| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ALPK1-NAA15 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ALPK1-NAA15 | FusionPDB ID: 4109 | FusionGDB2.0 ID: 4109 | Hgene | Tgene | Gene symbol | ALPK1 | NAA15 | Gene ID | 80216 | 80155 |
| Gene name | alpha kinase 1 | N-alpha-acetyltransferase 15, NatA auxiliary subunit | |
| Synonyms | 8430410J10Rik|LAK | Ga19|MRD50|NARG1|NAT1P|NATH|TBDN|TBDN100 | |
| Cytomap | 4q25 | 4q31.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | alpha-protein kinase 1chromosome 4 kinaselymphocyte alpha-kinaselymphocyte alpha-protein kinase | N-alpha-acetyltransferase 15, NatA auxiliary subunitN-terminal acetyltransferaseNMDA receptor regulated 1NMDA receptor-regulated protein 1gastric cancer antigen Ga19protein tubedown-1transcriptional coactivator tubedown-100tubedown-1 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | Q96QP1 | Q9BXJ9 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000177648, ENST00000458497, ENST00000504176, ENST00000505912, | ENST00000480277, ENST00000515576, ENST00000296543, ENST00000398947, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 6 X 6 X 3=108 | 14 X 11 X 8=1232 |
| # samples | 6 | 15 | |
| ** MAII score | log2(6/108*10)=-0.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/1232*10)=-3.03796785019902 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ALPK1 [Title/Abstract] AND NAA15 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ALPK1(113362261)-NAA15(140291365), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ALPK1-NAA15 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ALPK1-NAA15 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ALPK1-NAA15 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ALPK1-NAA15 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ALPK1 | GO:0002753 | cytoplasmic pattern recognition receptor signaling pathway | 28222186|28877472|30111836 |
| Hgene | ALPK1 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | 28222186|28877472|30111836 |
| Hgene | ALPK1 | GO:0045087 | innate immune response | 28222186|28877472|30111836 |
| Tgene | NAA15 | GO:0006474 | N-terminal protein amino acid acetylation | 15496142 |
