| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:IL17RC-TTLL3 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: IL17RC-TTLL3 | FusionPDB ID: 39448 | FusionGDB2.0 ID: 39448 | Hgene | Tgene | Gene symbol | IL17RC | TTLL3 | Gene ID | 84818 | 26140 |
| Gene name | interleukin 17 receptor C | tubulin tyrosine ligase like 3 | |
| Synonyms | CANDF9|IL17-RL|IL17RL | HOTTL | |
| Cytomap | 3p25.3|3p25.3-p24.1 | 3p25.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | interleukin-17 receptor CIL-17 receptor CIL-17RLIL17F receptorIL17Rhominterleukin-17 receptor homologinterleukin-17 receptor-like proteinzcytoR14 | tubulin monoglycylase TTLL3tubulin tyrosine ligase-like family, member 3tubulin--tyrosine ligase-like protein 3 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | Q8NAC3 | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000498214, ENST00000295981, ENST00000383812, ENST00000403601, ENST00000413608, ENST00000416074, ENST00000455057, | ENST00000426895, ENST00000547186, ENST00000383827, ENST00000397241, ENST00000427853, ENST00000430793, ENST00000455274, ENST00000466245, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 1 X 2 X 2=4 | 7 X 7 X 6=294 |
| # samples | 2 | 8 | |
| ** MAII score | log2(2/4*10)=2.32192809488736 | log2(8/294*10)=-1.877744249949 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: IL17RC [Title/Abstract] AND TTLL3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | IL17RC(9970314)-TTLL3(9857758), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | IL17RC-TTLL3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. IL17RC-TTLL3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. IL17RC-TTLL3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. IL17RC-TTLL3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | TTLL3 | GO:0018094 | protein polyglycylation | 19524510 |
 Fusion gene breakpoints across IL17RC (5'-gene) Fusion gene breakpoints across IL17RC (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
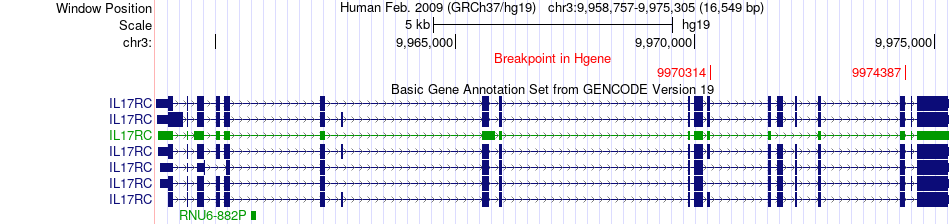 |
 Fusion gene breakpoints across TTLL3 (3'-gene) Fusion gene breakpoints across TTLL3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
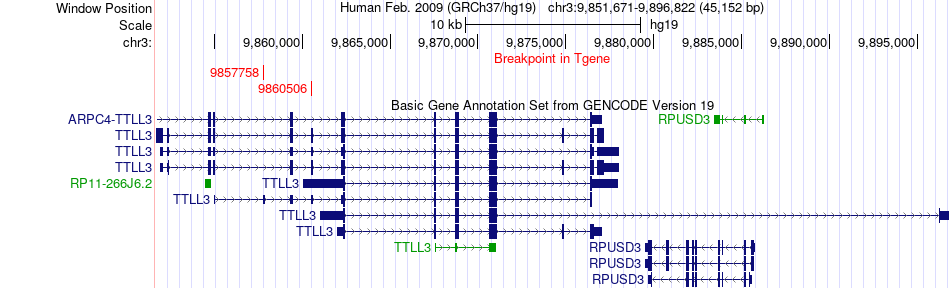 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LIHC | TCGA-GJ-A6C0-01A | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + |
| ChimerDB4 | SKCM | TCGA-FR-A44A-06A | IL17RC | chr3 | 9970314 | - | TTLL3 | chr3 | 9857758 | + |
| ChimerDB4 | SKCM | TCGA-FR-A44A-06A | IL17RC | chr3 | 9970314 | + | TTLL3 | chr3 | 9857758 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000383812 | IL17RC | chr3 | 9974387 | + | ENST00000426895 | TTLL3 | chr3 | 9860506 | + | 3569 | 1680 | 242 | 3568 | 1109 |
| ENST00000383812 | IL17RC | chr3 | 9974387 | + | ENST00000547186 | TTLL3 | chr3 | 9860506 | + | 4433 | 1680 | 242 | 3568 | 1108 |
| ENST00000295981 | IL17RC | chr3 | 9974387 | + | ENST00000426895 | TTLL3 | chr3 | 9860506 | + | 3803 | 1914 | 218 | 3802 | 1195 |
| ENST00000295981 | IL17RC | chr3 | 9974387 | + | ENST00000547186 | TTLL3 | chr3 | 9860506 | + | 4667 | 1914 | 218 | 3802 | 1194 |
| ENST00000403601 | IL17RC | chr3 | 9974387 | + | ENST00000426895 | TTLL3 | chr3 | 9860506 | + | 3569 | 1680 | 197 | 3568 | 1124 |
| ENST00000403601 | IL17RC | chr3 | 9974387 | + | ENST00000547186 | TTLL3 | chr3 | 9860506 | + | 4433 | 1680 | 197 | 3568 | 1123 |
| ENST00000416074 | IL17RC | chr3 | 9974387 | + | ENST00000426895 | TTLL3 | chr3 | 9860506 | + | 3329 | 1440 | 422 | 3328 | 969 |
| ENST00000416074 | IL17RC | chr3 | 9974387 | + | ENST00000547186 | TTLL3 | chr3 | 9860506 | + | 4193 | 1440 | 422 | 3328 | 968 |
| ENST00000455057 | IL17RC | chr3 | 9974387 | + | ENST00000426895 | TTLL3 | chr3 | 9860506 | + | 3429 | 1540 | 153 | 3428 | 1091 |
