| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:HSP90AA1-EIF4A2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: HSP90AA1-EIF4A2 | FusionPDB ID: 37749 | FusionGDB2.0 ID: 37749 | Hgene | Tgene | Gene symbol | HSP90AA1 | EIF4A2 | Gene ID | 3320 | 1974 |
| Gene name | heat shock protein 90 alpha family class A member 1 | eukaryotic translation initiation factor 4A2 | |
| Synonyms | EL52|HEL-S-65p|HSP86|HSP89A|HSP90A|HSP90N|HSPC1|HSPCA|HSPCAL1|HSPCAL4|HSPN|Hsp103|Hsp89|Hsp90|LAP-2|LAP2 | BM-010|DDX2B|EIF4A|EIF4F|eIF-4A-II|eIF4A-II | |
| Cytomap | 14q32.31 | 3q27.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | heat shock protein HSP 90-alphaHSP 86LPS-associated protein 2epididymis luminal secretory protein 52epididymis secretory sperm binding protein Li 65pheat shock 86 kDaheat shock 90kD protein 1, alphaheat shock 90kD protein 1, alpha-like 4heat shock | eukaryotic initiation factor 4A-IIATP-dependent RNA helicase eIF4A-2 | |
| Modification date | 20200327 | 20200322 | |
| UniProtAcc | P07900 | Q14240 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000216281, ENST00000334701, ENST00000441629, ENST00000558600, | ENST00000323963, ENST00000356531, ENST00000440191, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 38 X 40 X 8=12160 | 13 X 14 X 6=1092 |
| # samples | 43 | 16 | |
| ** MAII score | log2(43/12160*10)=-4.82166275874149 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/1092*10)=-2.77082904603249 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: HSP90AA1 [Title/Abstract] AND EIF4A2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | HSP90AA1(102549370)-EIF4A2(186504915), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | HSP90AA1-EIF4A2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HSP90AA1-EIF4A2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. HSP90AA1-EIF4A2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HSP90AA1-EIF4A2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. HSP90AA1-EIF4A2 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. HSP90AA1-EIF4A2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. HSP90AA1-EIF4A2 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. HSP90AA1-EIF4A2 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. HSP90AA1-EIF4A2 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. HSP90AA1-EIF4A2 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | HSP90AA1 | GO:0001934 | positive regulation of protein phosphorylation | 19363271 |
| Hgene | HSP90AA1 | GO:0007004 | telomere maintenance via telomerase | 10197982 |
| Hgene | HSP90AA1 | GO:0031396 | regulation of protein ubiquitination | 16809764 |
| Hgene | HSP90AA1 | GO:0032273 | positive regulation of protein polymerization | 19363271 |
| Hgene | HSP90AA1 | GO:0045040 | protein import into mitochondrial outer membrane | 15644312 |
| Hgene | HSP90AA1 | GO:0051131 | chaperone-mediated protein complex assembly | 15644312 |
| Hgene | HSP90AA1 | GO:0051973 | positive regulation of telomerase activity | 10197982 |
| Hgene | HSP90AA1 | GO:1902949 | positive regulation of tau-protein kinase activity | 19363271 |
| Hgene | HSP90AA1 | GO:1905323 | telomerase holoenzyme complex assembly | 10197982 |
| Tgene | EIF4A2 | GO:1900260 | negative regulation of RNA-directed 5'-3' RNA polymerase activity | 11922617 |
 Fusion gene breakpoints across HSP90AA1 (5'-gene) Fusion gene breakpoints across HSP90AA1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
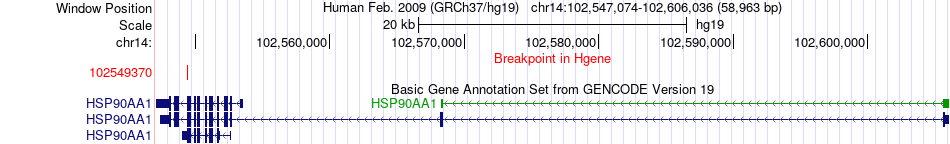 |
 Fusion gene breakpoints across EIF4A2 (3'-gene) Fusion gene breakpoints across EIF4A2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