| Tgene | NAA15 | GO:0045893 | positive regulation of transcription, DNA-templated | 12145306 |
 Fusion gene breakpoints across ALPK1 (5'-gene) Fusion gene breakpoints across ALPK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
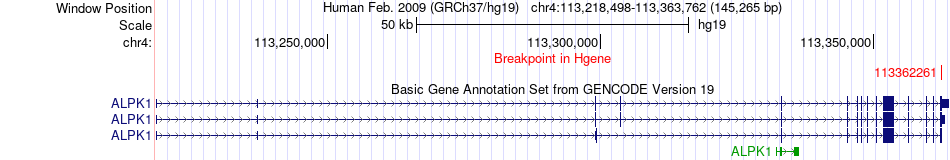 |
 Fusion gene breakpoints across NAA15 (3'-gene) Fusion gene breakpoints across NAA15 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
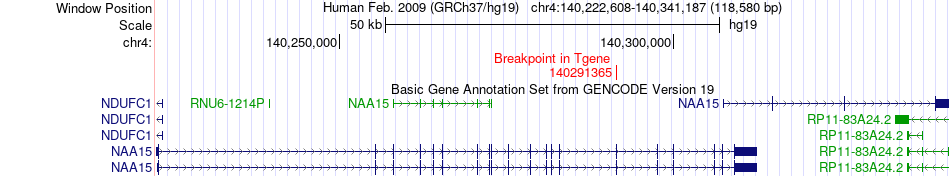 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-W3-A825-06A | ALPK1 | chr4 | 113362261 | - | NAA15 | chr4 | 140291365 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000458497 | ALPK1 | chr4 | 113362261 | - | ENST00000296543 | NAA15 | chr4 | 140291365 | + | 8152 | 4006 | 279 | 4853 | 1524 |
| ENST00000458497 | ALPK1 | chr4 | 113362261 | - | ENST00000398947 | NAA15 | chr4 | 140291365 | + | 8135 | 4006 | 279 | 4850 | 1523 |
| ENST00000177648 | ALPK1 | chr4 | 113362261 | - | ENST00000296543 | NAA15 | chr4 | 140291365 | + | 8073 | 3927 | 125 | 4774 | 1549 |
| ENST00000177648 | ALPK1 | chr4 | 113362261 | - | ENST00000398947 | NAA15 | chr4 | 140291365 | + | 8056 | 3927 | 125 | 4771 | 1548 |
| ENST00000504176 | ALPK1 | chr4 | 113362261 | - | ENST00000296543 | NAA15 | chr4 | 140291365 | + | 7908 | 3762 | 212 | 4609 | 1465 |
| ENST00000504176 | ALPK1 | chr4 | 113362261 | - | ENST00000398947 | NAA15 | chr4 | 140291365 | + | 7891 | 3762 | 212 | 4606 | 1464 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000458497 | ENST00000296543 | ALPK1 | chr4 | 113362261 | - | NAA15 | chr4 | 140291365 | + | 0.000201704 | 0.99979836 |
| ENST00000458497 | ENST00000398947 | ALPK1 | chr4 | 113362261 | - | NAA15 | chr4 | 140291365 | + | 0.000180267 | 0.9998198 |
| ENST00000177648 | ENST00000296543 | ALPK1 | chr4 | 113362261 | - | NAA15 | chr4 | 140291365 | + | 0.000186645 | 0.9998134 |
| ENST00000177648 | ENST00000398947 | ALPK1 | chr4 | 113362261 | - | NAA15 | chr4 | 140291365 | + | 0.000166989 | 0.999833 |
| ENST00000504176 | ENST00000296543 | ALPK1 | chr4 | 113362261 | - | NAA15 | chr4 | 140291365 | + | 0.000197065 | 0.999803 |
