| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:IGF1R-PRPS1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: IGF1R-PRPS1 | FusionPDB ID: 38489 | FusionGDB2.0 ID: 38489 | Hgene | Tgene | Gene symbol | IGF1R | PRPS1 | Gene ID | 3480 | 221823 |
| Gene name | insulin like growth factor 1 receptor | phosphoribosyl pyrophosphate synthetase 1 like 1 | |
| Synonyms | CD221|IGFIR|IGFR|JTK13 | PRPS1|PRPS3|PRPSL|PRS-III | |
| Cytomap | 15q26.3 | 7p21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | insulin-like growth factor 1 receptorIGF-I receptorsoluble IGF1R variant 1soluble IGF1R variant 2 | ribose-phosphate pyrophosphokinase 3PRPS1-like 1phosphoribosyl pyrophosphate synthase 1-like 1phosphoribosyl pyrophosphate synthase IIIphosphoribosylpyrophosphate synthetase subunit IIIribose-phosphate diphosphokinase catalytic chain IIIribose-phosp | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | P08069 | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000268035, ENST00000558762, ENST00000560432, | ENST00000372419, ENST00000372428, ENST00000543248, ENST00000372418, ENST00000372435, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 24 X 15 X 6=2160 | 3 X 1 X 3=9 |
| # samples | 25 | 3 | |
| ** MAII score | log2(25/2160*10)=-3.11103131238874 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(3/9*10)=1.73696559416621 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: IGF1R [Title/Abstract] AND PRPS1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | IGF1R(99486281)-PRPS1(106882525), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | IGF1R-PRPS1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. IGF1R-PRPS1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. IGF1R-PRPS1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. IGF1R-PRPS1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | IGF1R | GO:0043066 | negative regulation of apoptotic process | 12556535 |
| Hgene | IGF1R | GO:0046328 | regulation of JNK cascade | 12556535 |
| Hgene | IGF1R | GO:0046777 | protein autophosphorylation | 1846292|7679099|11162456 |
| Hgene | IGF1R | GO:0048009 | insulin-like growth factor receptor signaling pathway | 7679099 |
| Hgene | IGF1R | GO:0048015 | phosphatidylinositol-mediated signaling | 7692086 |
| Hgene | IGF1R | GO:0051389 | inactivation of MAPKK activity | 12556535 |
 Fusion gene breakpoints across IGF1R (5'-gene) Fusion gene breakpoints across IGF1R (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
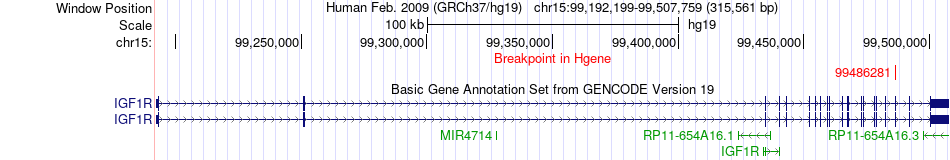 |
 Fusion gene breakpoints across PRPS1 (3'-gene) Fusion gene breakpoints across PRPS1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
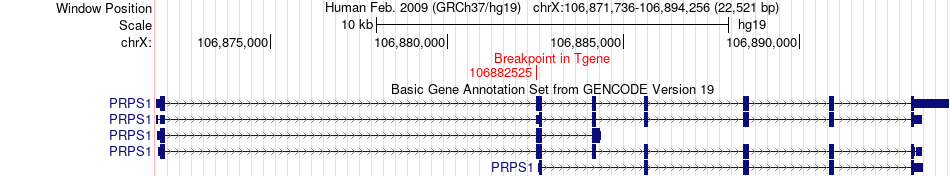 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A8-A09E-01A | IGF1R | chr15 | 99486281 | - | PRPS1 | chrX | 106882525 | + |
| ChimerDB4 | BRCA | TCGA-A8-A09E-01A | IGF1R | chr15 | 99486281 | + | PRPS1 | chrX | 106882525 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000268035 | IGF1R | chr15 | 99486281 | + | ENST00000372435 | PRPS1 | chrX | 106882525 | + | 6027 | 4198 | 611 | 5032 | 1473 |
