| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ITGA7-MIP |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ITGA7-MIP | FusionPDB ID: 40451 | FusionGDB2.0 ID: 40451 | Hgene | Tgene | Gene symbol | ITGA7 | MIP | Gene ID | 3679 | 4285 |
| Gene name | integrin subunit alpha 7 | mitochondrial intermediate peptidase | |
| Synonyms | - | COXPD31|HMIP|MIP | |
| Cytomap | 12q13.2 | 13q12.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | integrin alpha-7integrin alpha 7 chain | mitochondrial intermediate peptidase | |
| Modification date | 20200328 | 20200313 | |
| UniProtAcc | Q13683 | Q8TD10 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000257879, ENST00000257880, ENST00000347027, ENST00000394229, ENST00000394230, ENST00000452168, ENST00000553804, ENST00000555728, | ENST00000555551, ENST00000257979, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 7 X 8 X 5=280 | 3 X 2 X 2=12 |
| # samples | 8 | 4 | |
| ** MAII score | log2(8/280*10)=-1.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/12*10)=1.73696559416621 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ITGA7 [Title/Abstract] AND MIP [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ITGA7(56086627)-MIP(56847539), # samples:1 ITGA7(56086628)-MIP(56847539), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ITGA7-MIP seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ITGA7-MIP seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ITGA7-MIP seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ITGA7-MIP seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across ITGA7 (5'-gene) Fusion gene breakpoints across ITGA7 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
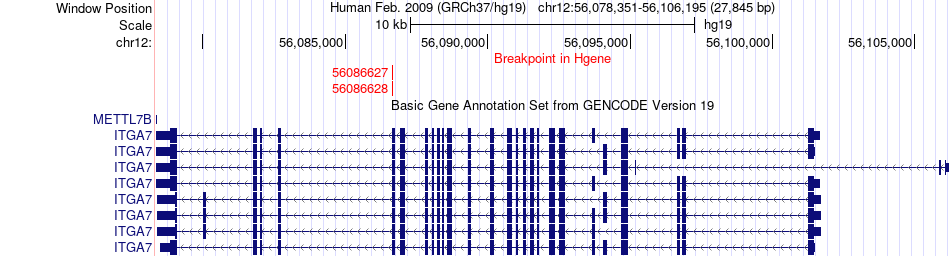 |
 Fusion gene breakpoints across MIP (3'-gene) Fusion gene breakpoints across MIP (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
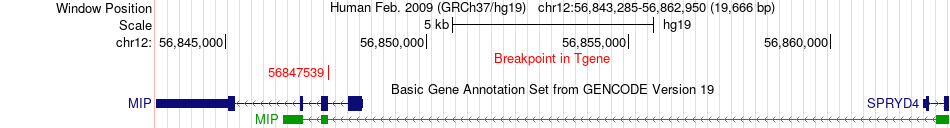 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-61-1724-01A | ITGA7 | chr12 | 56086628 | - | MIP | chr12 | 56847539 | - |
| ChimerDB4 | OV | TCGA-61-1724 | ITGA7 | chr12 | 56086627 | - | MIP | chr12 | 56847539 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000553804 | ITGA7 | chr12 | 56086627 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5085 | 2875 | 19 | 3306 | 1095 |
| ENST00000257879 | ITGA7 | chr12 | 56086627 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5270 | 3060 | 78 | 3491 | 1137 |
| ENST00000347027 | ITGA7 | chr12 | 56086627 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5238 | 3028 | 64 | 3459 | 1131 |
| ENST00000452168 | ITGA7 | chr12 | 56086627 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 4917 | 2707 | 31 | 3138 | 1035 |
| ENST00000394229 | ITGA7 | chr12 | 56086627 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5274 | 3064 | 82 | 3495 | 1137 |
| ENST00000257880 | ITGA7 | chr12 | 56086627 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5406 | 3196 | 82 | 3627 | 1181 |
| ENST00000394230 | ITGA7 | chr12 | 56086627 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5286 | 3076 | 82 | 3507 | 1141 |
| ENST00000555728 | ITGA7 | chr12 | 56086627 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5215 | 3005 | 29 | 3436 | 1135 |
| ENST00000553804 | ITGA7 | chr12 | 56086628 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5085 | 2875 | 19 | 3306 | 1095 |
| ENST00000257879 | ITGA7 | chr12 | 56086628 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5270 | 3060 | 78 | 3491 | 1137 |
| ENST00000347027 | ITGA7 | chr12 | 56086628 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5238 | 3028 | 64 | 3459 | 1131 |
| ENST00000452168 | ITGA7 | chr12 | 56086628 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 4917 | 2707 | 31 | 3138 | 1035 |
| ENST00000394229 | ITGA7 | chr12 | 56086628 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5274 | 3064 | 82 | 3495 | 1137 |
| ENST00000257880 | ITGA7 | chr12 | 56086628 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5406 | 3196 | 82 | 3627 | 1181 |
| ENST00000394230 | ITGA7 | chr12 | 56086628 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5286 | 3076 | 82 | 3507 | 1141 |
| ENST00000555728 | ITGA7 | chr12 | 56086628 | - | ENST00000257979 | MIP | chr12 | 56847539 | - | 5215 | 3005 | 29 | 3436 | 1135 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000553804 | ENST00000257979 | ITGA7 | chr12 | 56086627 | - | MIP | chr12 | 56847539 | - | 0.000604154 | 0.99939585 |
| ENST00000257879 | ENST00000257979 | ITGA7 | chr12 | 56086627 | - | MIP | chr12 | 56847539 | - | 0.000990451 | 0.9990095 |
| ENST00000347027 | ENST00000257979 | ITGA7 | chr12 | 56086627 | - | MIP | chr12 | 56847539 | - | 0.000758296 | 0.9992417 |
| ENST00000452168 | ENST00000257979 | ITGA7 | chr12 | 56086627 | - | MIP | chr12 | 56847539 | - | 0.000450703 | 0.9995493 |
| ENST00000394229 | ENST00000257979 | ITGA7 | chr12 | 56086627 | - | MIP | chr12 | 56847539 | - | 0.00098702 | 0.99901295 |
| ENST00000257880 | ENST00000257979 | ITGA7 | chr12 | 56086627 | - | MIP | chr12 | 56847539 | - | 0.000725597 | 0.99927443 |
| ENST00000394230 | ENST00000257979 | ITGA7 | chr12 | 56086627 | - | MIP | chr12 | 56847539 | - | 0.000794083 | 0.9992059 |
| ENST00000555728 | ENST00000257979 | ITGA7 | chr12 | 56086627 | - | MIP | chr12 | 56847539 | - | 0.000558713 | 0.9994413 |
| ENST00000553804 | ENST00000257979 | ITGA7 | chr12 | 56086628 | - | MIP | chr12 | 56847539 | - | 0.000604154 | 0.99939585 |
| ENST00000257879 | ENST00000257979 | ITGA7 | chr12 | 56086628 | - | MIP | chr12 | 56847539 | - | 0.000990451 | 0.9990095 |
| ENST00000347027 | ENST00000257979 | ITGA7 | chr12 | 56086628 | - | MIP | chr12 | 56847539 | - | 0.000758296 | 0.9992417 |
| ENST00000452168 | ENST00000257979 | ITGA7 | chr12 | 56086628 | - | MIP | chr12 | 56847539 | - | 0.000450703 | 0.9995493 |
| ENST00000394229 | ENST00000257979 | ITGA7 | chr12 | 56086628 | - | MIP | chr12 | 56847539 | - | 0.00098702 | 0.99901295 |
| ENST00000257880 | ENST00000257979 | ITGA7 | chr12 | 56086628 | - | MIP | chr12 | 56847539 | - | 0.000725597 | 0.99927443 |
| ENST00000394230 | ENST00000257979 | ITGA7 | chr12 | 56086628 | - | MIP | chr12 | 56847539 | - | 0.000794083 | 0.9992059 |