| ENST00000504176 | ENST00000398947 | ALPK1 | chr4 | 113362261 | - | NAA15 | chr4 | 140291365 | + | 0.000175144 | 0.99982494 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >4109_4109_1_ALPK1-NAA15_ALPK1_chr4_113362261_ENST00000177648_NAA15_chr4_140291365_ENST00000296543_length(amino acids)=1549AA_BP=1268 MKSNEPKLWVIDHPRPRDTQFIVIIMNNQKVVAVLLQECKQVLDQLLLEAPDVSEEDKSEDQRCRALLPSELRTLIQEAKEMKWPFVPEK WQYKQAVGPEDKTNLKDVIGAGLQQLLASLRASILARDCAAAAAIVFLVDRFLYGLDVSGKLLQVAKGLHKLQPATPIAPQVVIRQARIS VNSGKLLKAEYILSSLISNNGATGTWLYRNESDKVLVQSVCIQIRGQILQKLGMWYEAAELIWASIVGYLALPQPDKKGLSTSLGILADI FVSMSKNDYEKFKNNPQINLSLLKEFDHHLLSAAEACKLAAAFSAYTPLFVLTAVNIRGTCLLSYSSSNDCPPELKNLHLCEAKEAFEIG LLTKRDDEPVTGKQELHSFVKAAFGLTTVHRRLHGETGTVHAASQLCKEAMGKLYNFSTSSRSQDREALSQEVMSVIAQVKEHLQVQSFS NVDDRSYVPESFECRLDKLILHGQGDFQKILDTYSQHHTSVCEVFESDCGNNKNEQKDAKTGVCITALKTEIKNIDTVSTTQEKPHCQRD TGISSSLMGKNVQRELRRGGRRNWTHSDAFRVSLDQDVETETEPSDYSNGEGAVFNKSLSGSQTSSAWSNLSGFSSSASWEEVNYHVDDR SARKEPGKEHLVDTQCSTALSEELENDREGRAMHSLHSQLHDLSLQEPNNDNLEPSQNQPQQQMPLTPFSPHNTPGIFLAPGAGLLEGAP EGIQEVRNMGPRNTSAHSRPSYRSASWSSDSGRPKNMGTHPSVQKEEAFEIIVEFPETNCDVKDRQGKEQGEEISERGAGPTFKASPSWV DPEGETAESTEDAPLDFHRVLHNSLGNISMLPCSSFTPNWPVQNPDSRKSGGPVAEQGIDPDASTVDEEGQLLDSMDVPCTNGHGSHRLC ILRQPPGQRAETPNSSVSGNILFPVLSEDCTTTEEGNQPGNMLNCSQNSSSSSVWWLKSPAFSSGSSEGDSPWSYLNSSGSSWVSLPGKM RKEILEARTLQPDDFEKLLAGVRHDWLFQRLENTGVFKPSQLHRAHSALLLKYSKKSELWTAQETIVYLGDYLTVKKKGRQRNAFWVHHL HQEEILGRYVGKDYKEQKGLWHHFTDVERQMTAQHYVTEFNKRLYEQNIPTQIFYIPSTILLILEDKTIKGCISVEPYILGEFVKLSNNT KVVKTEYKATEYGLAYGHFSYEFSNHRDVVVDLQGWVTGNGKGLIYLTDPQIHSVDQKVFTTNFGKRGIFYFFNNQHVECNEICHRLSLT RPSMEKPSNMSDKELKKLRNKQRRAQKKAQIEEEKKNAEKEKQQRNQKKKKDDDDEEIGGPKEELIPEKLAKVETPLEEAIKFLTPLKNL VKNKIETHLFAFEIYFRKEKFLLMLQSVKRAFAIDSSHPWLHECMIRLFNTAVCESKDLSDTVRTVLKQEMNRLFGATNPKNFNETFLKR NSDSLPHRLSAAKMVYYLDPSSQKRAIELATTLDESLTNRNLQTCMEVLEALYDGSLGDCKEAAEIYRANCHKLFPYALAFMPPGYEEDM -------------------------------------------------------------- >4109_4109_2_ALPK1-NAA15_ALPK1_chr4_113362261_ENST00000177648_NAA15_chr4_140291365_ENST00000398947_length(amino acids)=1548AA_BP=1268 MKSNEPKLWVIDHPRPRDTQFIVIIMNNQKVVAVLLQECKQVLDQLLLEAPDVSEEDKSEDQRCRALLPSELRTLIQEAKEMKWPFVPEK WQYKQAVGPEDKTNLKDVIGAGLQQLLASLRASILARDCAAAAAIVFLVDRFLYGLDVSGKLLQVAKGLHKLQPATPIAPQVVIRQARIS VNSGKLLKAEYILSSLISNNGATGTWLYRNESDKVLVQSVCIQIRGQILQKLGMWYEAAELIWASIVGYLALPQPDKKGLSTSLGILADI