| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:IGF1R-ARID1B |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: IGF1R-ARID1B | FusionPDB ID: 38471 | FusionGDB2.0 ID: 38471 | Hgene | Tgene | Gene symbol | IGF1R | ARID1B | Gene ID | 3480 | 57492 |
| Gene name | insulin like growth factor 1 receptor | AT-rich interaction domain 1B | |
| Synonyms | CD221|IGFIR|IGFR|JTK13 | 6A3-5|BAF250B|BRIGHT|CSS1|DAN15|ELD/OSA1|MRD12|OSA2|P250R | |
| Cytomap | 15q26.3 | 6q25.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | insulin-like growth factor 1 receptorIGF-I receptorsoluble IGF1R variant 1soluble IGF1R variant 2 | AT-rich interactive domain-containing protein 1BARID domain-containing protein 1BAT rich interactive domain 1B (SWI1-like)BRG1-associated factor 250bBRG1-binding protein ELD/OSA1ELD (eyelid)/OSA protein | |
| Modification date | 20200329 | 20200320 | |
| UniProtAcc | P08069 | Q8NFD5 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000268035, ENST00000558762, ENST00000560432, | ENST00000478761, ENST00000275248, ENST00000346085, ENST00000350026, ENST00000367148, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 24 X 15 X 6=2160 | 10 X 12 X 6=720 |
| # samples | 25 | 12 | |
| ** MAII score | log2(25/2160*10)=-3.11103131238874 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/720*10)=-2.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: IGF1R [Title/Abstract] AND ARID1B [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | IGF1R(99251336)-ARID1B(157469758), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | IGF1R-ARID1B seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. IGF1R-ARID1B seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. IGF1R-ARID1B seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. IGF1R-ARID1B seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | IGF1R | GO:0043066 | negative regulation of apoptotic process | 12556535 |
| Hgene | IGF1R | GO:0046328 | regulation of JNK cascade | 12556535 |
| Hgene | IGF1R | GO:0046777 | protein autophosphorylation | 1846292|7679099|11162456 |
| Hgene | IGF1R | GO:0048009 | insulin-like growth factor receptor signaling pathway | 7679099 |
| Hgene | IGF1R | GO:0048015 | phosphatidylinositol-mediated signaling | 7692086 |
| Hgene | IGF1R | GO:0051389 | inactivation of MAPKK activity | 12556535 |
 Fusion gene breakpoints across IGF1R (5'-gene) Fusion gene breakpoints across IGF1R (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
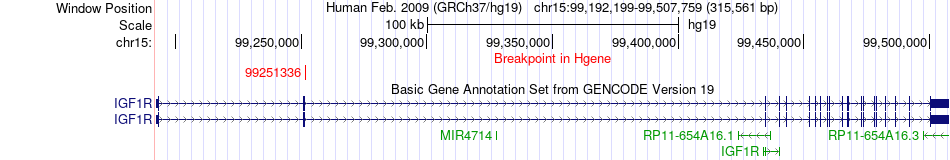 |
 Fusion gene breakpoints across ARID1B (3'-gene) Fusion gene breakpoints across ARID1B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
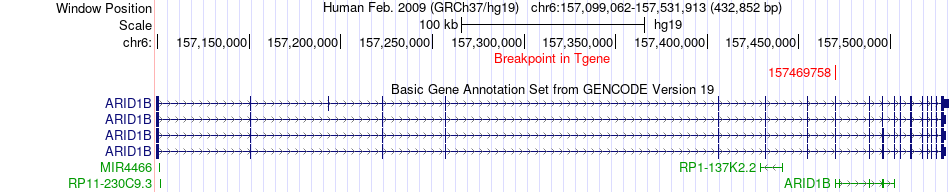 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-E9-A1NF-01A | IGF1R | chr15 | 99251336 | - | ARID1B | chr6 | 157469758 | + |
| ChimerDB4 | BRCA | TCGA-E9-A1NF-01A | IGF1R | chr15 | 99251336 | + | ARID1B | chr6 | 157469758 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000268035 | IGF1R | chr15 | 99251336 | + | ENST00000350026 | ARID1B | chr6 | 157469758 | + | 6709 | 1251 | 611 | 5449 | 1612 |
| ENST00000268035 | IGF1R | chr15 | 99251336 | + | ENST00000346085 | ARID1B | chr6 | 157469758 | + | 8338 | 1251 | 611 | 5449 | 1612 |
| ENST00000268035 | IGF1R | chr15 | 99251336 | + | ENST00000367148 | ARID1B | chr6 | 157469758 | + | 6985 | 1251 | 611 | 5608 | 1665 |
| ENST00000268035 | IGF1R | chr15 | 99251336 | + | ENST00000275248 | ARID1B | chr6 | 157469758 | + | 6985 | 1251 | 611 | 5608 | 1665 |
| ENST00000558762 | IGF1R | chr15 | 99251336 | + | ENST00000350026 | ARID1B | chr6 | 157469758 | + | 6636 | 1178 | 538 | 5376 | 1612 |