| ENST00000555728 | ENST00000257979 | ITGA7 | chr12 | 56086628 | - | MIP | chr12 | 56847539 | - | 0.000558713 | 0.9994413 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >40451_40451_1_ITGA7-MIP_ITGA7_chr12_56086627_ENST00000257879_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1137AA_BP=994 MAGRTSGRRLKDQRDFGDQRRAWGDLGLGACEISLAFAGSSRRDRPMAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRK EGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGGLFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPG GKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKG LLFVTNIDSSDPDQLVYKTLDPADRLPGPAGDLALNSYLGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGE RLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAP FDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPN CAGGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKD KLRAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTT ALFALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYL ILSTSGISIETTELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLR TLGSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKK NITLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWV -------------------------------------------------------------- >40451_40451_2_ITGA7-MIP_ITGA7_chr12_56086627_ENST00000257880_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1181AA_BP=1038 MAGRTSGRRLKDQRDFGDQRRAWGDLGLGACEISLAFAGSSRRDRPMAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRK EGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGGLFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPG GKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKG TARVELCAQGSADLAHLDDGPYEAGGEKEQDPRLIPVPANSYFGLLFVTNIDSSDPDQLVYKTLDPADRLPGPAGDLALNSYLGFSIDSG KGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGERLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLN QGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAPFDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMD GNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPNCAGGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQV PRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKDKLRAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEI HFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTTALFALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDS LHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYLILSTSGISIETTELEVELLLATISEQELHPVSARARVFIELPLSIA GMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLRTLGSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPN ILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKKNITLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVAL AVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWVGPIIGGGLGSLLYDFLLFPRLKSISERLSVLKGAKPDVSNGQPEVT -------------------------------------------------------------- >40451_40451_3_ITGA7-MIP_ITGA7_chr12_56086627_ENST00000347027_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1131AA_BP=988 MAGRTSGRRLKDQRDFGDQRRAWGDLGLGACEISLAFAGSSRRDRPMAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRK EGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGGLFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPG GKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKG LLFVTNIDSSDPDQLVYKTLDPADRLPGPAGDLALNSYLGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGE RLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAP FDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPN CAGGHSVCVDLRVCFSYIAVPSSYSPTVDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKDKLRAIV VTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTTALFALS GQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYLILSTSG ISIETTELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLRTLGSAF LNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKKNITLLH PAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWVGPIIGG -------------------------------------------------------------- >40451_40451_4_ITGA7-MIP_ITGA7_chr12_56086627_ENST00000394229_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1137AA_BP=994 MAGRTSGRRLKDQRDFGDQRRAWGDLGLGACEISLAFAGSSRRDRPMAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRK EGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGGLFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPG GKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKG LLFVTNIDSSDPDQLVYKTLDPADRLPGPAGDLALNSYLGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGE RLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAP FDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPN CAGGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKD KLRAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTT ALFALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYL ILSTSGISIETTELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLR TLGSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKK NITLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWV -------------------------------------------------------------- >40451_40451_5_ITGA7-MIP_ITGA7_chr12_56086627_ENST00000394230_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1141AA_BP=998 MAGRTSGRRLKDQRDFGDQRRAWGDLGLGACEISLAFAGSSRRDRPMAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRK EGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGGLFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPG GKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKG TARVELCAQGSADLAHLDDGPYEAGGEKEQDPRLIPVPANSYFGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVM LSGERLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIA VGAPFDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDL EQPNCAGGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQE NVKDKLRAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDV DGTTALFALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQV TFYLILSTSGISIETTELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQG QSLRTLGSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSA EKKKNITLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHW -------------------------------------------------------------- >40451_40451_6_ITGA7-MIP_ITGA7_chr12_56086627_ENST00000452168_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1035AA_BP=892 MGDLAGLQERGLVVVWLSSKCWPEASAQRKGALETVQMAPFATPMVQALTTTRIQRQAEGFQCWRECGTRRSPFEGKETCAHRYEARQRV DQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKGTARVELCAQGSADLAH LDDGPYEAGGEKEQDPRLIPVPANSYFGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGERLTSGFGYSLAV ADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAPFDGDGKVFIYHG SSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPNCAGGHSVCVDLR VCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKDKLRAIVVTLSYS LQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTTALFALSGQPVIG LELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYLILSTSGISIETT ELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLRTLGSAFLNIMWP HEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKKNITLLHPAVSVG QATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWVGPIIGGGLGSLL -------------------------------------------------------------- >40451_40451_7_ITGA7-MIP_ITGA7_chr12_56086627_ENST00000553804_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1095AA_BP=952 MAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRKEGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGG LFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPGGKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGE WKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKGTARVELCAQGSADLAHLDDGPYEAGGEKEQDPRLIPVPANSYFGFS IDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGERLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVY VYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAPFDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGS LDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPNCAGGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRL RGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKDKLRAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQ RAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTTALFALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVM LPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYLILSTSGISIETTELEVELLLATISEQELHPVSARARVFIELP LSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLRTLGSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCS PRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKKNITLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLG SVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWVGPIIGGGLGSLLYDFLLFPRLKSISERLSVLKGAKPDVSNGQ -------------------------------------------------------------- >40451_40451_8_ITGA7-MIP_ITGA7_chr12_56086627_ENST00000555728_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1135AA_BP=992 MAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRKEGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGG LFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPGGKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGE WKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKGTARVELCAQGSADLAHLDDGPYEAGGEKEQDPRLIPVPANSYFGLL FVTNIDSSDPDQLVYKTLDPADRLPGPAGDLALNSYLGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGERL TSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAPFD GDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPNCA GGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKDKL RAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTTAL FALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYLIL STSGISIETTELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLRTL GSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKKNI TLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWVGP -------------------------------------------------------------- >40451_40451_9_ITGA7-MIP_ITGA7_chr12_56086628_ENST00000257879_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1137AA_BP=994 MAGRTSGRRLKDQRDFGDQRRAWGDLGLGACEISLAFAGSSRRDRPMAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRK EGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGGLFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPG GKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKG LLFVTNIDSSDPDQLVYKTLDPADRLPGPAGDLALNSYLGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGE RLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAP FDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPN CAGGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKD KLRAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTT ALFALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYL ILSTSGISIETTELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLR TLGSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKK NITLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWV -------------------------------------------------------------- >40451_40451_10_ITGA7-MIP_ITGA7_chr12_56086628_ENST00000257880_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1181AA_BP=1038 MAGRTSGRRLKDQRDFGDQRRAWGDLGLGACEISLAFAGSSRRDRPMAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRK EGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGGLFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPG GKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKG TARVELCAQGSADLAHLDDGPYEAGGEKEQDPRLIPVPANSYFGLLFVTNIDSSDPDQLVYKTLDPADRLPGPAGDLALNSYLGFSIDSG KGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGERLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLN QGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAPFDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMD GNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPNCAGGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQV PRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKDKLRAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEI HFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTTALFALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDS LHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYLILSTSGISIETTELEVELLLATISEQELHPVSARARVFIELPLSIA GMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLRTLGSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPN ILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKKNITLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVAL AVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWVGPIIGGGLGSLLYDFLLFPRLKSISERLSVLKGAKPDVSNGQPEVT -------------------------------------------------------------- >40451_40451_11_ITGA7-MIP_ITGA7_chr12_56086628_ENST00000347027_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1131AA_BP=988 MAGRTSGRRLKDQRDFGDQRRAWGDLGLGACEISLAFAGSSRRDRPMAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRK EGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGGLFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPG GKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKG LLFVTNIDSSDPDQLVYKTLDPADRLPGPAGDLALNSYLGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGE RLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAP FDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPN CAGGHSVCVDLRVCFSYIAVPSSYSPTVDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKDKLRAIV VTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTTALFALS GQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYLILSTSG ISIETTELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLRTLGSAF LNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKKNITLLH PAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWVGPIIGG -------------------------------------------------------------- >40451_40451_12_ITGA7-MIP_ITGA7_chr12_56086628_ENST00000394229_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1137AA_BP=994 MAGRTSGRRLKDQRDFGDQRRAWGDLGLGACEISLAFAGSSRRDRPMAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRK EGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGGLFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPG GKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKG LLFVTNIDSSDPDQLVYKTLDPADRLPGPAGDLALNSYLGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGE RLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAP FDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPN CAGGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKD KLRAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTT ALFALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYL ILSTSGISIETTELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLR TLGSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKK NITLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWV -------------------------------------------------------------- >40451_40451_13_ITGA7-MIP_ITGA7_chr12_56086628_ENST00000394230_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1141AA_BP=998 MAGRTSGRRLKDQRDFGDQRRAWGDLGLGACEISLAFAGSSRRDRPMAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRK EGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGGLFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPG GKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKG TARVELCAQGSADLAHLDDGPYEAGGEKEQDPRLIPVPANSYFGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVM LSGERLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIA VGAPFDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDL EQPNCAGGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQE NVKDKLRAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDV DGTTALFALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQV TFYLILSTSGISIETTELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQG QSLRTLGSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSA EKKKNITLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHW -------------------------------------------------------------- >40451_40451_14_ITGA7-MIP_ITGA7_chr12_56086628_ENST00000452168_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1035AA_BP=892 MGDLAGLQERGLVVVWLSSKCWPEASAQRKGALETVQMAPFATPMVQALTTTRIQRQAEGFQCWRECGTRRSPFEGKETCAHRYEARQRV DQILETRDMIGRCFVLSQDLAIRDELDGGEWKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKGTARVELCAQGSADLAH LDDGPYEAGGEKEQDPRLIPVPANSYFGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGERLTSGFGYSLAV ADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAPFDGDGKVFIYHG SSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPNCAGGHSVCVDLR VCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKDKLRAIVVTLSYS LQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTTALFALSGQPVIG LELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYLILSTSGISIETT ELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLRTLGSAFLNIMWP HEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKKNITLLHPAVSVG QATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWVGPIIGGGLGSLL -------------------------------------------------------------- >40451_40451_15_ITGA7-MIP_ITGA7_chr12_56086628_ENST00000553804_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1095AA_BP=952 MAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRKEGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGG LFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPGGKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGE WKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKGTARVELCAQGSADLAHLDDGPYEAGGEKEQDPRLIPVPANSYFGFS IDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGERLTSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVY VYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAPFDGDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGS LDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPNCAGGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRL RGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKDKLRAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQ RAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTTALFALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVM LPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYLILSTSGISIETTELEVELLLATISEQELHPVSARARVFIELP LSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLRTLGSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCS PRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKKNITLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLG SVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWVGPIIGGGLGSLLYDFLLFPRLKSISERLSVLKGAKPDVSNGQ -------------------------------------------------------------- >40451_40451_16_ITGA7-MIP_ITGA7_chr12_56086628_ENST00000555728_MIP_chr12_56847539_ENST00000257979_length(amino acids)=1135AA_BP=992 MAGARSRDPWGASGICYLFGSLLVELLFSRAVAFNLDVMGALRKEGEPGSLFGFSVALHRQLQPRPQSWLLVGAPQALALPGQQANRTGG LFACPLSLEETDCYRVDIDQGADMQKESKENQWLGVSVRSQGPGGKIVTCAHRYEARQRVDQILETRDMIGRCFVLSQDLAIRDELDGGE WKFCEGRPQGHEQFGFCQQGTAAAFSPDSHYLLFGAPGTYNWKGTARVELCAQGSADLAHLDDGPYEAGGEKEQDPRLIPVPANSYFGLL FVTNIDSSDPDQLVYKTLDPADRLPGPAGDLALNSYLGFSIDSGKGLVRAEELSFVAGAPRANHKGAVVILRKDSASRLVPEVMLSGERL TSGFGYSLAVADLNSDGWPDLIVGAPYFFERQEELGGAVYVYLNQGGHWAGISPLRLCGSPDSMFGISLAVLGDLNQDGFPDIAVGAPFD GDGKVFIYHGSSLGVVAKPSQVLEGEAVGIKSFGYSLSGSLDMDGNQYPDLLVGSLADTAVLFRARPILHVSHEVSIAPRSIDLEQPNCA GGHSVCVDLRVCFSYIAVPSSYSPTVALDYVLDADTDRRLRGQVPRVTFLSRNLEEPKHQASGTVWLKHQHDRVCGDAMFQLQENVKDKL RAIVVTLSYSLQTPRLRRQAPGQGLPPVAPILNAHQPSTQRAEIHFLKQGCGEDKICQSNLQLVRARFCTRVSDTEFQPLPMDVDGTTAL FALSGQPVIGLELMVTNLPSDPAQPQADGDDAHEAQLLVMLPDSLHYSGVRALDPAEKPLCLSNENASHVECELGNPMKRGAQVTFYLIL STSGISIETTELEVELLLATISEQELHPVSARARVFIELPLSIAGMAIPQQLFFSGVVRGERAMQSERDVGSKVKYEVTVSNQGQSLRTL GSAFLNIMWPHEIANGKWLLYPMQVELEGGQGPGQKGLCSPRPNILHLDVDSRDRRRRELEPPEQQEPGERQEPSMSWWPVSSAEKKKNI TLLHPAVSVGQATTVEIFLTLQFVLCIFATYDERRNGQLGSVALAVGFSLALGHLFGMYYTGAGMNPARSFAPAILTGNFTNHWVYWVGP -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:56086627/chr12:56847539) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ITGA7 | MIP |