FVSMSKNDYEKFKNNPQINLSLLKEFDHHLLSAAEACKLAAAFSAYTPLFVLTAVNIRGTCLLSYSSSNDCPPELKNLHLCEAKEAFEIG LLTKRDDEPVTGKQELHSFVKAAFGLTTVHRRLHGETGTVHAASQLCKEAMGKLYNFSTSSRSQDREALSQEVMSVIAQVKEHLQVQSFS NVDDRSYVPESFECRLDKLILHGQGDFQKILDTYSQHHTSVCEVFESDCGNNKNEQKDAKTGVCITALKTEIKNIDTVSTTQEKPHCQRD TGISSSLMGKNVQRELRRGGRRNWTHSDAFRVSLDQDVETETEPSDYSNGEGAVFNKSLSGSQTSSAWSNLSGFSSSASWEEVNYHVDDR SARKEPGKEHLVDTQCSTALSEELENDREGRAMHSLHSQLHDLSLQEPNNDNLEPSQNQPQQQMPLTPFSPHNTPGIFLAPGAGLLEGAP EGIQEVRNMGPRNTSAHSRPSYRSASWSSDSGRPKNMGTHPSVQKEEAFEIIVEFPETNCDVKDRQGKEQGEEISERGAGPTFKASPSWV DPEGETAESTEDAPLDFHRVLHNSLGNISMLPCSSFTPNWPVQNPDSRKSGGPVAEQGIDPDASTVDEEGQLLDSMDVPCTNGHGSHRLC ILRQPPGQRAETPNSSVSGNILFPVLSEDCTTTEEGNQPGNMLNCSQNSSSSSVWWLKSPAFSSGSSEGDSPWSYLNSSGSSWVSLPGKM RKEILEARTLQPDDFEKLLAGVRHDWLFQRLENTGVFKPSQLHRAHSALLLKYSKKSELWTAQETIVYLGDYLTVKKKGRQRNAFWVHHL HQEEILGRYVGKDYKEQKGLWHHFTDVERQMTAQHYVTEFNKRLYEQNIPTQIFYIPSTILLILEDKTIKGCISVEPYILGEFVKLSNNT KVVKTEYKATEYGLAYGHFSYEFSNHRDVVVDLQGWVTGNGKGLIYLTDPQIHSVDQKVFTTNFGKRGIFYFFNNQHVECNEICHRLSLT RPSMEKPSNMSDKELKKLRNKQRRAQKKAQIEEEKKNAEKEKQQRNQKKKKDDDDEEIGGPKEELIPEKLAKVETPLEEAIKFLTPLKNL VKNKIETHLFAFEIYFRKEKFLLMLQSVKRAFAIDSSHPWLHECMIRLFNTVCESKDLSDTVRTVLKQEMNRLFGATNPKNFNETFLKRN SDSLPHRLSAAKMVYYLDPSSQKRAIELATTLDESLTNRNLQTCMEVLEALYDGSLGDCKEAAEIYRANCHKLFPYALAFMPPGYEEDMK -------------------------------------------------------------- >4109_4109_3_ALPK1-NAA15_ALPK1_chr4_113362261_ENST00000458497_NAA15_chr4_140291365_ENST00000296543_length(amino acids)=1524AA_BP=1243 MNNQKVVAVLLQECKQVLDQLLLEAPDVSEEDKSEDQRCRALLPSELRTLIQEAKEMKWPFVPEKWQYKQAVGPEDKTNLKDVIGAGLQQ LLASLRASILARDCAAAAAIVFLVDRFLYGLDVSGKLLQVAKGLHKLQPATPIAPQVVIRQARISVNSGKLLKAEYILSSLISNNGATGT WLYRNESDKVLVQSVCIQIRGQILQKLGMWYEAAELIWASIVGYLALPQPDKKGLSTSLGILADIFVSMSKNDYEKFKNNPQINLSLLKE FDHHLLSAAEACKLAAAFSAYTPLFVLTAVNIRGTCLLSYSSSNDCPPELKNLHLCEAKEAFEIGLLTKRDDEPVTGKQELHSFVKAAFG LTTVHRRLHGETGTVHAASQLCKEAMGKLYNFSTSSRSQDREALSQEVMSVIAQVKEHLQVQSFSNVDDRSYVPESFECRLDKLILHGQG DFQKILDTYSQHHTSVCEVFESDCGNNKNEQKDAKTGVCITALKTEIKNIDTVSTTQEKPHCQRDTGISSSLMGKNVQRELRRGGRRNWT HSDAFRVSLDQDVETETEPSDYSNGEGAVFNKSLSGSQTSSAWSNLSGFSSSASWEEVNYHVDDRSARKEPGKEHLVDTQCSTALSEELE NDREGRAMHSLHSQLHDLSLQEPNNDNLEPSQNQPQQQMPLTPFSPHNTPGIFLAPGAGLLEGAPEGIQEVRNMGPRNTSAHSRPSYRSA