| ENST00000558762 | IGF1R | chr15 | 99251336 | + | ENST00000346085 | ARID1B | chr6 | 157469758 | + | 8265 | 1178 | 538 | 5376 | 1612 |
| ENST00000558762 | IGF1R | chr15 | 99251336 | + | ENST00000367148 | ARID1B | chr6 | 157469758 | + | 6912 | 1178 | 538 | 5535 | 1665 |
| ENST00000558762 | IGF1R | chr15 | 99251336 | + | ENST00000275248 | ARID1B | chr6 | 157469758 | + | 6912 | 1178 | 538 | 5535 | 1665 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000268035 | ENST00000350026 | IGF1R | chr15 | 99251336 | + | ARID1B | chr6 | 157469758 | + | 0.002961082 | 0.99703896 |
| ENST00000268035 | ENST00000346085 | IGF1R | chr15 | 99251336 | + | ARID1B | chr6 | 157469758 | + | 0.001095424 | 0.9989046 |
| ENST00000268035 | ENST00000367148 | IGF1R | chr15 | 99251336 | + | ARID1B | chr6 | 157469758 | + | 0.002819619 | 0.99718034 |
| ENST00000268035 | ENST00000275248 | IGF1R | chr15 | 99251336 | + | ARID1B | chr6 | 157469758 | + | 0.002819619 | 0.99718034 |
| ENST00000558762 | ENST00000350026 | IGF1R | chr15 | 99251336 | + | ARID1B | chr6 | 157469758 | + | 0.002691637 | 0.9973084 |
| ENST00000558762 | ENST00000346085 | IGF1R | chr15 | 99251336 | + | ARID1B | chr6 | 157469758 | + | 0.001014489 | 0.99898547 |
| ENST00000558762 | ENST00000367148 | IGF1R | chr15 | 99251336 | + | ARID1B | chr6 | 157469758 | + | 0.002552976 | 0.997447 |
| ENST00000558762 | ENST00000275248 | IGF1R | chr15 | 99251336 | + | ARID1B | chr6 | 157469758 | + | 0.002552976 | 0.997447 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >38471_38471_1_IGF1R-ARID1B_IGF1R_chr15_99251336_ENST00000268035_ARID1B_chr6_157469758_ENST00000275248_length(amino acids)=1665AA_BP=214 MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYRFPKLTVITEYLLLFRV AGLESLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKSNYSRPPAYSGVPSASYSGPGPGMGISANNQMHGQGPSQPCGAVPLGRMPSAGMQNR PFPGNMSSMTPSSPGMSQQGGPGMGPPMPTVNRKAQEAAAAVMQAAANSAQSRQGSFPGMNQSGLMASSSPYSQPMNNSSSLMNTQAPPY SMAPAMVNSSAASVGLADMMSPGESKLPLPLKADGKEEGTPQPESKSKDSYSSQGISQPPTPGNLPVPSPMSPSSASISSFHGDESDSIS SPGWPKTPSSPKSSSSTTTGEKITKVYELGNEPERKLWVDRYLTFMEERGSPVSSLPAVGKKPLDLFRLYVCVKEIGGLAQVNKNKKWRE LATNLNVGTSSSAASSLKKQYIQYLFAFECKIERGEEPPPEVFSTGDTKKQPKLQPPSPANSGSLQGPQTPQSTGSNSMAEVPGDLKPPT PASTPHGQMTPMQGGRSSTISVHDPFSDVSDSSFPKRNSMTPNAPYQQGMSMPDVMGRMPYEPNKDPFGGMRKVPGSSEPFMTQGQMPNS SMQDMYNQSPSGAMSNLGMGQRQQFPYGASYDRRHEPYGQQYPGQGPPSGQPPYGGHQPGLYPQQPNYKRHMDGMYGPPAKRHEGDMYNM QYSSQQQEMYNQYGGSYSGPDRRPIQGQYPYPYSRERMQGPGQIQTHGIPPQMMGGPLQSSSSEGPQQNMWAARNDMPYPYQNRQGPGGP TQAPPYPGMNRTDDMMVPDQRINHESQWPSHVSQRQPYMSSSASMQPITRPPQPSYQTPPSLPNHISRAPSPASFQRSLENRMSPSKSPF LPSMKMQKVMPTVPTSQVTGPPPQPPPIRREITFPPGSVEASQPVLKQRRKITSKDIVTPEAWRVMMSLKSGLLAESTWALDTINILLYD DSTVATFNLSQLSGFLELLVEYFRKCLIDIFGILMEYEVGDPSQKALDHNAARKDDSQSLADDSGKEEEDAECIDDDEEDEEDEEEDSEK TESDEKSSIALTAPDAAADPKEKPKQASKFDKLPIKIVKKNNLFVVDRSDKLGRVQEFNSGLLHWQLGGGDTTEHIQTHFESKMEIPPRR RPPPPLSSAGRKKEQEGKGDSEEQQEKSIIATIDDVLSARPGALPEDANPGPQTESSKFPFGIQQAKSHRNIKLLEDEPRSRDETPLCTI AHWQDSLAKRCICVSNIVRSLSFVPGNDAEMSKHPGLVLILGKLILLHHEHPERKRAPQTYEKEEDEDKGVACSKDEWWWDCLEVLRDNT LVTLANISGQLDLSAYTESICLPILDGLLHWMVCPSAEAQDPFPTVGPNSVLSPQRLVLETLCKLSIQDNNVDLILATPPFSRQEKFYAT LVRYVGDRKNPVCREMSMALLSNLAQGDALAARAIAVQKGSIGNLISFLEDGVTMAQYQQSQHNLMHMQPPPLEPPSVDMMCRAAKALLA -------------------------------------------------------------- >38471_38471_2_IGF1R-ARID1B_IGF1R_chr15_99251336_ENST00000268035_ARID1B_chr6_157469758_ENST00000346085_length(amino acids)=1612AA_BP=214 MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYRFPKLTVITEYLLLFRV AGLESLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKSNYSRPPAYSGVPSASYSGPGPGMGISANNQMHGQGPSQPCGAVPLGRMPSAGMQNR