| FUNCTION: Integrin alpha-7/beta-1 is the primary laminin receptor on skeletal myoblasts and adult myofibers. During myogenic differentiation, it may induce changes in the shape and mobility of myoblasts, and facilitate their localization at laminin-rich sites of secondary fiber formation. It is involved in the maintenance of the myofibers cytoarchitecture as well as for their anchorage, viability and functional integrity. Isoform Alpha-7X2B and isoform Alpha-7X1B promote myoblast migration on laminin 1 and laminin 2/4, but isoform Alpha-7X1B is less active on laminin 1 (In vitro). Acts as Schwann cell receptor for laminin-2. Acts as a receptor of COMP and mediates its effect on vascular smooth muscle cells (VSMCs) maturation (By similarity). Required to promote contractile phenotype acquisition in differentiated airway smooth muscle (ASM) cells. {ECO:0000250, ECO:0000269|PubMed:10694445, ECO:0000269|PubMed:17641293, ECO:0000269|PubMed:9307969}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 372_380 | 948.0 | 1138.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 434_442 | 948.0 | 1138.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 492_500 | 948.0 | 1138.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 372_380 | 992.0 | 1163.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 434_442 | 992.0 | 1163.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 492_500 | 992.0 | 1163.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 372_380 | 855.0 | 1045.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 434_442 | 855.0 | 1045.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 492_500 | 855.0 | 1045.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 372_380 | 952.0 | 1142.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 434_442 | 952.0 | 1142.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 492_500 | 952.0 | 1142.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 372_380 | 992.0 | 1182.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 434_442 | 992.0 | 1182.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 492_500 | 992.0 | 1182.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 372_380 | 948.0 | 1138.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 434_442 | 948.0 | 1138.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 492_500 | 948.0 | 1138.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 372_380 | 992.0 | 1163.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 434_442 | 992.0 | 1163.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 492_500 | 992.0 | 1163.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 372_380 | 855.0 | 1045.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 434_442 | 855.0 | 1045.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 492_500 | 855.0 | 1045.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 372_380 | 952.0 | 1142.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 434_442 | 952.0 | 1142.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 492_500 | 952.0 | 1142.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 372_380 | 992.0 | 1182.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 434_442 | 992.0 | 1182.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 492_500 | 992.0 | 1182.0 | Calcium binding | Ontology_term=ECO:0000255 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 953_958 | 992.0 | 1163.0 | Compositional bias | Note=Poly-Arg |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 953_958 | 992.0 | 1182.0 | Compositional bias | Note=Poly-Arg |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 953_958 | 992.0 | 1163.0 | Compositional bias | Note=Poly-Arg |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 953_958 | 992.0 | 1182.0 | Compositional bias | Note=Poly-Arg |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 110_175 | 948.0 | 1138.0 | Repeat | FG-GAP 2 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 185_238 | 948.0 | 1138.0 | Repeat | FG-GAP 3 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 292_349 | 948.0 | 1138.0 | Repeat | FG-GAP 4 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 350_411 | 948.0 | 1138.0 | Repeat | FG-GAP 5 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 35_103 | 948.0 | 1138.0 | Repeat | FG-GAP 1 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 412_467 | 948.0 | 1138.0 | Repeat | FG-GAP 6 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 471_530 | 948.0 | 1138.0 | Repeat | FG-GAP 7 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 110_175 | 992.0 | 1163.0 | Repeat | FG-GAP 2 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 185_238 | 992.0 | 1163.0 | Repeat | FG-GAP 3 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 292_349 | 992.0 | 1163.0 | Repeat | FG-GAP 4 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 350_411 | 992.0 | 1163.0 | Repeat | FG-GAP 5 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 35_103 | 992.0 | 1163.0 | Repeat | FG-GAP 1 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 412_467 | 992.0 | 1163.0 | Repeat | FG-GAP 6 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 471_530 | 992.0 | 1163.0 | Repeat | FG-GAP 7 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 110_175 | 855.0 | 1045.0 | Repeat | FG-GAP 2 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 185_238 | 855.0 | 1045.0 | Repeat | FG-GAP 3 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 292_349 | 855.0 | 1045.0 | Repeat | FG-GAP 4 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 350_411 | 855.0 | 1045.0 | Repeat | FG-GAP 5 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 35_103 | 855.0 | 1045.0 | Repeat | FG-GAP 1 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 412_467 | 855.0 | 1045.0 | Repeat | FG-GAP 6 