SWSSDSGRPKNMGTHPSVQKEEAFEIIVEFPETNCDVKDRQGKEQGEEISERGAGPTFKASPSWVDPEGETAESTEDAPLDFHRVLHNSL GNISMLPCSSFTPNWPVQNPDSRKSGGPVAEQGIDPDASTVDEEGQLLDSMDVPCTNGHGSHRLCILRQPPGQRAETPNSSVSGNILFPV LSEDCTTTEEGNQPGNMLNCSQNSSSSSVWWLKSPAFSSGSSEGDSPWSYLNSSGSSWVSLPGKMRKEILEARTLQPDDFEKLLAGVRHD WLFQRLENTGVFKPSQLHRAHSALLLKYSKKSELWTAQETIVYLGDYLTVKKKGRQRNAFWVHHLHQEEILGRYVGKDYKEQKGLWHHFT DVERQMTAQHYVTEFNKRLYEQNIPTQIFYIPSTILLILEDKTIKGCISVEPYILGEFVKLSNNTKVVKTEYKATEYGLAYGHFSYEFSN HRDVVVDLQGWVTGNGKGLIYLTDPQIHSVDQKVFTTNFGKRGIFYFFNNQHVECNEICHRLSLTRPSMEKPSNMSDKELKKLRNKQRRA QKKAQIEEEKKNAEKEKQQRNQKKKKDDDDEEIGGPKEELIPEKLAKVETPLEEAIKFLTPLKNLVKNKIETHLFAFEIYFRKEKFLLML QSVKRAFAIDSSHPWLHECMIRLFNTAVCESKDLSDTVRTVLKQEMNRLFGATNPKNFNETFLKRNSDSLPHRLSAAKMVYYLDPSSQKR -------------------------------------------------------------- >4109_4109_4_ALPK1-NAA15_ALPK1_chr4_113362261_ENST00000458497_NAA15_chr4_140291365_ENST00000398947_length(amino acids)=1523AA_BP=1243 MNNQKVVAVLLQECKQVLDQLLLEAPDVSEEDKSEDQRCRALLPSELRTLIQEAKEMKWPFVPEKWQYKQAVGPEDKTNLKDVIGAGLQQ LLASLRASILARDCAAAAAIVFLVDRFLYGLDVSGKLLQVAKGLHKLQPATPIAPQVVIRQARISVNSGKLLKAEYILSSLISNNGATGT WLYRNESDKVLVQSVCIQIRGQILQKLGMWYEAAELIWASIVGYLALPQPDKKGLSTSLGILADIFVSMSKNDYEKFKNNPQINLSLLKE FDHHLLSAAEACKLAAAFSAYTPLFVLTAVNIRGTCLLSYSSSNDCPPELKNLHLCEAKEAFEIGLLTKRDDEPVTGKQELHSFVKAAFG LTTVHRRLHGETGTVHAASQLCKEAMGKLYNFSTSSRSQDREALSQEVMSVIAQVKEHLQVQSFSNVDDRSYVPESFECRLDKLILHGQG DFQKILDTYSQHHTSVCEVFESDCGNNKNEQKDAKTGVCITALKTEIKNIDTVSTTQEKPHCQRDTGISSSLMGKNVQRELRRGGRRNWT HSDAFRVSLDQDVETETEPSDYSNGEGAVFNKSLSGSQTSSAWSNLSGFSSSASWEEVNYHVDDRSARKEPGKEHLVDTQCSTALSEELE NDREGRAMHSLHSQLHDLSLQEPNNDNLEPSQNQPQQQMPLTPFSPHNTPGIFLAPGAGLLEGAPEGIQEVRNMGPRNTSAHSRPSYRSA SWSSDSGRPKNMGTHPSVQKEEAFEIIVEFPETNCDVKDRQGKEQGEEISERGAGPTFKASPSWVDPEGETAESTEDAPLDFHRVLHNSL GNISMLPCSSFTPNWPVQNPDSRKSGGPVAEQGIDPDASTVDEEGQLLDSMDVPCTNGHGSHRLCILRQPPGQRAETPNSSVSGNILFPV LSEDCTTTEEGNQPGNMLNCSQNSSSSSVWWLKSPAFSSGSSEGDSPWSYLNSSGSSWVSLPGKMRKEILEARTLQPDDFEKLLAGVRHD WLFQRLENTGVFKPSQLHRAHSALLLKYSKKSELWTAQETIVYLGDYLTVKKKGRQRNAFWVHHLHQEEILGRYVGKDYKEQKGLWHHFT DVERQMTAQHYVTEFNKRLYEQNIPTQIFYIPSTILLILEDKTIKGCISVEPYILGEFVKLSNNTKVVKTEYKATEYGLAYGHFSYEFSN HRDVVVDLQGWVTGNGKGLIYLTDPQIHSVDQKVFTTNFGKRGIFYFFNNQHVECNEICHRLSLTRPSMEKPSNMSDKELKKLRNKQRRA