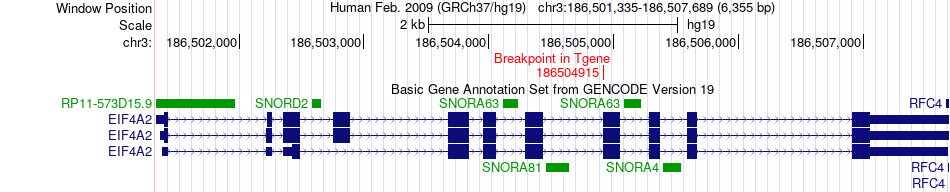 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-61-1900 | HSP90AA1 | chr14 | 102549370 | - | EIF4A2 | chr3 | 186504915 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000334701 | HSP90AA1 | chr14 | 102549370 | - | ENST00000323963 | EIF4A2 | chr3 | 186504915 | + | 3487 | 2403 | 282 | 2855 | 857 |
| ENST00000334701 | HSP90AA1 | chr14 | 102549370 | - | ENST00000440191 | EIF4A2 | chr3 | 186504915 | + | 3484 | 2403 | 282 | 2855 | 857 |
| ENST00000334701 | HSP90AA1 | chr14 | 102549370 | - | ENST00000356531 | EIF4A2 | chr3 | 186504915 | + | 3481 | 2403 | 282 | 2855 | 857 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000334701 | ENST00000323963 | HSP90AA1 | chr14 | 102549370 | - | EIF4A2 | chr3 | 186504915 | + | 0.000720128 | 0.9992799 |
| ENST00000334701 | ENST00000440191 | HSP90AA1 | chr14 | 102549370 | - | EIF4A2 | chr3 | 186504915 | + | 0.000724851 | 0.99927515 |
| ENST00000334701 | ENST00000356531 | HSP90AA1 | chr14 | 102549370 | - | EIF4A2 | chr3 | 186504915 | + | 0.000743102 | 0.9992569 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >37749_37749_1_HSP90AA1-EIF4A2_HSP90AA1_chr14_102549370_ENST00000334701_EIF4A2_chr3_186504915_ENST00000323963_length(amino acids)=857AA_BP=123 MPPCSGGDGSTPPGPSLRDRDCPAQSAEYPRDRLDPRPGSPSEASSPPFLRSRAPVNWYQEKAQVFLWHLMVSGSTTLLCLWKQPFHVSA FPVTASLAFRQSQGAGQHLYKDLQPFILLRLLMPEETQTQDQPMEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDK IRYESLTDPSKLDSGKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQFGVGFYSAYLVAEKV TVITKHNDDEQYAWESSAGGSFTVRTDTGEPMGRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITLFVEKERDKEVSDDEAEEK EDKEEEKEKEEKESEDKPEIEDVGSDEEEEKKDGDKKKKKKIKEKYIDQEELNKTKPIWTRNPDDITNEEYGEFYKSLTNDWEDHLAVKH FSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNLVK KCLELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSASGDEMVSLKDYCTRMKENQKHIYYITGETKDQVANSAFV ERLRKHGLEVIYMIEPIDEYCVQQLKEFEGKTLVSVTKEGLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKEWKLDTLCDLYET LTITQAVIFLNTRRKVDWLTEKMHARDFTVSALHGDMDQKERDVIMREFRSGSSRVLITTDLLARGIDVQQVSLVINYDLPTNRENYIHR -------------------------------------------------------------- >37749_37749_2_HSP90AA1-EIF4A2_HSP90AA1_chr14_102549370_ENST00000334701_EIF4A2_chr3_186504915_ENST00000356531_length(amino acids)=857AA_BP=123 MPPCSGGDGSTPPGPSLRDRDCPAQSAEYPRDRLDPRPGSPSEASSPPFLRSRAPVNWYQEKAQVFLWHLMVSGSTTLLCLWKQPFHVSA FPVTASLAFRQSQGAGQHLYKDLQPFILLRLLMPEETQTQDQPMEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDK IRYESLTDPSKLDSGKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQFGVGFYSAYLVAEKV TVITKHNDDEQYAWESSAGGSFTVRTDTGEPMGRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITLFVEKERDKEVSDDEAEEK EDKEEEKEKEEKESEDKPEIEDVGSDEEEEKKDGDKKKKKKIKEKYIDQEELNKTKPIWTRNPDDITNEEYGEFYKSLTNDWEDHLAVKH FSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNLVK KCLELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSASGDEMVSLKDYCTRMKENQKHIYYITGETKDQVANSAFV ERLRKHGLEVIYMIEPIDEYCVQQLKEFEGKTLVSVTKEGLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKEWKLDTLCDLYET LTITQAVIFLNTRRKVDWLTEKMHARDFTVSALHGDMDQKERDVIMREFRSGSSRVLITTDLLARGIDVQQVSLVINYDLPTNRENYIHR -------------------------------------------------------------- >37749_37749_3_HSP90AA1-EIF4A2_HSP90AA1_chr14_102549370_ENST00000334701_EIF4A2_chr3_186504915_ENST00000440191_length(amino acids)=857AA_BP=123 MPPCSGGDGSTPPGPSLRDRDCPAQSAEYPRDRLDPRPGSPSEASSPPFLRSRAPVNWYQEKAQVFLWHLMVSGSTTLLCLWKQPFHVSA FPVTASLAFRQSQGAGQHLYKDLQPFILLRLLMPEETQTQDQPMEEEEVETFAFQAEIAQLMSLIINTFYSNKEIFLRELISNSSDALDK