| ENST00000455057 | IL17RC | chr3 | 9974387 | + | ENST00000547186 | TTLL3 | chr3 | 9860506 | + | 4293 | 1540 | 153 | 3428 | 1091 |
| ENST00000413608 | IL17RC | chr3 | 9974387 | + | ENST00000426895 | TTLL3 | chr3 | 9860506 | + | 3372 | 1483 | 0 | 3371 | 1123 |
| ENST00000413608 | IL17RC | chr3 | 9974387 | + | ENST00000547186 | TTLL3 | chr3 | 9860506 | + | 4236 | 1483 | 0 | 3371 | 1123 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000383812 | ENST00000426895 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.00488313 | 0.99511683 |
| ENST00000383812 | ENST00000547186 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.003142157 | 0.9968579 |
| ENST00000295981 | ENST00000426895 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.006160275 | 0.9938398 |
| ENST00000295981 | ENST00000547186 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.004555993 | 0.99544406 |
| ENST00000403601 | ENST00000426895 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.006334799 | 0.9936652 |
| ENST00000403601 | ENST00000547186 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.004365144 | 0.99563485 |
| ENST00000416074 | ENST00000426895 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.004597048 | 0.99540293 |
| ENST00000416074 | ENST00000547186 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.002922659 | 0.99707735 |
| ENST00000455057 | ENST00000426895 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.004133519 | 0.99586654 |
| ENST00000455057 | ENST00000547186 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.002676453 | 0.9973236 |
| ENST00000413608 | ENST00000426895 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.005267644 | 0.9947324 |
| ENST00000413608 | ENST00000547186 | IL17RC | chr3 | 9974387 | + | TTLL3 | chr3 | 9860506 | + | 0.003475914 | 0.99652404 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >39448_39448_1_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000295981_TTLL3_chr3_9860506_ENST00000426895_length(amino acids)=1195AA_BP=565 MPVPWFLLSLALGRSPVVLSLERLVGPQDATHCSPVSLEPWGDEERLRVQFLAQQSLSLAPVTAATARTALSGLSGADGRREERGRGKSW VCLSLGGSGNTEPQKKGLSCRLWDSDILCLPGDIVPAPGPVLAPTHLQTELVLRCQKETDCDLCLRVAVHLAVHGHWEEPEDEEKFGGAA DSGVEEPRNASLQAQVVLSFQAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPRYEKELNHTQQLPDCRGLEV WNSIPSCWALPWLNVSADGDNVHLVLNVSEEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPCLCIQVWPLEPDSVRTNICP FREDPRAHQNLWQAARLQLLTLQSWLLDAPCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDKVLEFPLLKGHPNLCVQVNSSEKLQL QECLWADSLGPLKDDVLLLETRGPQDNRSLCALEPSGCTSLPSKASTRAARLGEYLLQDLQSGQCLQLWDDDLGALWACPMDKYIHKRWA LVWLACLLFAAALSLILLLKKDHAKEDFWLTAARNVLKLVVKSEWKSYPIQAVEEEASGDKQPKKQEKNPVLVSPEFVDEALCACEEYLS NLAHMDIDKDLEAPLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQVQRCEDILQQLQAVVPQIDMEGDRNIWIVKPGAKSRGRGIMCMDH LEEMLKLVNGNPVVMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVTDWNPLTVWFYRDSYIRFSTQPFSLKNLDNSVHLCNNSIQKHLEN SCHRHPLLPPDNMWSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIHALQTSQDTVQCRKASFELYGADFVFGEDFQPWLIEINASPTMAP STAVTARLCAGVQADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVPQYVGIRLLVEGFTIKKPMAMCHRRMGVRPAVPLLTQRGSGEARH HFPSLHTKAQLPSPHVLRHQGQVLRRQHSKLVGTKALSTTGKALRTLPTAKVFISLPPNLDFKVAPSILKPRKAPALLCLRGPQLEVPCC LCPLKSEQFLAPVGRSRPKANSRPDCDKPRAEACPMKRLSPLKPLPLVGTFQRRRGLGDMKLGKPLLRFPTALVLDPTPNKKKQVKYLGL -------------------------------------------------------------- >39448_39448_2_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000295981_TTLL3_chr3_9860506_ENST00000547186_length(amino acids)=1194AA_BP=565 MPVPWFLLSLALGRSPVVLSLERLVGPQDATHCSPVSLEPWGDEERLRVQFLAQQSLSLAPVTAATARTALSGLSGADGRREERGRGKSW