QKKAQIEEEKKNAEKEKQQRNQKKKKDDDDEEIGGPKEELIPEKLAKVETPLEEAIKFLTPLKNLVKNKIETHLFAFEIYFRKEKFLLML QSVKRAFAIDSSHPWLHECMIRLFNTVCESKDLSDTVRTVLKQEMNRLFGATNPKNFNETFLKRNSDSLPHRLSAAKMVYYLDPSSQKRA -------------------------------------------------------------- >4109_4109_5_ALPK1-NAA15_ALPK1_chr4_113362261_ENST00000504176_NAA15_chr4_140291365_ENST00000296543_length(amino acids)=1465AA_BP=1184 MCYCKSASKCWISSCWKRQMCRKRTRARTSAAEASLRASILARDCAAAAAIVFLVDRFLYGLDVSGKLLQVAKGLHKLQPATPIAPQVVI RQARISVNSGKLLKAEYILSSLISNNGATGTWLYRNESDKVLVQSVCIQIRGQILQKLGMWYEAAELIWASIVGYLALPQPDKKGLSTSL GILADIFVSMSKNDYEKFKNNPQINLSLLKEFDHHLLSAAEACKLAAAFSAYTPLFVLTAVNIRGTCLLSYSSSNDCPPELKNLHLCEAK EAFEIGLLTKRDDEPVTGKQELHSFVKAAFGLTTVHRRLHGETGTVHAASQLCKEAMGKLYNFSTSSRSQDREALSQEVMSVIAQVKEHL QVQSFSNVDDRSYVPESFECRLDKLILHGQGDFQKILDTYSQHHTSVCEVFESDCGNNKNEQKDAKTGVCITALKTEIKNIDTVSTTQEK PHCQRDTGISSSLMGKNVQRELRRGGRRNWTHSDAFRVSLDQDVETETEPSDYSNGEGAVFNKSLSGSQTSSAWSNLSGFSSSASWEEVN YHVDDRSARKEPGKEHLVDTQCSTALSEELENDREGRAMHSLHSQLHDLSLQEPNNDNLEPSQNQPQQQMPLTPFSPHNTPGIFLAPGAG LLEGAPEGIQEVRNMGPRNTSAHSRPSYRSASWSSDSGRPKNMGTHPSVQKEEAFEIIVEFPETNCDVKDRQGKEQGEEISERGAGPTFK ASPSWVDPEGETAESTEDAPLDFHRVLHNSLGNISMLPCSSFTPNWPVQNPDSRKSGGPVAEQGIDPDASTVDEEGQLLDSMDVPCTNGH GSHRLCILRQPPGQRAETPNSSVSGNILFPVLSEDCTTTEEGNQPGNMLNCSQNSSSSSVWWLKSPAFSSGSSEGDSPWSYLNSSGSSWV SLPGKMRKEILEARTLQPDDFEKLLAGVRHDWLFQRLENTGVFKPSQLHRAHSALLLKYSKKSELWTAQETIVYLGDYLTVKKKGRQRNA FWVHHLHQEEILGRYVGKDYKEQKGLWHHFTDVERQMTAQHYVTEFNKRLYEQNIPTQIFYIPSTILLILEDKTIKGCISVEPYILGEFV KLSNNTKVVKTEYKATEYGLAYGHFSYEFSNHRDVVVDLQGWVTGNGKGLIYLTDPQIHSVDQKVFTTNFGKRGIFYFFNNQHVECNEIC HRLSLTRPSMEKPSNMSDKELKKLRNKQRRAQKKAQIEEEKKNAEKEKQQRNQKKKKDDDDEEIGGPKEELIPEKLAKVETPLEEAIKFL TPLKNLVKNKIETHLFAFEIYFRKEKFLLMLQSVKRAFAIDSSHPWLHECMIRLFNTAVCESKDLSDTVRTVLKQEMNRLFGATNPKNFN ETFLKRNSDSLPHRLSAAKMVYYLDPSSQKRAIELATTLDESLTNRNLQTCMEVLEALYDGSLGDCKEAAEIYRANCHKLFPYALAFMPP -------------------------------------------------------------- >4109_4109_6_ALPK1-NAA15_ALPK1_chr4_113362261_ENST00000504176_NAA15_chr4_140291365_ENST00000398947_length(amino acids)=1464AA_BP=1184 MCYCKSASKCWISSCWKRQMCRKRTRARTSAAEASLRASILARDCAAAAAIVFLVDRFLYGLDVSGKLLQVAKGLHKLQPATPIAPQVVI RQARISVNSGKLLKAEYILSSLISNNGATGTWLYRNESDKVLVQSVCIQIRGQILQKLGMWYEAAELIWASIVGYLALPQPDKKGLSTSL