PFPGNMSSMTPSSPGMSQQGGPGMGPPMPTVNRKAQEAAAAVMQAAANSAQSRQGSFPGMNQSGLMASSSPYSQPMNNSSSLMNTQAPPY SMAPAMVNSSAASVGLADMMSPGESKLPLPLKADGKEEGTPQPESKSKKSSSSTTTGEKITKVYELGNEPERKLWVDRYLTFMEERGSPV SSLPAVGKKPLDLFRLYVCVKEIGGLAQVNKNKKWRELATNLNVGTSSSAASSLKKQYIQYLFAFECKIERGEEPPPEVFSTGDTKKQPK LQPPSPANSGSLQGPQTPQSTGSNSMAEVPGDLKPPTPASTPHGQMTPMQGGRSSTISVHDPFSDVSDSSFPKRNSMTPNAPYQQGMSMP DVMGRMPYEPNKDPFGGMRKVPGSSEPFMTQGQMPNSSMQDMYNQSPSGAMSNLGMGQRQQFPYGASYDRRHEPYGQQYPGQGPPSGQPP YGGHQPGLYPQQPNYKRHMDGMYGPPAKRHEGDMYNMQYSSQQQEMYNQYGGSYSGPDRRPIQGQYPYPYSRERMQGPGQIQTHGIPPQM MGGPLQSSSSEGPQQNMWAARNDMPYPYQNRQGPGGPTQAPPYPGMNRTDDMMVPDQRINHESQWPSHVSQRQPYMSSSASMQPITRPPQ PSYQTPPSLPNHISRAPSPASFQRSLENRMSPSKSPFLPSMKMQKVMPTVPTSQVTGPPPQPPPIRREITFPPGSVEASQPVLKQRRKIT SKDIVTPEAWRVMMSLKSGLLAESTWALDTINILLYDDSTVATFNLSQLSGFLELLVEYFRKCLIDIFGILMEYEVGDPSQKALDHNAAR KDDSQSLADDSGKEEEDAECIDDDEEDEEDEEEDSEKTESDEKSSIALTAPDAAADPKEKPKQASKFDKLPIKIVKKNNLFVVDRSDKLG RVQEFNSGLLHWQLGGGDTTEHIQTHFESKMEIPPRRRPPPPLSSAGRKKEQEGKGDSEEQQEKSIIATIDDVLSARPGALPEDANPGPQ TESSKFPFGIQQAKSHRNIKLLEDEPRSRDETPLCTIAHWQDSLAKRCICVSNIVRSLSFVPGNDAEMSKHPGLVLILGKLILLHHEHPE RKRAPQTYEKEEDEDKGVACSKDEWWWDCLEVLRDNTLVTLANISGQLDLSAYTESICLPILDGLLHWMVCPSAEAQDPFPTVGPNSVLS PQRLVLETLCKLSIQDNNVDLILATPPFSRQEKFYATLVRYVGDRKNPVCREMSMALLSNLAQGDALAARAIAVQKGSIGNLISFLEDGV -------------------------------------------------------------- >38471_38471_3_IGF1R-ARID1B_IGF1R_chr15_99251336_ENST00000268035_ARID1B_chr6_157469758_ENST00000350026_length(amino acids)=1612AA_BP=214 MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYRFPKLTVITEYLLLFRV AGLESLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKSNYSRPPAYSGVPSASYSGPGPGMGISANNQMHGQGPSQPCGAVPLGRMPSAGMQNR PFPGNMSSMTPSSPGMSQQGGPGMGPPMPTVNRKAQEAAAAVMQAAANSAQSRQGSFPGMNQSGLMASSSPYSQPMNNSSSLMNTQAPPY SMAPAMVNSSAASVGLADMMSPGESKLPLPLKADGKEEGTPQPESKSKKSSSSTTTGEKITKVYELGNEPERKLWVDRYLTFMEERGSPV SSLPAVGKKPLDLFRLYVCVKEIGGLAQVNKNKKWRELATNLNVGTSSSAASSLKKQYIQYLFAFECKIERGEEPPPEVFSTGDTKKQPK LQPPSPANSGSLQGPQTPQSTGSNSMAEVPGDLKPPTPASTPHGQMTPMQGGRSSTISVHDPFSDVSDSSFPKRNSMTPNAPYQQGMSMP DVMGRMPYEPNKDPFGGMRKVPGSSEPFMTQGQMPNSSMQDMYNQSPSGAMSNLGMGQRQQFPYGASYDRRHEPYGQQYPGQGPPSGQPP YGGHQPGLYPQQPNYKRHMDGMYGPPAKRHEGDMYNMQYSSQQQEMYNQYGGSYSGPDRRPIQGQYPYPYSRERMQGPGQIQTHGIPPQM MGGPLQSSSSEGPQQNMWAARNDMPYPYQNRQGPGGPTQAPPYPGMNRTDDMMVPDQRINHESQWPSHVSQRQPYMSSSASMQPITRPPQ PSYQTPPSLPNHISRAPSPASFQRSLENRMSPSKSPFLPSMKMQKVMPTVPTSQVTGPPPQPPPIRREITFPPGSVEASQPVLKQRRKIT SKDIVTPEAWRVMMSLKSGLLAESTWALDTINILLYDDSTVATFNLSQLSGFLELLVEYFRKCLIDIFGILMEYEVGDPSQKALDHNAAR KDDSQSLADDSGKEEEDAECIDDDEEDEEDEEEDSEKTESDEKSSIALTAPDAAADPKEKPKQASKFDKLPIKIVKKNNLFVVDRSDKLG RVQEFNSGLLHWQLGGGDTTEHIQTHFESKMEIPPRRRPPPPLSSAGRKKEQEGKGDSEEQQEKSIIATIDDVLSARPGALPEDANPGPQ TESSKFPFGIQQAKSHRNIKLLEDEPRSRDETPLCTIAHWQDSLAKRCICVSNIVRSLSFVPGNDAEMSKHPGLVLILGKLILLHHEHPE RKRAPQTYEKEEDEDKGVACSKDEWWWDCLEVLRDNTLVTLANISGQLDLSAYTESICLPILDGLLHWMVCPSAEAQDPFPTVGPNSVLS PQRLVLETLCKLSIQDNNVDLILATPPFSRQEKFYATLVRYVGDRKNPVCREMSMALLSNLAQGDALAARAIAVQKGSIGNLISFLEDGV -------------------------------------------------------------- >38471_38471_4_IGF1R-ARID1B_IGF1R_chr15_99251336_ENST00000268035_ARID1B_chr6_157469758_ENST00000367148_length(amino acids)=1665AA_BP=214 MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYRFPKLTVITEYLLLFRV AGLESLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKSNYSRPPAYSGVPSASYSGPGPGMGISANNQMHGQGPSQPCGAVPLGRMPSAGMQNR PFPGNMSSMTPSSPGMSQQGGPGMGPPMPTVNRKAQEAAAAVMQAAANSAQSRQGSFPGMNQSGLMASSSPYSQPMNNSSSLMNTQAPPY SMAPAMVNSSAASVGLADMMSPGESKLPLPLKADGKEEGTPQPESKSKDSYSSQGISQPPTPGNLPVPSPMSPSSASISSFHGDESDSIS SPGWPKTPSSPKSSSSTTTGEKITKVYELGNEPERKLWVDRYLTFMEERGSPVSSLPAVGKKPLDLFRLYVCVKEIGGLAQVNKNKKWRE LATNLNVGTSSSAASSLKKQYIQYLFAFECKIERGEEPPPEVFSTGDTKKQPKLQPPSPANSGSLQGPQTPQSTGSNSMAEVPGDLKPPT