IRYESLTDPSKLDSGKELHINLIPNKQDRTLTIVDTGIGMTKADLINNLGTIAKSGTKAFMEALQAGADISMIGQFGVGFYSAYLVAEKV TVITKHNDDEQYAWESSAGGSFTVRTDTGEPMGRGTKVILHLKEDQTEYLEERRIKEIVKKHSQFIGYPITLFVEKERDKEVSDDEAEEK EDKEEEKEKEEKESEDKPEIEDVGSDEEEEKKDGDKKKKKKIKEKYIDQEELNKTKPIWTRNPDDITNEEYGEFYKSLTNDWEDHLAVKH FSVEGQLEFRALLFVPRRAPFDLFENRKKKNNIKLYVRRVFIMDNCEELIPEYLNFIRGVVDSEDLPLNISREMLQQSKILKVIRKNLVK KCLELFTELAEDKENYKKFYEQFSKNIKLGIHEDSQNRKKLSELLRYYTSASGDEMVSLKDYCTRMKENQKHIYYITGETKDQVANSAFV ERLRKHGLEVIYMIEPIDEYCVQQLKEFEGKTLVSVTKEGLELPEDEEEKKKQEEKKTKFENLCKIMKDILEKKVEKEWKLDTLCDLYET LTITQAVIFLNTRRKVDWLTEKMHARDFTVSALHGDMDQKERDVIMREFRSGSSRVLITTDLLARGIDVQQVSLVINYDLPTNRENYIHR -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:102549370/chr3:186504915) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| HSP90AA1 | EIF4A2 |
| FUNCTION: Molecular chaperone that promotes the maturation, structural maintenance and proper regulation of specific target proteins involved for instance in cell cycle control and signal transduction. Undergoes a functional cycle that is linked to its ATPase activity which is essential for its chaperone activity. This cycle probably induces conformational changes in the client proteins, thereby causing their activation. Interacts dynamically with various co-chaperones that modulate its substrate recognition, ATPase cycle and chaperone function (PubMed:11274138, PubMed:15577939, PubMed:15937123, PubMed:27353360, PubMed:29127155, PubMed:12526792). Engages with a range of client protein classes via its interaction with various co-chaperone proteins or complexes, that act as adapters, simultaneously able to interact with the specific client and the central chaperone itself (PubMed:29127155). Recruitment of ATP and co-chaperone followed by client protein forms a functional chaperone. After the completion of the chaperoning process, properly folded client protein and co-chaperone leave HSP90 in an ADP-bound partially open conformation and finally, ADP is released from HSP90 which acquires an open conformation for the next cycle (PubMed:27295069, PubMed:26991466). Plays a critical role in mitochondrial import, delivers preproteins to the mitochondrial import receptor TOMM70 (PubMed:12526792). Apart from its chaperone activity, it also plays a role in the regulation of the transcription machinery. HSP90 and its co-chaperones modulate transcription at least at three different levels (PubMed:25973397). In the first place, they alter the steady-state levels of certain transcription factors in response to various physiological cues(PubMed:25973397). Second, they modulate the activity of certain epigenetic modifiers, such as histone deacetylases or DNA methyl transferases, and thereby respond to the change in the environment (PubMed:25973397). Third, they participate in the eviction of histones from the promoter region of certain genes and thereby turn on gene expression (PubMed:25973397). Binds bacterial lipopolysaccharide (LPS) and mediates LPS-induced inflammatory response, including TNF secretion by monocytes (PubMed:11276205). Antagonizes STUB1-mediated inhibition of TGF-beta signaling via inhibition of STUB1-mediated SMAD3 ubiquitination and degradation (PubMed:24613385). Mediates the association of TOMM70 with IRF3 or TBK1 in mitochodria outer membrane which promotes host antiviral response (PubMed:20628368, PubMed:25609812). {ECO:0000269|PubMed:11274138, ECO:0000269|PubMed:11276205, ECO:0000269|PubMed:12526792, ECO:0000269|PubMed:15577939, ECO:0000269|PubMed:15937123, ECO:0000269|PubMed:20628368, ECO:0000269|PubMed:24613385, ECO:0000269|PubMed:25609812, ECO:0000269|PubMed:27353360, ECO:0000269|PubMed:29127155, ECO:0000303|PubMed:25973397, ECO:0000303|PubMed:26991466, ECO:0000303|PubMed:27295069}. | FUNCTION: ATP-dependent RNA helicase which is a subunit of the eIF4F complex involved in cap recognition and is required for mRNA binding to ribosome. In the current model of translation initiation, eIF4A unwinds RNA secondary structures in the 5'-UTR of mRNAs which is necessary to allow efficient binding of the small ribosomal subunit, and subsequent scanning for the initiator codon. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | HSP90AA1 | chr14:102549370 | chr3:186504915 | ENST00000216281 | - | 9 | 11 | 723_732 | 585.0 | 733.0 | Motif | TPR repeat-binding |
| Hgene | HSP90AA1 | chr14:102549370 | chr3:186504915 | ENST00000334701 | - | 10 | 12 | 723_732 | 707.0 | 855.0 | Motif | TPR repeat-binding |
| Hgene | HSP90AA1 | chr14:102549370 | chr3:186504915 | ENST00000216281 | - | 9 | 11 | 682_732 | 585.0 | 733.0 | Region | Required for homodimerization |
| Hgene | HSP90AA1 | chr14:102549370 | chr3:186504915 | ENST00000334701 | - | 10 | 12 | 682_732 | 707.0 | 855.0 | Region | Required for homodimerization |
| Tgene | EIF4A2 | chr14:102549370 | chr3:186504915 | ENST00000323963 | 6 | 11 | 246_407 | 257.0 | 408.0 | Domain | Helicase C-terminal | |
| Tgene | EIF4A2 | chr14:102549370 | chr3:186504915 | ENST00000323963 | 6 | 11 | 64_235 | 257.0 | 408.0 | Domain | Helicase ATP-binding | |
| Tgene | EIF4A2 | chr14:102549370 | chr3:186504915 | ENST00000440191 | 6 | 11 | 246_407 | 258.0 | 409.0 | Domain | Helicase C-terminal | |
| Tgene | EIF4A2 | chr14:102549370 | chr3:186504915 | ENST00000440191 | 6 | 11 | 64_235 | 258.0 | 409.0 | Domain | Helicase ATP-binding | |
| Tgene | EIF4A2 | chr14:102549370 | chr3:186504915 | ENST00000323963 | 6 | 11 | 183_186 | 257.0 | 408.0 | Motif | Note=DEAD box | |
| Tgene | EIF4A2 | chr14:102549370 | chr3:186504915 | ENST00000323963 | 6 | 11 | 33_61 | 257.0 | 408.0 | Motif | Note=Q motif | |
| Tgene | EIF4A2 | chr14:102549370 | chr3:186504915 | ENST00000440191 | 6 | 11 | 183_186 | 258.0 | 409.0 | Motif | Note=DEAD box | |
| Tgene | EIF4A2 | chr14:102549370 | chr3:186504915 | ENST00000440191 | 6 | 11 | 33_61 | 258.0 | 409.0 | Motif | Note=Q motif | |
| Tgene | EIF4A2 | chr14:102549370 | chr3:186504915 | ENST00000323963 | 6 | 11 | 77_84 | 257.0 | 408.0 | Nucleotide binding | ATP | |
| Tgene | EIF4A2 | chr14:102549370 | chr3:186504915 | ENST00000440191 | 6 | 11 | 77_84 | 258.0 | 409.0 | Nucleotide binding | ATP |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| HSP90AA1 | |
| EIF4A2 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | HSP90AA1 | chr14:102549370 | chr3:186504915 | ENST00000216281 | - | 9 | 11 | 284_732 | 585.0 | 733.0 | FLCN and FNIP1 |
| Hgene | HSP90AA1 | chr14:102549370 | chr3:186504915 | ENST00000334701 | - | 10 | 12 | 284_732 | 707.0 | 855.0 | FLCN and FNIP1 |
| Hgene | HSP90AA1 | chr14:102549370 | chr3:186504915 | ENST00000216281 | - | 9 | 11 | 284_620 | 585.0 | 733.0 | FNIP2 and TSC1 |
| Hgene | HSP90AA1 | chr14:102549370 | chr3:186504915 | ENST00000216281 | - | 9 | 11 | 628_731 | 585.0 | 733.0 | NR1D1 |
| Hgene | HSP90AA1 | chr14:102549370 | chr3:186504915 | ENST00000334701 | - | 10 | 12 | 628_731 | 707.0 | 855.0 | NR1D1 |
| Hgene | HSP90AA1 | chr14:102549370 | chr3:186504915 | ENST00000216281 | - | 9 | 11 | 271_616 | 585.0 | 733.0 | NR3C1 |
Top |
Related Drugs to HSP90AA1-EIF4A2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to HSP90AA1-EIF4A2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