GILADIFVSMSKNDYEKFKNNPQINLSLLKEFDHHLLSAAEACKLAAAFSAYTPLFVLTAVNIRGTCLLSYSSSNDCPPELKNLHLCEAK EAFEIGLLTKRDDEPVTGKQELHSFVKAAFGLTTVHRRLHGETGTVHAASQLCKEAMGKLYNFSTSSRSQDREALSQEVMSVIAQVKEHL QVQSFSNVDDRSYVPESFECRLDKLILHGQGDFQKILDTYSQHHTSVCEVFESDCGNNKNEQKDAKTGVCITALKTEIKNIDTVSTTQEK PHCQRDTGISSSLMGKNVQRELRRGGRRNWTHSDAFRVSLDQDVETETEPSDYSNGEGAVFNKSLSGSQTSSAWSNLSGFSSSASWEEVN YHVDDRSARKEPGKEHLVDTQCSTALSEELENDREGRAMHSLHSQLHDLSLQEPNNDNLEPSQNQPQQQMPLTPFSPHNTPGIFLAPGAG LLEGAPEGIQEVRNMGPRNTSAHSRPSYRSASWSSDSGRPKNMGTHPSVQKEEAFEIIVEFPETNCDVKDRQGKEQGEEISERGAGPTFK ASPSWVDPEGETAESTEDAPLDFHRVLHNSLGNISMLPCSSFTPNWPVQNPDSRKSGGPVAEQGIDPDASTVDEEGQLLDSMDVPCTNGH GSHRLCILRQPPGQRAETPNSSVSGNILFPVLSEDCTTTEEGNQPGNMLNCSQNSSSSSVWWLKSPAFSSGSSEGDSPWSYLNSSGSSWV SLPGKMRKEILEARTLQPDDFEKLLAGVRHDWLFQRLENTGVFKPSQLHRAHSALLLKYSKKSELWTAQETIVYLGDYLTVKKKGRQRNA FWVHHLHQEEILGRYVGKDYKEQKGLWHHFTDVERQMTAQHYVTEFNKRLYEQNIPTQIFYIPSTILLILEDKTIKGCISVEPYILGEFV KLSNNTKVVKTEYKATEYGLAYGHFSYEFSNHRDVVVDLQGWVTGNGKGLIYLTDPQIHSVDQKVFTTNFGKRGIFYFFNNQHVECNEIC HRLSLTRPSMEKPSNMSDKELKKLRNKQRRAQKKAQIEEEKKNAEKEKQQRNQKKKKDDDDEEIGGPKEELIPEKLAKVETPLEEAIKFL TPLKNLVKNKIETHLFAFEIYFRKEKFLLMLQSVKRAFAIDSSHPWLHECMIRLFNTVCESKDLSDTVRTVLKQEMNRLFGATNPKNFNE TFLKRNSDSLPHRLSAAKMVYYLDPSSQKRAIELATTLDESLTNRNLQTCMEVLEALYDGSLGDCKEAAEIYRANCHKLFPYALAFMPPG -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:113362261/chr4:140291365) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ALPK1 | NAA15 |
| FUNCTION: Serine/threonine-protein kinase that detects bacterial pathogen-associated molecular pattern metabolites (PAMPs) and initiates an innate immune response, a critical step for pathogen elimination and engagement of adaptive immunity (PubMed:28877472, PubMed:28222186, PubMed:30111836). Specifically recognizes and binds ADP-D-glycero-beta-D-manno-heptose (ADP-Heptose), a potent PAMP present in all Gram-negative and some Gram-positive bacteria (PubMed:30111836). ADP-Heptose-binding stimulates its kinase activity to phosphorylate and activate TIFA, triggering proinflammatory NF-kappa-B signaling (PubMed:30111836). May be involved in monosodium urate monohydrate (MSU)-induced inflammation by mediating phosphorylation of unconventional