PASTPHGQMTPMQGGRSSTISVHDPFSDVSDSSFPKRNSMTPNAPYQQGMSMPDVMGRMPYEPNKDPFGGMRKVPGSSEPFMTQGQMPNS SMQDMYNQSPSGAMSNLGMGQRQQFPYGASYDRRHEPYGQQYPGQGPPSGQPPYGGHQPGLYPQQPNYKRHMDGMYGPPAKRHEGDMYNM QYSSQQQEMYNQYGGSYSGPDRRPIQGQYPYPYSRERMQGPGQIQTHGIPPQMMGGPLQSSSSEGPQQNMWAARNDMPYPYQNRQGPGGP TQAPPYPGMNRTDDMMVPDQRINHESQWPSHVSQRQPYMSSSASMQPITRPPQPSYQTPPSLPNHISRAPSPASFQRSLENRMSPSKSPF LPSMKMQKVMPTVPTSQVTGPPPQPPPIRREITFPPGSVEASQPVLKQRRKITSKDIVTPEAWRVMMSLKSGLLAESTWALDTINILLYD DSTVATFNLSQLSGFLELLVEYFRKCLIDIFGILMEYEVGDPSQKALDHNAARKDDSQSLADDSGKEEEDAECIDDDEEDEEDEEEDSEK TESDEKSSIALTAPDAAADPKEKPKQASKFDKLPIKIVKKNNLFVVDRSDKLGRVQEFNSGLLHWQLGGGDTTEHIQTHFESKMEIPPRR RPPPPLSSAGRKKEQEGKGDSEEQQEKSIIATIDDVLSARPGALPEDANPGPQTESSKFPFGIQQAKSHRNIKLLEDEPRSRDETPLCTI AHWQDSLAKRCICVSNIVRSLSFVPGNDAEMSKHPGLVLILGKLILLHHEHPERKRAPQTYEKEEDEDKGVACSKDEWWWDCLEVLRDNT LVTLANISGQLDLSAYTESICLPILDGLLHWMVCPSAEAQDPFPTVGPNSVLSPQRLVLETLCKLSIQDNNVDLILATPPFSRQEKFYAT LVRYVGDRKNPVCREMSMALLSNLAQGDALAARAIAVQKGSIGNLISFLEDGVTMAQYQQSQHNLMHMQPPPLEPPSVDMMCRAAKALLA -------------------------------------------------------------- >38471_38471_5_IGF1R-ARID1B_IGF1R_chr15_99251336_ENST00000558762_ARID1B_chr6_157469758_ENST00000275248_length(amino acids)=1665AA_BP=214 MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYRFPKLTVITEYLLLFRV AGLESLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKSNYSRPPAYSGVPSASYSGPGPGMGISANNQMHGQGPSQPCGAVPLGRMPSAGMQNR PFPGNMSSMTPSSPGMSQQGGPGMGPPMPTVNRKAQEAAAAVMQAAANSAQSRQGSFPGMNQSGLMASSSPYSQPMNNSSSLMNTQAPPY SMAPAMVNSSAASVGLADMMSPGESKLPLPLKADGKEEGTPQPESKSKDSYSSQGISQPPTPGNLPVPSPMSPSSASISSFHGDESDSIS SPGWPKTPSSPKSSSSTTTGEKITKVYELGNEPERKLWVDRYLTFMEERGSPVSSLPAVGKKPLDLFRLYVCVKEIGGLAQVNKNKKWRE LATNLNVGTSSSAASSLKKQYIQYLFAFECKIERGEEPPPEVFSTGDTKKQPKLQPPSPANSGSLQGPQTPQSTGSNSMAEVPGDLKPPT PASTPHGQMTPMQGGRSSTISVHDPFSDVSDSSFPKRNSMTPNAPYQQGMSMPDVMGRMPYEPNKDPFGGMRKVPGSSEPFMTQGQMPNS SMQDMYNQSPSGAMSNLGMGQRQQFPYGASYDRRHEPYGQQYPGQGPPSGQPPYGGHQPGLYPQQPNYKRHMDGMYGPPAKRHEGDMYNM QYSSQQQEMYNQYGGSYSGPDRRPIQGQYPYPYSRERMQGPGQIQTHGIPPQMMGGPLQSSSSEGPQQNMWAARNDMPYPYQNRQGPGGP TQAPPYPGMNRTDDMMVPDQRINHESQWPSHVSQRQPYMSSSASMQPITRPPQPSYQTPPSLPNHISRAPSPASFQRSLENRMSPSKSPF LPSMKMQKVMPTVPTSQVTGPPPQPPPIRREITFPPGSVEASQPVLKQRRKITSKDIVTPEAWRVMMSLKSGLLAESTWALDTINILLYD DSTVATFNLSQLSGFLELLVEYFRKCLIDIFGILMEYEVGDPSQKALDHNAARKDDSQSLADDSGKEEEDAECIDDDEEDEEDEEEDSEK TESDEKSSIALTAPDAAADPKEKPKQASKFDKLPIKIVKKNNLFVVDRSDKLGRVQEFNSGLLHWQLGGGDTTEHIQTHFESKMEIPPRR RPPPPLSSAGRKKEQEGKGDSEEQQEKSIIATIDDVLSARPGALPEDANPGPQTESSKFPFGIQQAKSHRNIKLLEDEPRSRDETPLCTI AHWQDSLAKRCICVSNIVRSLSFVPGNDAEMSKHPGLVLILGKLILLHHEHPERKRAPQTYEKEEDEDKGVACSKDEWWWDCLEVLRDNT LVTLANISGQLDLSAYTESICLPILDGLLHWMVCPSAEAQDPFPTVGPNSVLSPQRLVLETLCKLSIQDNNVDLILATPPFSRQEKFYAT LVRYVGDRKNPVCREMSMALLSNLAQGDALAARAIAVQKGSIGNLISFLEDGVTMAQYQQSQHNLMHMQPPPLEPPSVDMMCRAAKALLA -------------------------------------------------------------- >38471_38471_6_IGF1R-ARID1B_IGF1R_chr15_99251336_ENST00000558762_ARID1B_chr6_157469758_ENST00000346085_length(amino acids)=1612AA_BP=214 MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYRFPKLTVITEYLLLFRV AGLESLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKSNYSRPPAYSGVPSASYSGPGPGMGISANNQMHGQGPSQPCGAVPLGRMPSAGMQNR PFPGNMSSMTPSSPGMSQQGGPGMGPPMPTVNRKAQEAAAAVMQAAANSAQSRQGSFPGMNQSGLMASSSPYSQPMNNSSSLMNTQAPPY SMAPAMVNSSAASVGLADMMSPGESKLPLPLKADGKEEGTPQPESKSKKSSSSTTTGEKITKVYELGNEPERKLWVDRYLTFMEERGSPV SSLPAVGKKPLDLFRLYVCVKEIGGLAQVNKNKKWRELATNLNVGTSSSAASSLKKQYIQYLFAFECKIERGEEPPPEVFSTGDTKKQPK LQPPSPANSGSLQGPQTPQSTGSNSMAEVPGDLKPPTPASTPHGQMTPMQGGRSSTISVHDPFSDVSDSSFPKRNSMTPNAPYQQGMSMP DVMGRMPYEPNKDPFGGMRKVPGSSEPFMTQGQMPNSSMQDMYNQSPSGAMSNLGMGQRQQFPYGASYDRRHEPYGQQYPGQGPPSGQPP