| ENST00000558762 | IGF1R | chr15 | 99486281 | + | ENST00000372435 | PRPS1 | chrX | 106882525 | + | 5951 | 4122 | 538 | 4956 | 1472 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000268035 | ENST00000372435 | IGF1R | chr15 | 99486281 | + | PRPS1 | chrX | 106882525 | + | 0.001538465 | 0.9984616 |
| ENST00000558762 | ENST00000372435 | IGF1R | chr15 | 99486281 | + | PRPS1 | chrX | 106882525 | + | 0.001583534 | 0.9984164 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >38489_38489_1_IGF1R-PRPS1_IGF1R_chr15_99486281_ENST00000268035_PRPS1_chrX_106882525_ENST00000372435_length(amino acids)=1473AA_BP=1196 MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYRFPKLTVITEYLLLFRV AGLESLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKMCPSTCGKRACTENNECCHPECLGSCSAPDNDTACVACRHYYYAGVCVPACPPNTYR FEGWRCVDRDFCANILSAESSDSEGFVIHDGECMQECPSGFIRNGSQSMYCIPCEGPCPKVCEEEKKTKTIDSVTSAQMLQGCTIFKGNL LINIRRGNNIASELENFMGLIEVVTGYVKIRHSHALVSLSFLKNLRLILGEEQLEGNYSFYVLDNQNLQQLWDWDHRNLTIKAGKMYFAF NPKLCVSEIYRMEEVTGTKGRQSKGDINTRNNGERASCESDVLHFTSTTTSKNRIIITWHRYRPPDYRDLISFTVYYKEAPFKNVTEYDG QDACGSNSWNMVDVDLPPNKDVEPGILLHGLKPWTQYAVYVKAVTLTMVENDHIRGAKSEILYIRTNASVPSIPLDVLSASNSSSQLIVK WNPPSLPNGNLSYYIVRWQRQPQDGYLYRHNYCSKDKIPIRKYADGTIDIEEVTENPKTEVCGGEKGPCCACPKTEAEKQAEKEEAEYRK VFENFLHNSIFVPRPERKRRDVMQVANTTMSSRSRNTTAADTYNITDPEELETEYPFFESRVDNKERTVISNLRPFTLYRIDIHSCNHEA EKLGCSASNFVFARTMPAEGADDIPGPVTWEPRPENSIFLKWPEPENPNGLILMYEIKYGSQVEDQRECVSRQEYRKYGGAKLNRLNPGN YTARIQATSLSGNGSWTDPVFFYVQAKTGYENFIHLIIALPVAVLLIVGGLVIMLYVFHRKRNNSRLGNGVLYASVNPEYFSAADVYVPD EWEVAREKITMSRELGQGSFGMVYEGVAKGVVKDEPETRVAIKTVNEAASMRERIEFLNEASVMKEFNCHHVVRLLGVVSQGQPTLVIME LMTRGDLKSYLRSLRPEMENNPVLAPPSLSKMIQMAGEIADGMAYLNANKFVHRDLAARNCMVAEDFTVKIGDFGMTRDIYETDYYRKGG KGLLPVRWMSPESLKDGVFTTYSDVCVEIGESVRGEDVYIVQSGCGEINDNLMELLIMINACKIASASRVTAVIPCFPYARQDKKDKSRA PISAKLVANMLSVAGADHIITMDLHASQIQGFFDIPVDNLYAEPAVLKWIRENISEWRNCTIVSPDAGGAKRVTSIADRLNVDFALIHKE RKKANEVDRMVLVGDVKDRVAILVDDMADTCGTICHAADKLLSAGATRVYAILTHGIFSGPAISRINNACFEAVVVTNTIPQEDKMKHCS -------------------------------------------------------------- >38489_38489_2_IGF1R-PRPS1_IGF1R_chr15_99486281_ENST00000558762_PRPS1_chrX_106882525_ENST00000372435_length(amino acids)=1472AA_BP=1195 MKSGSGGGSPTSLWGLLFLSAALSLWPTSGEICGPGIDIRNDYQQLKRLENCTVIEGYLHILLISKAEDYRSYRFPKLTVITEYLLLFRV AGLESLGDLFPNLTVIRGWKLFYNYALVIFEMTNLKDIGLYNLRNITRGAIRIEKNADLCYLSTVDWSLILDAVSNNYIVGNKPPKECGD LCPGTMEEKPMCEKTTINNEYNYRCWTTNRCQKMCPSTCGKRACTENNECCHPECLGSCSAPDNDTACVACRHYYYAGVCVPACPPNTYR FEGWRCVDRDFCANILSAESSDSEGFVIHDGECMQECPSGFIRNGSQSMYCIPCEGPCPKVCEEEKKTKTIDSVTSAQMLQGCTIFKGNL LINIRRGNNIASELENFMGLIEVVTGYVKIRHSHALVSLSFLKNLRLILGEEQLEGNYSFYVLDNQNLQQLWDWDHRNLTIKAGKMYFAF NPKLCVSEIYRMEEVTGTKGRQSKGDINTRNNGERASCESDVLHFTSTTTSKNRIIITWHRYRPPDYRDLISFTVYYKEAPFKNVTEYDG QDACGSNSWNMVDVDLPPNKDVEPGILLHGLKPWTQYAVYVKAVTLTMVENDHIRGAKSEILYIRTNASVPSIPLDVLSASNSSSQLIVK WNPPSLPNGNLSYYIVRWQRQPQDGYLYRHNYCSKDKIPIRKYADGTIDIEEVTENPKTEVCGGEKGPCCACPKTEAEKQAEKEEAEYRK VFENFLHNSIFVPRPERKRRDVMQVANTTMSSRSRNTTAADTYNITDPEELETEYPFFESRVDNKERTVISNLRPFTLYRIDIHSCNHEA