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 471_530 | 855.0 | 1045.0 | Repeat | FG-GAP 7 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 110_175 | 952.0 | 1142.0 | Repeat | FG-GAP 2 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 185_238 | 952.0 | 1142.0 | Repeat | FG-GAP 3 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 292_349 | 952.0 | 1142.0 | Repeat | FG-GAP 4 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 350_411 | 952.0 | 1142.0 | Repeat | FG-GAP 5 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 35_103 | 952.0 | 1142.0 | Repeat | FG-GAP 1 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 412_467 | 952.0 | 1142.0 | Repeat | FG-GAP 6 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 471_530 | 952.0 | 1142.0 | Repeat | FG-GAP 7 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 110_175 | 992.0 | 1182.0 | Repeat | FG-GAP 2 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 185_238 | 992.0 | 1182.0 | Repeat | FG-GAP 3 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 292_349 | 992.0 | 1182.0 | Repeat | FG-GAP 4 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 350_411 | 992.0 | 1182.0 | Repeat | FG-GAP 5 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 35_103 | 992.0 | 1182.0 | Repeat | FG-GAP 1 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 412_467 | 992.0 | 1182.0 | Repeat | FG-GAP 6 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 471_530 | 992.0 | 1182.0 | Repeat | FG-GAP 7 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 110_175 | 948.0 | 1138.0 | Repeat | FG-GAP 2 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 185_238 | 948.0 | 1138.0 | Repeat | FG-GAP 3 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 292_349 | 948.0 | 1138.0 | Repeat | FG-GAP 4 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 350_411 | 948.0 | 1138.0 | Repeat | FG-GAP 5 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 35_103 | 948.0 | 1138.0 | Repeat | FG-GAP 1 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 412_467 | 948.0 | 1138.0 | Repeat | FG-GAP 6 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 471_530 | 948.0 | 1138.0 | Repeat | FG-GAP 7 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 110_175 | 992.0 | 1163.0 | Repeat | FG-GAP 2 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 185_238 | 992.0 | 1163.0 | Repeat | FG-GAP 3 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 292_349 | 992.0 | 1163.0 | Repeat | FG-GAP 4 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 350_411 | 992.0 | 1163.0 | Repeat | FG-GAP 5 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 35_103 | 992.0 | 1163.0 | Repeat | FG-GAP 1 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 412_467 | 992.0 | 1163.0 | Repeat | FG-GAP 6 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 471_530 | 992.0 | 1163.0 | Repeat | FG-GAP 7 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 110_175 | 855.0 | 1045.0 | Repeat | FG-GAP 2 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 185_238 | 855.0 | 1045.0 | Repeat | FG-GAP 3 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 292_349 | 855.0 | 1045.0 | Repeat | FG-GAP 4 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 350_411 | 855.0 | 1045.0 | Repeat | FG-GAP 5 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 35_103 | 855.0 | 1045.0 | Repeat | FG-GAP 1 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 412_467 | 855.0 | 1045.0 | Repeat | FG-GAP 6 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 471_530 | 855.0 | 1045.0 | Repeat | FG-GAP 7 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 110_175 | 952.0 | 1142.0 | Repeat | FG-GAP 2 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 185_238 | 952.0 | 1142.0 | Repeat | FG-GAP 3 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 292_349 | 952.0 | 1142.0 | Repeat | FG-GAP 4 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 350_411 | 952.0 | 1142.0 | Repeat | FG-GAP 5 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 35_103 | 952.0 | 1142.0 | Repeat | FG-GAP 1 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 412_467 | 952.0 | 1142.0 | Repeat | FG-GAP 6 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 471_530 | 952.0 | 1142.0 | Repeat | FG-GAP 7 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 110_175 | 992.0 | 1182.0 | Repeat | FG-GAP 2 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 185_238 | 992.0 | 1182.0 | Repeat | FG-GAP 3 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 292_349 | 992.0 | 1182.0 | Repeat | FG-GAP 4 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 350_411 | 992.0 | 1182.0 | Repeat | FG-GAP 5 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 35_103 | 992.0 | 1182.0 | Repeat | FG-GAP 1 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 412_467 | 992.0 | 1182.0 | Repeat | FG-GAP 6 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 471_530 | 992.0 | 1182.0 | Repeat | FG-GAP 7 |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 177_183 | 120.0 | 264.0 | Intramembrane | Ontology_term=ECO:0000250 | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 184_194 | 120.0 | 264.0 | Intramembrane | Helical | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 177_183 | 120.0 | 264.0 | Intramembrane | Ontology_term=ECO:0000250 | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 184_194 | 120.0 | 264.0 | Intramembrane | Helical | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 184_186 | 120.0 | 264.0 | Motif | Note=NPA 2 | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 184_186 | 120.0 | 264.0 | Motif | Note=NPA 2 | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 148_159 | 120.0 | 264.0 | Topological domain | Cytoplasmic | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 195_200 | 120.0 | 264.0 | Topological domain | Extracellular | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 220_263 | 120.0 | 264.0 | Topological domain | Cytoplasmic | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 148_159 | 120.0 | 264.0 | Topological domain | Cytoplasmic | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 195_200 | 120.0 | 264.0 | Topological domain | Extracellular | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 220_263 | 120.0 | 264.0 | Topological domain | Cytoplasmic | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 