YGGHQPGLYPQQPNYKRHMDGMYGPPAKRHEGDMYNMQYSSQQQEMYNQYGGSYSGPDRRPIQGQYPYPYSRERMQGPGQIQTHGIPPQM MGGPLQSSSSEGPQQNMWAARNDMPYPYQNRQGPGGPTQAPPYPGMNRTDDMMVPDQRINHESQWPSHVSQRQPYMSSSASMQPITRPPQ PSYQTPPSLPNHISRAPSPASFQRSLENRMSPSKSPFLPSMKMQKVMPTVPTSQVTGPPPQPPPIRREITFPPGSVEASQPVLKQRRKIT SKDIVTPEAWRVMMSLKSGLLAESTWALDTINILLYDDSTVATFNLSQLSGFLELLVEYFRKCLIDIFGILMEYEVGDPSQKALDHNAAR KDDSQSLADDSGKEEEDAECIDDDEEDEEDEEEDSEKTESDEKSSIALTAPDAAADPKEKPKQASKFDKLPIKIVKKNNLFVVDRSDKLG RVQEFNSGLLHWQLGGGDTTEHIQTHFESKMEIPPRRRPPPPLSSAGRKKEQEGKGDSEEQQEKSIIATIDDVLSARPGALPEDANPGPQ TESSKFPFGIQQAKSHRNIKLLEDEPRSRDETPLCTIAHWQDSLAKRCICVSNIVRSLSFVPGNDAEMSKHPGLVLILGKLILLHHEHPE RKRAPQTYEKEEDEDKGVACSKDEWWWDCLEVLRDNTLVTLANISGQLDLSAYTESICLPILDGLLHWMVCPSAEAQDPFPTVGPNSVLS PQRLVLETLCKLSIQDNNVDLILATPPFSRQEKFYATLVRYVGDRKNPVCREMSMALLSNLAQGDALAARAIAVQKGSIGNLISFLEDGV -------------------------------------------------------------- >38471_38471_7_IGF1R-ARID1B_IGF1R_chr15_99251336_ENST00000558762_ARID1B_chr6_157469758_ENST00000350026_length(amino acids)=1612AA_BP=214 MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYRFPKLTVITEYLLLFRV AGLESLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKSNYSRPPAYSGVPSASYSGPGPGMGISANNQMHGQGPSQPCGAVPLGRMPSAGMQNR PFPGNMSSMTPSSPGMSQQGGPGMGPPMPTVNRKAQEAAAAVMQAAANSAQSRQGSFPGMNQSGLMASSSPYSQPMNNSSSLMNTQAPPY SMAPAMVNSSAASVGLADMMSPGESKLPLPLKADGKEEGTPQPESKSKKSSSSTTTGEKITKVYELGNEPERKLWVDRYLTFMEERGSPV SSLPAVGKKPLDLFRLYVCVKEIGGLAQVNKNKKWRELATNLNVGTSSSAASSLKKQYIQYLFAFECKIERGEEPPPEVFSTGDTKKQPK LQPPSPANSGSLQGPQTPQSTGSNSMAEVPGDLKPPTPASTPHGQMTPMQGGRSSTISVHDPFSDVSDSSFPKRNSMTPNAPYQQGMSMP DVMGRMPYEPNKDPFGGMRKVPGSSEPFMTQGQMPNSSMQDMYNQSPSGAMSNLGMGQRQQFPYGASYDRRHEPYGQQYPGQGPPSGQPP YGGHQPGLYPQQPNYKRHMDGMYGPPAKRHEGDMYNMQYSSQQQEMYNQYGGSYSGPDRRPIQGQYPYPYSRERMQGPGQIQTHGIPPQM MGGPLQSSSSEGPQQNMWAARNDMPYPYQNRQGPGGPTQAPPYPGMNRTDDMMVPDQRINHESQWPSHVSQRQPYMSSSASMQPITRPPQ PSYQTPPSLPNHISRAPSPASFQRSLENRMSPSKSPFLPSMKMQKVMPTVPTSQVTGPPPQPPPIRREITFPPGSVEASQPVLKQRRKIT SKDIVTPEAWRVMMSLKSGLLAESTWALDTINILLYDDSTVATFNLSQLSGFLELLVEYFRKCLIDIFGILMEYEVGDPSQKALDHNAAR KDDSQSLADDSGKEEEDAECIDDDEEDEEDEEEDSEKTESDEKSSIALTAPDAAADPKEKPKQASKFDKLPIKIVKKNNLFVVDRSDKLG RVQEFNSGLLHWQLGGGDTTEHIQTHFESKMEIPPRRRPPPPLSSAGRKKEQEGKGDSEEQQEKSIIATIDDVLSARPGALPEDANPGPQ TESSKFPFGIQQAKSHRNIKLLEDEPRSRDETPLCTIAHWQDSLAKRCICVSNIVRSLSFVPGNDAEMSKHPGLVLILGKLILLHHEHPE RKRAPQTYEKEEDEDKGVACSKDEWWWDCLEVLRDNTLVTLANISGQLDLSAYTESICLPILDGLLHWMVCPSAEAQDPFPTVGPNSVLS PQRLVLETLCKLSIQDNNVDLILATPPFSRQEKFYATLVRYVGDRKNPVCREMSMALLSNLAQGDALAARAIAVQKGSIGNLISFLEDGV -------------------------------------------------------------- >38471_38471_8_IGF1R-ARID1B_IGF1R_chr15_99251336_ENST00000558762_ARID1B_chr6_157469758_ENST00000367148_length(amino acids)=1665AA_BP=214 MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYRFPKLTVITEYLLLFRV AGLESLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKSNYSRPPAYSGVPSASYSGPGPGMGISANNQMHGQGPSQPCGAVPLGRMPSAGMQNR PFPGNMSSMTPSSPGMSQQGGPGMGPPMPTVNRKAQEAAAAVMQAAANSAQSRQGSFPGMNQSGLMASSSPYSQPMNNSSSLMNTQAPPY SMAPAMVNSSAASVGLADMMSPGESKLPLPLKADGKEEGTPQPESKSKDSYSSQGISQPPTPGNLPVPSPMSPSSASISSFHGDESDSIS SPGWPKTPSSPKSSSSTTTGEKITKVYELGNEPERKLWVDRYLTFMEERGSPVSSLPAVGKKPLDLFRLYVCVKEIGGLAQVNKNKKWRE LATNLNVGTSSSAASSLKKQYIQYLFAFECKIERGEEPPPEVFSTGDTKKQPKLQPPSPANSGSLQGPQTPQSTGSNSMAEVPGDLKPPT PASTPHGQMTPMQGGRSSTISVHDPFSDVSDSSFPKRNSMTPNAPYQQGMSMPDVMGRMPYEPNKDPFGGMRKVPGSSEPFMTQGQMPNS SMQDMYNQSPSGAMSNLGMGQRQQFPYGASYDRRHEPYGQQYPGQGPPSGQPPYGGHQPGLYPQQPNYKRHMDGMYGPPAKRHEGDMYNM QYSSQQQEMYNQYGGSYSGPDRRPIQGQYPYPYSRERMQGPGQIQTHGIPPQMMGGPLQSSSSEGPQQNMWAARNDMPYPYQNRQGPGGP TQAPPYPGMNRTDDMMVPDQRINHESQWPSHVSQRQPYMSSSASMQPITRPPQPSYQTPPSLPNHISRAPSPASFQRSLENRMSPSKSPF LPSMKMQKVMPTVPTSQVTGPPPQPPPIRREITFPPGSVEASQPVLKQRRKITSKDIVTPEAWRVMMSLKSGLLAESTWALDTINILLYD