VCLSLGGSGNTEPQKKGLSCRLWDSDILCLPGDIVPAPGPVLAPTHLQTELVLRCQKETDCDLCLRVAVHLAVHGHWEEPEDEEKFGGAA DSGVEEPRNASLQAQVVLSFQAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPRYEKELNHTQQLPDCRGLEV WNSIPSCWALPWLNVSADGDNVHLVLNVSEEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPCLCIQVWPLEPDSVRTNICP FREDPRAHQNLWQAARLQLLTLQSWLLDAPCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDKVLEFPLLKGHPNLCVQVNSSEKLQL QECLWADSLGPLKDDVLLLETRGPQDNRSLCALEPSGCTSLPSKASTRAARLGEYLLQDLQSGQCLQLWDDDLGALWACPMDKYIHKRWA LVWLACLLFAAALSLILLLKKDHAKEDFWLTAARNVLKLVVKSEWKSYPIQAVEEEASGDKQPKKQEKNPVLVSPEFVDEALCACEEYLS NLAHMDIDKDLEAPLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQVQRCEDILQQLQAVVPQIDMEGDRNIWIVKPGAKSRGRGIMCMDH LEEMLKLVNGNPVVMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVTDWNPLTVWFYRDSYIRFSTQPFSLKNLDNSVHLCNNSIQKHLEN SCHRHPLLPPDNMWSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIHALQTSQDTVQCRKASFELYGADFVFGEDFQPWLIEINASPTMAP STAVTARLCAGVQADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVPQYVGIRLLVEGFTIKKPMAMCHRRMGVRPAVPLLTQRGSGEARH HFPSLHTKAQLPSPHVLRHQGQVLRRQHSKLVGTKALSTTGKALRTLPTAKVFISLPPNLDFKVAPSILKPRKAPALLCLRGPQLEVPCC LCPLKSEQFLAPVGRSRPKANSRPDCDKPRAEACPMKRLSPLKPLPLVGTFQRRRGLGDMKLGKPLLRFPTALVLDPTPNKKKQVKYLGL -------------------------------------------------------------- >39448_39448_3_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000383812_TTLL3_chr3_9860506_ENST00000426895_length(amino acids)=1109AA_BP=479 MPVPWFLLSLALGRSPVVLSLERLVGPQDATHCSPGLSCRLWDSDILCLPGDIVPAPGPVLAPTHLQTELVLRCQKETDCDLCLRVAVHL AVHGHWEEPEDEEKFGGAADSGVEEPRNASLQAQVVLSFQAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPR YEKELNHTQQLPALPWLNVSADGDNVHLVLNVSEEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPCLCIQVWPLEPDSVRT NICPFREDPRAHQNLWQAARLQLLTLQSWLLDAPCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDKVLEFPLLKGHPNLCVQVNSSE KLQLQECLWADSLGPLKDDVLLLETRGPQDNRSLCALEPSGCTSLPSKASTRAARLGEYLLQDLQSGQCLQLWDDDLGALWACPMDKYIH KRWALVWLACLLFAAALSLILLLKKDHAKEDFWLTAARNVLKLVVKSEWKSYPIQAVEEEASGDKQPKKQEKNPVLVSPEFVDEALCACE EYLSNLAHMDIDKDLEAPLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQVQRCEDILQQLQAVVPQIDMEGDRNIWIVKPGAKSRGRGIM CMDHLEEMLKLVNGNPVVMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVTDWNPLTVWFYRDSYIRFSTQPFSLKNLDNSVHLCNNSIQK HLENSCHRHPLLPPDNMWSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIHALQTSQDTVQCRKASFELYGADFVFGEDFQPWLIEINASP TMAPSTAVTARLCAGVQADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVPQYVGIRLLVEGFTIKKPMAMCHRRMGVRPAVPLLTQRGSG EARHHFPSLHTKAQLPSPHVLRHQGQVLRRQHSKLVGTKALSTTGKALRTLPTAKVFISLPPNLDFKVAPSILKPRKAPALLCLRGPQLE VPCCLCPLKSEQFLAPVGRSRPKANSRPDCDKPRAEACPMKRLSPLKPLPLVGTFQRRRGLGDMKLGKPLLRFPTALVLDPTPNKKKQVK -------------------------------------------------------------- >39448_39448_4_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000383812_TTLL3_chr3_9860506_ENST00000547186_length(amino acids)=1108AA_BP=479 MPVPWFLLSLALGRSPVVLSLERLVGPQDATHCSPGLSCRLWDSDILCLPGDIVPAPGPVLAPTHLQTELVLRCQKETDCDLCLRVAVHL AVHGHWEEPEDEEKFGGAADSGVEEPRNASLQAQVVLSFQAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPR YEKELNHTQQLPALPWLNVSADGDNVHLVLNVSEEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPCLCIQVWPLEPDSVRT NICPFREDPRAHQNLWQAARLQLLTLQSWLLDAPCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDKVLEFPLLKGHPNLCVQVNSSE KLQLQECLWADSLGPLKDDVLLLETRGPQDNRSLCALEPSGCTSLPSKASTRAARLGEYLLQDLQSGQCLQLWDDDLGALWACPMDKYIH KRWALVWLACLLFAAALSLILLLKKDHAKEDFWLTAARNVLKLVVKSEWKSYPIQAVEEEASGDKQPKKQEKNPVLVSPEFVDEALCACE EYLSNLAHMDIDKDLEAPLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQVQRCEDILQQLQAVVPQIDMEGDRNIWIVKPGAKSRGRGIM CMDHLEEMLKLVNGNPVVMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVTDWNPLTVWFYRDSYIRFSTQPFSLKNLDNSVHLCNNSIQK HLENSCHRHPLLPPDNMWSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIHALQTSQDTVQCRKASFELYGADFVFGEDFQPWLIEINASP TMAPSTAVTARLCAGVQADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVPQYVGIRLLVEGFTIKKPMAMCHRRMGVRPAVPLLTQRGSG EARHHFPSLHTKAQLPSPHVLRHQGQVLRRQHSKLVGTKALSTTGKALRTLPTAKVFISLPPNLDFKVAPSILKPRKAPALLCLRGPQLE VPCCLCPLKSEQFLAPVGRSRPKANSRPDCDKPRAEACPMKRLSPLKPLPLVGTFQRRRGLGDMKLGKPLLRFPTALVLDPTPNKKKQVK -------------------------------------------------------------- >39448_39448_5_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000403601_TTLL3_chr3_9860506_ENST00000426895_length(amino acids)=1124AA_BP=494 MPVPWFLLSLALGRSPVVLSLERLVGPQDATHCSPGLSCRLWDSDILCLPGDIVPAPGPVLAPTHLQTELVLRCQKETDCDLCLRVAVHL AVHGHWEEPEDEEKFGGAADSGVEEPRNASLQAQVVLSFQAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPR YEKELNHTQQLPDCRGLEVWNSIPSCWALPWLNVSADGDNVHLVLNVSEEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPC LCIQVWPLEPDSVRTNICPFREDPRAHQNLWQAARLQLLTLQSWLLDAPCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDKVLEFPL LKGHPNLCVQVNSSEKLQLQECLWADSLGPLKDDVLLLETRGPQDNRSLCALEPSGCTSLPSKASTRAARLGEYLLQDLQSGQCLQLWDD DLGALWACPMDKYIHKRWALVWLACLLFAAALSLILLLKKDHAKEDFWLTAARNVLKLVVKSEWKSYPIQAVEEEASGDKQPKKQEKNPV LVSPEFVDEALCACEEYLSNLAHMDIDKDLEAPLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQVQRCEDILQQLQAVVPQIDMEGDRNI WIVKPGAKSRGRGIMCMDHLEEMLKLVNGNPVVMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVTDWNPLTVWFYRDSYIRFSTQPFSLK NLDNSVHLCNNSIQKHLENSCHRHPLLPPDNMWSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIHALQTSQDTVQCRKASFELYGADFVF GEDFQPWLIEINASPTMAPSTAVTARLCAGVQADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVPQYVGIRLLVEGFTIKKPMAMCHRRM GVRPAVPLLTQRGSGEARHHFPSLHTKAQLPSPHVLRHQGQVLRRQHSKLVGTKALSTTGKALRTLPTAKVFISLPPNLDFKVAPSILKP RKAPALLCLRGPQLEVPCCLCPLKSEQFLAPVGRSRPKANSRPDCDKPRAEACPMKRLSPLKPLPLVGTFQRRRGLGDMKLGKPLLRFPT -------------------------------------------------------------- >39448_39448_6_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000403601_TTLL3_chr3_9860506_ENST00000547186_length(amino acids)=1123AA_BP=494 MPVPWFLLSLALGRSPVVLSLERLVGPQDATHCSPGLSCRLWDSDILCLPGDIVPAPGPVLAPTHLQTELVLRCQKETDCDLCLRVAVHL AVHGHWEEPEDEEKFGGAADSGVEEPRNASLQAQVVLSFQAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPR YEKELNHTQQLPDCRGLEVWNSIPSCWALPWLNVSADGDNVHLVLNVSEEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPC LCIQVWPLEPDSVRTNICPFREDPRAHQNLWQAARLQLLTLQSWLLDAPCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDKVLEFPL LKGHPNLCVQVNSSEKLQLQECLWADSLGPLKDDVLLLETRGPQDNRSLCALEPSGCTSLPSKASTRAARLGEYLLQDLQSGQCLQLWDD DLGALWACPMDKYIHKRWALVWLACLLFAAALSLILLLKKDHAKEDFWLTAARNVLKLVVKSEWKSYPIQAVEEEASGDKQPKKQEKNPV LVSPEFVDEALCACEEYLSNLAHMDIDKDLEAPLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQVQRCEDILQQLQAVVPQIDMEGDRNI WIVKPGAKSRGRGIMCMDHLEEMLKLVNGNPVVMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVTDWNPLTVWFYRDSYIRFSTQPFSLK NLDNSVHLCNNSIQKHLENSCHRHPLLPPDNMWSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIHALQTSQDTVQCRKASFELYGADFVF GEDFQPWLIEINASPTMAPSTAVTARLCAGVQADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVPQYVGIRLLVEGFTIKKPMAMCHRRM GVRPAVPLLTQRGSGEARHHFPSLHTKAQLPSPHVLRHQGQVLRRQHSKLVGTKALSTTGKALRTLPTAKVFISLPPNLDFKVAPSILKP RKAPALLCLRGPQLEVPCCLCPLKSEQFLAPVGRSRPKANSRPDCDKPRAEACPMKRLSPLKPLPLVGTFQRRRGLGDMKLGKPLLRFPT -------------------------------------------------------------- >39448_39448_7_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000413608_TTLL3_chr3_9860506_ENST00000426895_length(amino acids)=1123AA_BP=494 MPVPWFLLSLALGRSPVVLSLERLVGPQDATHCSPGLSCRLWDSDILCLPGDIVPAPGPVLAPTHLQTELVLRCQKETDCDLCLRVAVHL AVHGHWEEPEDEEKFGGAADSGVEEPRNASLQAQVVLSFQAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPR YEKELNHTQQLPDCRGLEVWNSIPSCWALPWLNVSADGDNVHLVLNVSEEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPC LCIQVWPLEPDSVRTNICPFREDPRAHQNLWQAARLQLLTLQSWLLDAPCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDKVLEFPL LKGHPNLCVQVNSSEKLQLQECLWADSLGPLKDDVLLLETRGPQDNRSLCALEPSGCTSLPSKASTRAARLGEYLLQDLQSGQCLQLWDD DLGALWACPMDKYIHKRWALVWLACLLFAAALSLILLLKKDHAKEDFWLTAARNVLKLVVKSEWKSYPIQAVEEEASGDKQPKKQEKNPV LVSPEFVDEALCACEEYLSNLAHMDIDKDLEAPLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQVQRCEDILQQLQAVVPQIDMEGDRNI WIVKPGAKSRGRGIMCMDHLEEMLKLVNGNPVVMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVTDWNPLTVWFYRDSYIRFSTQPFSLK NLDNSVHLCNNSIQKHLENSCHRHPLLPPDNMWSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIHALQTSQDTVQCRKASFELYGADFVF GEDFQPWLIEINASPTMAPSTAVTARLCAGVQADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVPQYVGIRLLVEGFTIKKPMAMCHRRM GVRPAVPLLTQRGSGEARHHFPSLHTKAQLPSPHVLRHQGQVLRRQHSKLVGTKALSTTGKALRTLPTAKVFISLPPNLDFKVAPSILKP RKAPALLCLRGPQLEVPCCLCPLKSEQFLAPVGRSRPKANSRPDCDKPRAEACPMKRLSPLKPLPLVGTFQRRRGLGDMKLGKPLLRFPT -------------------------------------------------------------- >39448_39448_8_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000413608_TTLL3_chr3_9860506_ENST00000547186_length(amino acids)=1123AA_BP=494 MPVPWFLLSLALGRSPVVLSLERLVGPQDATHCSPGLSCRLWDSDILCLPGDIVPAPGPVLAPTHLQTELVLRCQKETDCDLCLRVAVHL AVHGHWEEPEDEEKFGGAADSGVEEPRNASLQAQVVLSFQAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPR YEKELNHTQQLPDCRGLEVWNSIPSCWALPWLNVSADGDNVHLVLNVSEEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPC LCIQVWPLEPDSVRTNICPFREDPRAHQNLWQAARLQLLTLQSWLLDAPCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDKVLEFPL LKGHPNLCVQVNSSEKLQLQECLWADSLGPLKDDVLLLETRGPQDNRSLCALEPSGCTSLPSKASTRAARLGEYLLQDLQSGQCLQLWDD DLGALWACPMDKYIHKRWALVWLACLLFAAALSLILLLKKDHAKEDFWLTAARNVLKLVVKSEWKSYPIQAVEEEASGDKQPKKQEKNPV LVSPEFVDEALCACEEYLSNLAHMDIDKDLEAPLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQVQRCEDILQQLQAVVPQIDMEGDRNI WIVKPGAKSRGRGIMCMDHLEEMLKLVNGNPVVMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVTDWNPLTVWFYRDSYIRFSTQPFSLK NLDNSVHLCNNSIQKHLENSCHRHPLLPPDNMWSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIHALQTSQDTVQCRKASFELYGADFVF GEDFQPWLIEINASPTMAPSTAVTARLCAGVQADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVPQYVGIRLLVEGFTIKKPMAMCHRRM GVRPAVPLLTQRGSGEARHHFPSLHTKAQLPSPHVLRHQGQVLRRQHSKLVGTKALSTTGKALRTLPTAKVFISLPPNLDFKVAPSILKP RKAPALLCLRGPQLEVPCCLCPLKSEQFLAPVGRSRPKANSRPDCDKPRAEACPMKRLSPLKPLPLVGTFQRRRGLGDMKLGKPLLRFPT -------------------------------------------------------------- >39448_39448_9_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000416074_TTLL3_chr3_9860506_ENST00000426895_length(amino acids)=969AA_BP=339 MSTWPCMAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPRYEKELNHTQQLPALPWLNVSADGDNVHLVLNVS EEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPCLCIQVWPLEPDSVRTNICPFREDPRAHQNLWQAARLQLLTLQSWLLDA PCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDVNSSEKLQLQECLWADSLGPLKDDVLLLETRGPQDNRSLCALEPSGCTSLPSKAS TRAARLGEYLLQDLQSGQCLQLWDDDLGALWACPMDKYIHKRWALVWLACLLFAAALSLILLLKKDHAKEDFWLTAARNVLKLVVKSEWK SYPIQAVEEEASGDKQPKKQEKNPVLVSPEFVDEALCACEEYLSNLAHMDIDKDLEAPLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQV QRCEDILQQLQAVVPQIDMEGDRNIWIVKPGAKSRGRGIMCMDHLEEMLKLVNGNPVVMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVT DWNPLTVWFYRDSYIRFSTQPFSLKNLDNSVHLCNNSIQKHLENSCHRHPLLPPDNMWSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIH ALQTSQDTVQCRKASFELYGADFVFGEDFQPWLIEINASPTMAPSTAVTARLCAGVQADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVP QYVGIRLLVEGFTIKKPMAMCHRRMGVRPAVPLLTQRGSGEARHHFPSLHTKAQLPSPHVLRHQGQVLRRQHSKLVGTKALSTTGKALRT LPTAKVFISLPPNLDFKVAPSILKPRKAPALLCLRGPQLEVPCCLCPLKSEQFLAPVGRSRPKANSRPDCDKPRAEACPMKRLSPLKPLP -------------------------------------------------------------- >39448_39448_10_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000416074_TTLL3_chr3_9860506_ENST00000547186_length(amino acids)=968AA_BP=339 MSTWPCMAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPRYEKELNHTQQLPALPWLNVSADGDNVHLVLNVS EEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPCLCIQVWPLEPDSVRTNICPFREDPRAHQNLWQAARLQLLTLQSWLLDA PCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDVNSSEKLQLQECLWADSLGPLKDDVLLLETRGPQDNRSLCALEPSGCTSLPSKAS TRAARLGEYLLQDLQSGQCLQLWDDDLGALWACPMDKYIHKRWALVWLACLLFAAALSLILLLKKDHAKEDFWLTAARNVLKLVVKSEWK SYPIQAVEEEASGDKQPKKQEKNPVLVSPEFVDEALCACEEYLSNLAHMDIDKDLEAPLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQV QRCEDILQQLQAVVPQIDMEGDRNIWIVKPGAKSRGRGIMCMDHLEEMLKLVNGNPVVMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVT DWNPLTVWFYRDSYIRFSTQPFSLKNLDNSVHLCNNSIQKHLENSCHRHPLLPPDNMWSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIH ALQTSQDTVQCRKASFELYGADFVFGEDFQPWLIEINASPTMAPSTAVTARLCAGVQADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVP QYVGIRLLVEGFTIKKPMAMCHRRMGVRPAVPLLTQRGSGEARHHFPSLHTKAQLPSPHVLRHQGQVLRRQHSKLVGTKALSTTGKALRT LPTAKVFISLPPNLDFKVAPSILKPRKAPALLCLRGPQLEVPCCLCPLKSEQFLAPVGRSRPKANSRPDCDKPRAEACPMKRLSPLKPLP -------------------------------------------------------------- >39448_39448_11_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000455057_TTLL3_chr3_9860506_ENST00000426895_length(amino acids)=1091AA_BP=462 MPVPWFLLSLALGRSPVVLSLERLVGPQDATHCSPGLSCRLWDSDILCLPGDIVPAPGPVLAPTHLQTELVLRCQKETDCDLCLRVAVHL AVHGHWEEPEDEEKFGGAADSGVEEPRNASLQAQVVLSFQAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPR YEKELNHTQQLPALPWLNVSADGDNVHLVLNVSEEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPCLCIQVWPLEPDSVRT NICPFREDPRAHQNLWQAARLQLLTLQSWLLDAPCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDVNSSEKLQLQECLWADSLGPLK DDVLLLETRGPQDNRSLCALEPSGCTSLPSKASTRAARLGEYLLQDLQSGQCLQLWDDDLGALWACPMDKYIHKRWALVWLACLLFAAAL SLILLLKKDHAKEDFWLTAARNVLKLVVKSEWKSYPIQAVEEEASGDKQPKKQEKNPVLVSPEFVDEALCACEEYLSNLAHMDIDKDLEA PLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQVQRCEDILQQLQAVVPQIDMEGDRNIWIVKPGAKSRGRGIMCMDHLEEMLKLVNGNPV VMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVTDWNPLTVWFYRDSYIRFSTQPFSLKNLDNSVHLCNNSIQKHLENSCHRHPLLPPDNM WSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIHALQTSQDTVQCRKASFELYGADFVFGEDFQPWLIEINASPTMAPSTAVTARLCAGVQ ADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVPQYVGIRLLVEGFTIKKPMAMCHRRMGVRPAVPLLTQRGSGEARHHFPSLHTKAQLPS PHVLRHQGQVLRRQHSKLVGTKALSTTGKALRTLPTAKVFISLPPNLDFKVAPSILKPRKAPALLCLRGPQLEVPCCLCPLKSEQFLAPV GRSRPKANSRPDCDKPRAEACPMKRLSPLKPLPLVGTFQRRRGLGDMKLGKPLLRFPTALVLDPTPNKKKQVKYLGLDSIAVGGSRVDGA -------------------------------------------------------------- >39448_39448_12_IL17RC-TTLL3_IL17RC_chr3_9974387_ENST00000455057_TTLL3_chr3_9860506_ENST00000547186_length(amino acids)=1091AA_BP=462 MPVPWFLLSLALGRSPVVLSLERLVGPQDATHCSPGLSCRLWDSDILCLPGDIVPAPGPVLAPTHLQTELVLRCQKETDCDLCLRVAVHL AVHGHWEEPEDEEKFGGAADSGVEEPRNASLQAQVVLSFQAYPTARCVLLEVQVPAALVQFGQSVGSVVYDCFEAALGSEVRIWSYTQPR YEKELNHTQQLPALPWLNVSADGDNVHLVLNVSEEQHFGLSLYWNQVQGPPKPRWHKNLTGPQIITLNHTDLVPCLCIQVWPLEPDSVRT NICPFREDPRAHQNLWQAARLQLLTLQSWLLDAPCSLPAEAALCWRAPGGDPCQPLVPPLSWENVTVDVNSSEKLQLQECLWADSLGPLK DDVLLLETRGPQDNRSLCALEPSGCTSLPSKASTRAARLGEYLLQDLQSGQCLQLWDDDLGALWACPMDKYIHKRWALVWLACLLFAAAL SLILLLKKDHAKEDFWLTAARNVLKLVVKSEWKSYPIQAVEEEASGDKQPKKQEKNPVLVSPEFVDEALCACEEYLSNLAHMDIDKDLEA PLYLTPEGWSLFLQRYYQVVHEGAELRHLDTQVQRCEDILQQLQAVVPQIDMEGDRNIWIVKPGAKSRGRGIMCMDHLEEMLKLVNGNPV VMKDGKWVVQKYIERPLLIFGTKFDLRQWFLVTDWNPLTVWFYRDSYIRFSTQPFSLKNLDNSVHLCNNSIQKHLENSCHRHPLLPPDNM WSSQRFQAHLQEMGAPNAWSTIIVPGMKDAVIHALQTSQDTVQCRKASFELYGADFVFGEDFQPWLIEINASPTMAPSTAVTARLCAGVQ ADTLRVVIDRMLDRNCDTGAFELIYKQPAVEVPQYVGIRLLVEGFTIKKPMAMCHRRMGVRPAVPLLTQRGSGEARHHFPSLHTKAQLPS PHVLRHQGQVLRRQHSKLVGTKALSTTGKALRTLPTAKVFISLPPNLDFKVAPSILKPRKAPALLCLRGPQLEVPCCLCPLKSEQFLAPV GRSRPKANSRPDCDKPRAEACPMKRLSPLKPLPLVGTFQRRRGLGDMKLGKPLLRFPTALVLDPTPNKKKQVKYLGLDSIAVGGSRVDGA -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:9970314/chr3:9857758) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| IL17RC | . |
