472 | Hypertrophic obstructive cardiomyopathy | 2 | CTD_human |
| Tgene | RAF1 | C4551602 | Noonan Syndrome 1 | 2 | CTD_human |
| Tgene | RAF1 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | RAF1 | C0007131 | Non-Small Cell Lung Carcinoma | 1 | CTD_human |
| Tgene | RAF1 | C0007193 | Cardiomyopathy, Dilated | 1 | CTD_human |
| Tgene | RAF1 | C0017638 | Glioma | 1 | CTD_human |
| Tgene | RAF1 | C0020429 | Hyperalgesia | 1 | CTD_human |
| Tgene | RAF1 | C0022665 | Kidney Neoplasm | 1 | CTD_human |
| Tgene | RAF1 | C0023903 | Liver neoplasms | 1 | CTD_human |
| Tgene | RAF1 | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Tgene | RAF1 | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Tgene | RAF1 | C0259783 | mixed gliomas | 1 | CTD_human |
| Tgene | RAF1 | C0340427 | Familial dilated cardiomyopathy | 1 | ORPHANET |
| Tgene | RAF1 | C0345904 | Malignant neoplasm of liver | 1 | CTD_human |
| Tgene | RAF1 | C0345967 | Malignant mesothelioma | 1 | CTD_human |
| Tgene | RAF1 | C0458247 | Allodynia | 1 | CTD_human |
| Tgene | RAF1 | C0555198 | Malignant Glioma | 1 | CTD_human |
| Tgene | RAF1 | C0587248 | Costello syndrome (disorder) | 1 | CLINGEN |
| Tgene | RAF1 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | RAF1 | C0740457 | Malignant neoplasm of kidney | 1 | CTD_human |
| Tgene | RAF1 | C0751211 | Hyperalgesia, Primary | 1 | CTD_human |
| Tgene | RAF1 | C0751212 | Hyperalgesia, Secondary | 1 | CTD_human |
| Tgene | RAF1 | C0751213 | Tactile Allodynia | 1 | CTD_human |
| Tgene | RAF1 | C0751214 | Hyperalgesia, Thermal | 1 | CTD_human |
| Tgene | RAF1 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | RAF1 | C1275081 | Cardio-facio-cutaneous syndrome | 1 | CLINGEN |
| Tgene | RAF1 | C1449563 | Cardiomyopathy, Familial Idiopathic | 1 | CTD_human |
| Tgene | RAF1 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | RAF1 | C2936719 | Mechanical Allodynia | 1 | CTD_human |
| Tgene | RAF1 | C4014656 | CARDIOMYOPATHY, DILATED, 1NN | 1 | CTD_human;UNIPROT |
| Tgene | RAF1 | C4551484 | Leopard Syndrome 1 | 1 | CTD_human;GENOMICS_ENGLAND |
| Tgene | RAF1 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |
| Tgene | RAF1 | C4721610 | Carcinoma, Ovarian Epithelial | 1 | CTD_human |