DSTVATFNLSQLSGFLELLVEYFRKCLIDIFGILMEYEVGDPSQKALDHNAARKDDSQSLADDSGKEEEDAECIDDDEEDEEDEEEDSEK TESDEKSSIALTAPDAAADPKEKPKQASKFDKLPIKIVKKNNLFVVDRSDKLGRVQEFNSGLLHWQLGGGDTTEHIQTHFESKMEIPPRR RPPPPLSSAGRKKEQEGKGDSEEQQEKSIIATIDDVLSARPGALPEDANPGPQTESSKFPFGIQQAKSHRNIKLLEDEPRSRDETPLCTI AHWQDSLAKRCICVSNIVRSLSFVPGNDAEMSKHPGLVLILGKLILLHHEHPERKRAPQTYEKEEDEDKGVACSKDEWWWDCLEVLRDNT LVTLANISGQLDLSAYTESICLPILDGLLHWMVCPSAEAQDPFPTVGPNSVLSPQRLVLETLCKLSIQDNNVDLILATPPFSRQEKFYAT LVRYVGDRKNPVCREMSMALLSNLAQGDALAARAIAVQKGSIGNLISFLEDGVTMAQYQQSQHNLMHMQPPPLEPPSVDMMCRAAKALLA -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:99251336/chr6:157469758) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| IGF1R | ARID1B |
| FUNCTION: Receptor tyrosine kinase which mediates actions of insulin-like growth factor 1 (IGF1). Binds IGF1 with high affinity and IGF2 and insulin (INS) with a lower affinity. The activated IGF1R is involved in cell growth and survival control. IGF1R is crucial for tumor transformation and survival of malignant cell. Ligand binding activates the receptor kinase, leading to receptor autophosphorylation, and tyrosines phosphorylation of multiple substrates, that function as signaling adapter proteins including, the insulin-receptor substrates (IRS1/2), Shc and 14-3-3 proteins. Phosphorylation of IRSs proteins lead to the activation of two main signaling pathways: the PI3K-AKT/PKB pathway and the Ras-MAPK pathway. The result of activating the MAPK pathway is increased cellular proliferation, whereas activating the PI3K pathway inhibits apoptosis and stimulates protein synthesis. Phosphorylated IRS1 can activate the 85 kDa regulatory subunit of PI3K (PIK3R1), leading to activation of several downstream substrates, including protein AKT/PKB. AKT phosphorylation, in turn, enhances protein synthesis through mTOR activation and triggers the antiapoptotic effects of IGFIR through phosphorylation and inactivation of BAD. In parallel to PI3K-driven signaling, recruitment of Grb2/SOS by phosphorylated IRS1 or Shc leads to recruitment of Ras and activation of the ras-MAPK pathway. In addition to these two main signaling pathways IGF1R signals also through the Janus kinase/signal transducer and activator of transcription pathway (JAK/STAT). Phosphorylation of JAK proteins can lead to phosphorylation/activation of signal transducers and activators of transcription (STAT) proteins. In particular activation of STAT3, may be essential for the transforming activity of IGF1R. The JAK/STAT pathway activates gene transcription and may be responsible for the transforming activity. JNK kinases can also be activated by the IGF1R. IGF1 exerts inhibiting activities on JNK activation via phosphorylation and inhibition of MAP3K5/ASK1, which is able to directly associate with the IGF1R.; FUNCTION: When present in a hybrid receptor with INSR, binds IGF1. PubMed:12138094 shows that hybrid receptors composed of IGF1R and INSR isoform Long are activated with a high affinity by IGF1, with low affinity by IGF2 and not significantly activated by insulin, and that hybrid receptors composed of IGF1R and INSR isoform Short are activated by IGF1, IGF2 and insulin. In contrast, PubMed:16831875 shows that hybrid receptors composed of IGF1R and INSR isoform Long and hybrid receptors composed of IGF1R and INSR isoform Short have similar binding characteristics, both bind IGF1 and have a low affinity for insulin. | FUNCTION: Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Component of SWI/SNF chromatin remodeling complexes that carry out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth (By similarity). Binds DNA