127_147 | 120.0 | 264.0 | Transmembrane | Helical | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 160_176 | 120.0 | 264.0 | Transmembrane | Helical | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 201_219 | 120.0 | 264.0 | Transmembrane | Helical | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 127_147 | 120.0 | 264.0 | Transmembrane | Helical | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 160_176 | 120.0 | 264.0 | Transmembrane | Helical | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 201_219 | 120.0 | 264.0 | Transmembrane | Helical |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 953_958 | 948.0 | 1138.0 | Compositional bias | Note=Poly-Arg |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 953_958 | 855.0 | 1045.0 | Compositional bias | Note=Poly-Arg |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 953_958 | 952.0 | 1142.0 | Compositional bias | Note=Poly-Arg |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 953_958 | 948.0 | 1138.0 | Compositional bias | Note=Poly-Arg |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 953_958 | 855.0 | 1045.0 | Compositional bias | Note=Poly-Arg |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 953_958 | 952.0 | 1142.0 | Compositional bias | Note=Poly-Arg |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1107_1111 | 948.0 | 1138.0 | Motif | Note=GFFKR motif |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1107_1111 | 992.0 | 1163.0 | Motif | Note=GFFKR motif |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1107_1111 | 855.0 | 1045.0 | Motif | Note=GFFKR motif |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1107_1111 | 952.0 | 1142.0 | Motif | Note=GFFKR motif |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1107_1111 | 992.0 | 1182.0 | Motif | Note=GFFKR motif |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1107_1111 | 948.0 | 1138.0 | Motif | Note=GFFKR motif |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1107_1111 | 992.0 | 1163.0 | Motif | Note=GFFKR motif |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1107_1111 | 855.0 | 1045.0 | Motif | Note=GFFKR motif |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1107_1111 | 952.0 | 1142.0 | Motif | Note=GFFKR motif |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1107_1111 | 992.0 | 1182.0 | Motif | Note=GFFKR motif |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1157_1176 | 948.0 | 1138.0 | Region | Note=3 X 4 AA repeats of D-X-H-P |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1157_1176 | 992.0 | 1163.0 | Region | Note=3 X 4 AA repeats of D-X-H-P |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1157_1176 | 855.0 | 1045.0 | Region | Note=3 X 4 AA repeats of D-X-H-P |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1157_1176 | 952.0 | 1142.0 | Region | Note=3 X 4 AA repeats of D-X-H-P |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1157_1176 | 992.0 | 1182.0 | Region | Note=3 X 4 AA repeats of D-X-H-P |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1157_1176 | 948.0 | 1138.0 | Region | Note=3 X 4 AA repeats of D-X-H-P |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1157_1176 | 992.0 | 1163.0 | Region | Note=3 X 4 AA repeats of D-X-H-P |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1157_1176 | 855.0 | 1045.0 | Region | Note=3 X 4 AA repeats of D-X-H-P |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1157_1176 | 952.0 | 1142.0 | Region | Note=3 X 4 AA repeats of D-X-H-P |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1157_1176 | 992.0 | 1182.0 | Region | Note=3 X 4 AA repeats of D-X-H-P |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1157_1160 | 948.0 | 1138.0 | Repeat | Note=1 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1165_1168 | 948.0 | 1138.0 | Repeat | Note=2 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1173_1176 | 948.0 | 1138.0 | Repeat | Note=3 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1157_1160 | 992.0 | 1163.0 | Repeat | Note=1 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1165_1168 | 992.0 | 1163.0 | Repeat | Note=2 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1173_1176 | 992.0 | 1163.0 | Repeat | Note=3 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1157_1160 | 855.0 | 1045.0 | Repeat | Note=1 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1165_1168 | 855.0 | 1045.0 | Repeat | Note=2 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1173_1176 | 855.0 | 1045.0 | Repeat | Note=3 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1157_1160 | 952.0 | 1142.0 | Repeat | Note=1 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1165_1168 | 952.0 | 1142.0 | Repeat | Note=2 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1173_1176 | 952.0 | 1142.0 | Repeat | Note=3 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1157_1160 | 992.0 | 1182.0 | Repeat | Note=1 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1165_1168 | 992.0 | 1182.0 | Repeat | Note=2 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1173_1176 | 992.0 | 1182.0 | Repeat | Note=3 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1157_1160 | 948.0 | 1138.0 | Repeat | Note=1 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1165_1168 | 948.0 | 1138.0 | Repeat | Note=2 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1173_1176 | 948.0 | 1138.0 | Repeat | Note=3 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1157_1160 | 992.0 | 1163.0 | Repeat | Note=1 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1165_1168 | 992.0 | 1163.0 | Repeat | Note=2 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1173_1176 | 992.0 | 1163.0 | Repeat | Note=3 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1157_1160 | 855.0 | 1045.0 | Repeat | Note=1 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1165_1168 | 855.0 | 1045.0 | Repeat | Note=2 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1173_1176 | 855.0 | 1045.0 | Repeat | Note=3 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1157_1160 | 952.0 | 1142.0 | Repeat | Note=1 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1165_1168 | 952.0 | 1142.0 | Repeat | Note=2 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1173_1176 | 952.0 | 1142.0 | Repeat | Note=3 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1157_1160 | 992.0 | 1182.0 | Repeat | Note=1 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1165_1168 | 992.0 | 1182.0 | Repeat | Note=2 |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1173_1176 | 992.0 | 1182.0 | Repeat | Note=3 |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1104_1181 | 948.0 | 1138.0 | Topological domain | Cytoplasmic |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 34_1082 | 948.0 | 1138.0 | Topological domain | Extracellular |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1104_1181 | 992.0 | 1163.0 | Topological domain | Cytoplasmic |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 34_1082 | 992.0 | 1163.0 | Topological domain | Extracellular |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1104_1181 | 855.0 | 1045.0 | Topological domain | Cytoplasmic |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 34_1082 | 855.0 | 1045.0 | Topological domain | Extracellular |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1104_1181 | 952.0 | 1142.0 | Topological domain | Cytoplasmic |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 34_1082 | 952.0 | 1142.0 | Topological domain | Extracellular |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1104_1181 | 992.0 | 1182.0 | Topological domain | Cytoplasmic |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 34_1082 | 992.0 | 1182.0 | Topological domain | Extracellular |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1104_1181 | 948.0 | 1138.0 | Topological domain | Cytoplasmic |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 34_1082 | 948.0 | 1138.0 | Topological domain | Extracellular |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1104_1181 | 992.0 | 1163.0 | Topological domain | Cytoplasmic |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 34_1082 | 992.0 | 1163.0 | Topological domain | Extracellular |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1104_1181 | 855.0 | 1045.0 | Topological domain | Cytoplasmic |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 34_1082 | 855.0 | 1045.0 | Topological domain | Extracellular |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1104_1181 | 952.0 | 1142.0 | Topological domain | Cytoplasmic |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 34_1082 | 952.0 | 1142.0 | Topological domain | Extracellular |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1104_1181 | 992.0 | 1182.0 | Topological domain | Cytoplasmic |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 34_1082 | 992.0 | 1182.0 | Topological domain | Extracellular |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1083_1103 | 948.0 | 1138.0 | Transmembrane | Helical |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1083_1103 | 992.0 | 1163.0 | Transmembrane | Helical |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1083_1103 | 855.0 | 1045.0 | Transmembrane | Helical |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1083_1103 | 952.0 | 1142.0 | Transmembrane | Helical |
| Hgene | ITGA7 | chr12:56086627 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1083_1103 | 992.0 | 1182.0 | Transmembrane | Helical |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257879 | - | 21 | 25 | 1083_1103 | 948.0 | 1138.0 | Transmembrane | Helical |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000257880 | - | 22 | 27 | 1083_1103 | 992.0 | 1163.0 | Transmembrane | Helical |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000452168 | - | 21 | 25 | 1083_1103 | 855.0 | 1045.0 | Transmembrane | Helical |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000553804 | - | 21 | 25 | 1083_1103 | 952.0 | 1142.0 | Transmembrane | Helical |
| Hgene | ITGA7 | chr12:56086628 | chr12:56847539 | ENST00000555728 | - | 22 | 26 | 1083_1103 | 992.0 | 1182.0 | Transmembrane | Helical |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 62_67 | 120.0 | 264.0 | Intramembrane | Ontology_term=ECO:0000250 | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 68_78 | 120.0 | 264.0 | Intramembrane | Helical | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 62_67 | 120.0 | 264.0 | Intramembrane | Ontology_term=ECO:0000250 | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 68_78 | 120.0 | 264.0 | Intramembrane | Helical | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 68_70 | 120.0 | 264.0 | Motif | Note=NPA 1 | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 68_70 | 120.0 | 264.0 | Motif | Note=NPA 1 | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 108_126 | 120.0 | 264.0 | Topological domain | Extracellular | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 1_8 | 120.0 | 264.0 | Topological domain | Cytoplasmic | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 33_38 | 120.0 | 264.0 | Topological domain | Extracellular | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 79_84 | 120.0 | 264.0 | Topological domain | Cytoplasmic | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 108_126 | 120.0 | 264.0 | Topological domain | Extracellular | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 1_8 | 120.0 | 264.0 | Topological domain | Cytoplasmic | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 33_38 | 120.0 | 264.0 | Topological domain | Extracellular | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 79_84 | 120.0 | 264.0 | Topological domain | Cytoplasmic | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 39_61 | 120.0 | 264.0 | Transmembrane | Helical | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 85_107 | 120.0 | 264.0 | Transmembrane | Helical | |
| Tgene | MIP | chr12:56086627 | chr12:56847539 | ENST00000257979 | 0 | 4 | 9_32 | 120.0 | 264.0 | Transmembrane | Helical | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 39_61 | 120.0 | 264.0 | Transmembrane | Helical | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 85_107 | 120.0 | 264.0 | Transmembrane | Helical | |
| Tgene | MIP | chr12:56086628 | chr12:56847539 | ENST00000257979 | 0 | 4 | 9_32 | 120.0 | 264.0 | Transmembrane | Helical |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ITGA7 | |
| MIP |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to ITGA7-MIP |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ITGA7-MIP |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