| FUNCTION: Receptor for IL17A and IL17F, major effector cytokines of innate and adaptive immune system involved in antimicrobial host defense and maintenance of tissue integrity (By similarity). Receptor for IL17A and IL17F, major effector cytokines of innate and adaptive immune system involved in antimicrobial host defense and maintenance of tissue integrity. Receptor for IL17A and IL17F homodimers as part of a heterodimeric complex with IL17RA (PubMed:16785495). Receptor for the heterodimer formed by IL17A and IL17B as part of a heterodimeric complex with IL17RA (PubMed:18684971). Has also been shown to be the cognate receptor for IL17F and to bind IL17A with high affinity without the need for IL17RA (PubMed:17911633). Upon binding of IL17F homodimer triggers downstream activation of TRAF6 and NF-kappa-B signaling pathway (PubMed:16785495, PubMed:32187518). Induces transcriptional activation of IL33, a potent cytokine that stimulates group 2 innate lymphoid cells and adaptive T-helper 2 cells involved in pulmonary allergic response to fungi (By similarity). Promotes sympathetic innervation of peripheral organs by coordinating the communication between gamma-delta T cells and parenchymal cells. Stimulates sympathetic innervation of thermogenic adipose tissue by driving TGFB1 expression (By similarity). Binding of IL17A-IL17F to IL17RA-IL17RC heterodimeric receptor complex triggers homotypic interaction of IL17RA and IL17RC chains with TRAF3IP2 adapter through SEFIR domains. This leads to downstream TRAF6-mediated activation of NF-kappa-B and MAPkinase pathways ultimately resulting in transcriptional activation of cytokines, chemokines, antimicrobial peptides and matrix metalloproteinases, with potential strong immune inflammation (PubMed:18684971, PubMed:17911633). Primarily induces neutrophil activation and recruitment at infection and inflammatory sites (By similarity). Stimulates the production of antimicrobial beta-defensins DEFB1, DEFB103A, and DEFB104A by mucosal epithelial cells, limiting the entry of microbes through the epithelial barriers (By similarity). {ECO:0000250|UniProtKB:Q8K4C2, ECO:0000269|PubMed:16785495, ECO:0000269|PubMed:17911633, ECO:0000269|PubMed:18684971, ECO:0000269|PubMed:32187518}.; FUNCTION: [Isoform 5]: Receptor for both IL17A and IL17F. {ECO:0000269|PubMed:16785495}.; FUNCTION: [Isoform 6]: Does not bind IL17A or IL17F. {ECO:0000269|PubMed:16785495}.; FUNCTION: [Isoform 7]: Does not bind IL17A or IL17F. {ECO:0000269|PubMed:16785495}.; FUNCTION: [Isoform 8]: Receptor for both IL17A and IL17F. {ECO:0000269|PubMed:16785495}. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000295981 | + | 17 | 19 | 21_538 | 565.3333333333334 | 792.0 | Topological domain | Extracellular |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000295981 | + | 17 | 19 | 539_559 | 565.3333333333334 | 792.0 | Transmembrane | Helical |
| Tgene | TTLL3 | chr3:9974387 | chr3:9860506 | ENST00000383827 | 0 | 5 | 151_510 | 0 | 353.0 | Domain | TTL | |
| Tgene | TTLL3 | chr3:9974387 | chr3:9860506 | ENST00000397241 | 4 | 12 | 151_510 | 0 | 714.6666666666666 | Domain | TTL | |
| Tgene | TTLL3 | chr3:9974387 | chr3:9860506 | ENST00000427853 | 2 | 9 | 151_510 | 0 | 353.0 | Domain | TTL | |