non-specifically (PubMed:14982958, PubMed:15170388). {ECO:0000250|UniProtKB:E9Q4N7, ECO:0000269|PubMed:14982958, ECO:0000269|PubMed:15170388, ECO:0000303|PubMed:12672490, ECO:0000303|PubMed:22952240, ECO:0000303|PubMed:26601204}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 1034_1037 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 1441_1444 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 1459_1597 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 1833_1836 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 932_935 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 1034_1037 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 1441_1444 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 1459_1597 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 1833_1836 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 932_935 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 1034_1037 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 1441_1444 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 1459_1597 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Pro-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 1833_1836 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 932_935 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 1053_1144 | 850.3333333333334 | 2250.0 | Domain | ARID | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 1053_1144 | 837.3333333333334 | 2237.0 | Domain | ARID | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 1053_1144 | 837.3333333333334 | 2290.0 | Domain | ARID | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 1358_1377 | 850.3333333333334 | 2250.0 | Motif | Nuclear localization signal | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 2036_2040 | 850.3333333333334 | 2250.0 | Motif | Note=LXXLL | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 1358_1377 | 837.3333333333334 | 2237.0 | Motif | Nuclear localization signal | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 2036_2040 | 837.3333333333334 | 2237.0 | Motif | Note=LXXLL | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 1358_1377 | 837.3333333333334 | 2290.0 | Motif | Nuclear localization signal | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 2036_2040 | 837.3333333333334 | 2290.0 | Motif | Note=LXXLL |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IGF1R | chr15:99251336 | chr6:157469758 | ENST00000268035 | + | 2 | 21 | 491_609 | 213.33333333333334 | 1368.0 | Domain | Fibronectin type-III 1 |
| Hgene | IGF1R | chr15:99251336 | chr6:157469758 | ENST00000268035 | + | 2 | 21 | 610_708 | 213.33333333333334 | 1368.0 | Domain | Fibronectin type-III 2 |
| Hgene | IGF1R | chr15:99251336 | chr6:157469758 | ENST00000268035 | + | 2 | 21 | 735_828 | 213.33333333333334 | 1368.0 | Domain | Fibronectin type-III 3 |
| Hgene | IGF1R | chr15:99251336 | chr6:157469758 | ENST00000268035 | + | 2 | 21 | 834_927 | 213.33333333333334 | 1368.0 | Domain | Fibronectin type-III 4 |
| Hgene | IGF1R | chr15:99251336 | chr6:157469758 | ENST00000268035 | + | 2 | 21 | 999_1274 | 213.33333333333334 | 1368.0 | Domain | Protein kinase |
| Hgene | IGF1R | chr15:99251336 | chr6:157469758 | ENST00000268035 | + | 2 | 21 | 977_980 | 213.33333333333334 | 1368.0 | Motif | Note=IRS1- and SHC1-binding |
| Hgene | IGF1R | chr15:99251336 | chr6:157469758 | ENST00000268035 | + | 2 | 21 | 1005_1013 | 213.33333333333334 | 1368.0 | Nucleotide binding | ATP |
| Hgene | IGF1R | chr15:99251336 | chr6:157469758 | ENST00000268035 | + | 2 | 21 | 741_935 | 213.33333333333334 | 1368.0 | Topological domain | Extracellular |