EKLGCSASNFVFARTMPAEGADDIPGPVTWEPRPENSIFLKWPEPENPNGLILMYEIKYGSQVEDQRECVSRQEYRKYGGAKLNRLNPGN YTARIQATSLSGNGSWTDPVFFYVQAKRYENFIHLIIALPVAVLLIVGGLVIMLYVFHRKRNNSRLGNGVLYASVNPEYFSAADVYVPDE WEVAREKITMSRELGQGSFGMVYEGVAKGVVKDEPETRVAIKTVNEAASMRERIEFLNEASVMKEFNCHHVVRLLGVVSQGQPTLVIMEL MTRGDLKSYLRSLRPEMENNPVLAPPSLSKMIQMAGEIADGMAYLNANKFVHRDLAARNCMVAEDFTVKIGDFGMTRDIYETDYYRKGGK GLLPVRWMSPESLKDGVFTTYSDVCVEIGESVRGEDVYIVQSGCGEINDNLMELLIMINACKIASASRVTAVIPCFPYARQDKKDKSRAP ISAKLVANMLSVAGADHIITMDLHASQIQGFFDIPVDNLYAEPAVLKWIRENISEWRNCTIVSPDAGGAKRVTSIADRLNVDFALIHKER KKANEVDRMVLVGDVKDRVAILVDDMADTCGTICHAADKLLSAGATRVYAILTHGIFSGPAISRINNACFEAVVVTNTIPQEDKMKHCSK -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:99486281/chrX:106882525) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| IGF1R | . |
| FUNCTION: Receptor tyrosine kinase which mediates actions of insulin-like growth factor 1 (IGF1). Binds IGF1 with high affinity and IGF2 and insulin (INS) with a lower affinity. The activated IGF1R is involved in cell growth and survival control. IGF1R is crucial for tumor transformation and survival of malignant cell. Ligand binding activates the receptor kinase, leading to receptor autophosphorylation, and tyrosines phosphorylation of multiple substrates, that function as signaling adapter proteins including, the insulin-receptor substrates (IRS1/2), Shc and 14-3-3 proteins. Phosphorylation of IRSs proteins lead to the activation of two main signaling pathways: the PI3K-AKT/PKB pathway and the Ras-MAPK pathway. The result of activating the MAPK pathway is increased cellular proliferation, whereas activating the PI3K pathway inhibits apoptosis and stimulates protein synthesis. Phosphorylated IRS1 can activate the 85 kDa regulatory subunit of PI3K (PIK3R1), leading to activation of several downstream substrates, including protein AKT/PKB. AKT phosphorylation, in turn, enhances protein synthesis through mTOR activation and triggers the antiapoptotic effects of IGFIR through phosphorylation and inactivation of BAD. In parallel to PI3K-driven signaling, recruitment of Grb2/SOS by phosphorylated IRS1 or Shc leads to recruitment of Ras and activation of the ras-MAPK pathway. In addition to these two main signaling pathways IGF1R signals also through the Janus kinase/signal transducer and activator of transcription pathway (JAK/STAT). Phosphorylation of JAK proteins can lead to phosphorylation/activation of signal transducers and activators of transcription (STAT) proteins. In particular activation of STAT3, may be essential for the transforming activity of IGF1R. The JAK/STAT pathway activates gene transcription and may be responsible for the transforming activity. JNK kinases can also be activated by the IGF1R. IGF1 exerts inhibiting activities on JNK activation via phosphorylation and inhibition of MAP3K5/ASK1, which is able to directly associate with the IGF1R.; FUNCTION: When present in a hybrid receptor with INSR, binds IGF1. PubMed:12138094 shows that hybrid receptors composed of IGF1R and INSR isoform Long