| Tgene | TTLL3 | chr3:9974387 | chr3:9860506 | ENST00000430793 | 0 | 6 | 151_510 | 0 | 435.0 | Domain | TTL | |
| Tgene | TTLL3 | chr3:9974387 | chr3:9860506 | ENST00000547186 | 4 | 13 | 151_510 | 143.33333333333334 | 773.0 | Domain | TTL | |
| Tgene | TTLL3 | chr3:9974387 | chr3:9860506 | ENST00000383827 | 0 | 5 | 321_324 | 0 | 353.0 | Nucleotide binding | ATP binding | |
| Tgene | TTLL3 | chr3:9974387 | chr3:9860506 | ENST00000397241 | 4 | 12 | 321_324 | 0 | 714.6666666666666 | Nucleotide binding | ATP binding | |
| Tgene | TTLL3 | chr3:9974387 | chr3:9860506 | ENST00000427853 | 2 | 9 | 321_324 | 0 | 353.0 | Nucleotide binding | ATP binding | |
| Tgene | TTLL3 | chr3:9974387 | chr3:9860506 | ENST00000430793 | 0 | 6 | 321_324 | 0 | 435.0 | Nucleotide binding | ATP binding | |
| Tgene | TTLL3 | chr3:9974387 | chr3:9860506 | ENST00000547186 | 4 | 13 | 321_324 | 143.33333333333334 | 773.0 | Nucleotide binding | ATP binding |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000295981 | + | 17 | 19 | 583_735 | 565.3333333333334 | 792.0 | Domain | SEFIR |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000383812 | + | 16 | 18 | 583_735 | 479.3333333333333 | 706.0 | Domain | SEFIR |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000403601 | + | 17 | 19 | 583_735 | 494.3333333333333 | 721.0 | Domain | SEFIR |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000413608 | + | 17 | 18 | 583_735 | 494.3333333333333 | 708.0 | Domain | SEFIR |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000455057 | + | 15 | 17 | 583_735 | 462.3333333333333 | 689.0 | Domain | SEFIR |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000295981 | + | 17 | 19 | 560_791 | 565.3333333333334 | 792.0 | Topological domain | Cytoplasmic |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000383812 | + | 16 | 18 | 21_538 | 479.3333333333333 | 706.0 | Topological domain | Extracellular |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000383812 | + | 16 | 18 | 560_791 | 479.3333333333333 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000403601 | + | 17 | 19 | 21_538 | 494.3333333333333 | 721.0 | Topological domain | Extracellular |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000403601 | + | 17 | 19 | 560_791 | 494.3333333333333 | 721.0 | Topological domain | Cytoplasmic |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000413608 | + | 17 | 18 | 21_538 | 494.3333333333333 | 708.0 | Topological domain | Extracellular |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000413608 | + | 17 | 18 | 560_791 | 494.3333333333333 | 708.0 | Topological domain | Cytoplasmic |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000455057 | + | 15 | 17 | 21_538 | 462.3333333333333 | 689.0 | Topological domain | Extracellular |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000455057 | + | 15 | 17 | 560_791 | 462.3333333333333 | 689.0 | Topological domain | Cytoplasmic |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000383812 | + | 16 | 18 | 539_559 | 479.3333333333333 | 706.0 | Transmembrane | Helical |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000403601 | + | 17 | 19 | 539_559 | 494.3333333333333 | 721.0 | Transmembrane | Helical |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000413608 | + | 17 | 18 | 539_559 | 494.3333333333333 | 708.0 | Transmembrane | Helical |
| Hgene | IL17RC | chr3:9974387 | chr3:9860506 | ENST00000455057 | + | 15 | 17 | 539_559 | 462.3333333333333 | 689.0 | Transmembrane | Helical |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| IL17RC | |
| TTLL3 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to IL17RC-TTLL3 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to IL17RC-TTLL3 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