| Hgene | IGF1R | chr15:99251336 | chr6:157469758 | ENST00000268035 | + | 2 | 21 | 960_1367 | 213.33333333333334 | 1368.0 | Topological domain | Cytoplasmic |
| Hgene | IGF1R | chr15:99251336 | chr6:157469758 | ENST00000268035 | + | 2 | 21 | 936_959 | 213.33333333333334 | 1368.0 | Transmembrane | Helical |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 107_131 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Gln-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 114_131 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 141_401 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 2_47 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Ala-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 329_493 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Ala-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 35_57 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 574_633 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Gln-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 684_771 | 850.3333333333334 | 2250.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 81_104 | 850.3333333333334 | 2250.0 | Compositional bias | Note=His-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 107_131 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Gln-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 114_131 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 141_401 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 2_47 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Ala-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 329_493 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Ala-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 35_57 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 574_633 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Gln-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 684_771 | 837.3333333333334 | 2237.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 81_104 | 837.3333333333334 | 2237.0 | Compositional bias | Note=His-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 107_131 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Gln-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 114_131 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Poly-Gln | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 141_401 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 2_47 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Ala-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 329_493 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Ala-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 35_57 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 574_633 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Gln-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 684_771 | 837.3333333333334 | 2290.0 | Compositional bias | Note=Ser-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 81_104 | 837.3333333333334 | 2290.0 | Compositional bias | Note=His-rich | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000346085 | 7 | 20 | 419_423 | 850.3333333333334 | 2250.0 | Motif | Note=LXXLL | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000350026 | 6 | 19 | 419_423 | 837.3333333333334 | 2237.0 | Motif | Note=LXXLL | |
| Tgene | ARID1B | chr15:99251336 | chr6:157469758 | ENST00000367148 | 6 | 20 | 419_423 | 837.3333333333334 | 2290.0 | Motif | Note=LXXLL |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| IGF1R | |
| ARID1B |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to IGF1R-ARID1B |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to IGF1R-ARID1B |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