are activated with a high affinity by IGF1, with low affinity by IGF2 and not significantly activated by insulin, and that hybrid receptors composed of IGF1R and INSR isoform Short are activated by IGF1, IGF2 and insulin. In contrast, PubMed:16831875 shows that hybrid receptors composed of IGF1R and INSR isoform Long and hybrid receptors composed of IGF1R and INSR isoform Short have similar binding characteristics, both bind IGF1 and have a low affinity for insulin. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IGF1R | chr15:99486281 | chrX:106882525 | ENST00000268035 | + | 19 | 21 | 491_609 | 1195.6666666666667 | 1368.0 | Domain | Fibronectin type-III 1 |
| Hgene | IGF1R | chr15:99486281 | chrX:106882525 | ENST00000268035 | + | 19 | 21 | 610_708 | 1195.6666666666667 | 1368.0 | Domain | Fibronectin type-III 2 |
| Hgene | IGF1R | chr15:99486281 | chrX:106882525 | ENST00000268035 | + | 19 | 21 | 735_828 | 1195.6666666666667 | 1368.0 | Domain | Fibronectin type-III 3 |
| Hgene | IGF1R | chr15:99486281 | chrX:106882525 | ENST00000268035 | + | 19 | 21 | 834_927 | 1195.6666666666667 | 1368.0 | Domain | Fibronectin type-III 4 |
| Hgene | IGF1R | chr15:99486281 | chrX:106882525 | ENST00000268035 | + | 19 | 21 | 977_980 | 1195.6666666666667 | 1368.0 | Motif | Note=IRS1- and SHC1-binding |
| Hgene | IGF1R | chr15:99486281 | chrX:106882525 | ENST00000268035 | + | 19 | 21 | 1005_1013 | 1195.6666666666667 | 1368.0 | Nucleotide binding | ATP |
| Hgene | IGF1R | chr15:99486281 | chrX:106882525 | ENST00000268035 | + | 19 | 21 | 741_935 | 1195.6666666666667 | 1368.0 | Topological domain | Extracellular |
| Hgene | IGF1R | chr15:99486281 | chrX:106882525 | ENST00000268035 | + | 19 | 21 | 936_959 | 1195.6666666666667 | 1368.0 | Transmembrane | Helical |
| Tgene | PRPS1 | chr15:99486281 | chrX:106882525 | ENST00000372435 | 0 | 7 | 96_101 | 40.666666666666664 | 319.0 | Nucleotide binding | Note=ATP | |
| Tgene | PRPS1 | chr15:99486281 | chrX:106882525 | ENST00000543248 | 0 | 8 | 96_101 | 40.666666666666664 | 312.3333333333333 | Nucleotide binding | Note=ATP | |
| Tgene | PRPS1 | chr15:99486281 | chrX:106882525 | ENST00000372435 | 0 | 7 | 212_227 | 40.666666666666664 | 319.0 | Region | Binding of phosphoribosylpyrophosphate | |
| Tgene | PRPS1 | chr15:99486281 | chrX:106882525 | ENST00000543248 | 0 | 8 | 212_227 | 40.666666666666664 | 312.3333333333333 | Region | Binding of phosphoribosylpyrophosphate |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IGF1R | chr15:99486281 | chrX:106882525 | ENST00000268035 | + | 19 | 21 | 999_1274 | 1195.6666666666667 | 1368.0 | Domain | Protein kinase |
| Hgene | IGF1R | chr15:99486281 | chrX:106882525 | ENST00000268035 | + | 19 | 21 | 960_1367 | 1195.6666666666667 | 1368.0 | Topological domain | Cytoplasmic |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| IGF1R | |
| PRPS1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to IGF1R-PRPS1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to IGF1R-PRPS1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