myosin MYO9A (PubMed:27169898). May also play a role in apical protein transport by mediating phosphorylation of unconventional myosin MYO1A (PubMed:15883161). {ECO:0000269|PubMed:15883161, ECO:0000269|PubMed:27169898, ECO:0000269|PubMed:28222186, ECO:0000269|PubMed:28877472, ECO:0000269|PubMed:30111836}. | FUNCTION: Auxillary subunit of the N-terminal acetyltransferase A (NatA) complex which displays alpha (N-terminal) acetyltransferase activity. The NAT activity may be important for vascular, hematopoietic and neuronal growth and development. Required to control retinal neovascularization in adult ocular endothelial cells. In complex with XRCC6 and XRCC5 (Ku80), up-regulates transcription from the osteocalcin promoter. {ECO:0000269|PubMed:11687548, ECO:0000269|PubMed:12145306, ECO:0000269|PubMed:15496142}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000177648 | - | 15 | 16 | 105_109 | 1242.3333333333333 | 1245.0 | Compositional bias | Note=Poly-Ala |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000177648 | - | 15 | 16 | 921_960 | 1242.3333333333333 | 1245.0 | Compositional bias | Note=Ser-rich |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000458497 | - | 15 | 16 | 105_109 | 1242.3333333333333 | 1245.0 | Compositional bias | Note=Poly-Ala |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000458497 | - | 15 | 16 | 921_960 | 1242.3333333333333 | 1245.0 | Compositional bias | Note=Ser-rich |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000504176 | - | 14 | 15 | 105_109 | 1164.3333333333333 | 1167.0 | Compositional bias | Note=Poly-Ala |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000504176 | - | 14 | 15 | 921_960 | 1164.3333333333333 | 1167.0 | Compositional bias | Note=Ser-rich |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000177648 | - | 15 | 16 | 1017_1237 | 1242.3333333333333 | 1245.0 | Domain | Alpha-type protein kinase |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000458497 | - | 15 | 16 | 1017_1237 | 1242.3333333333333 | 1245.0 | Domain | Alpha-type protein kinase |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000177648 | - | 15 | 16 | 150_153 | 1242.3333333333333 | 1245.0 | Region | ADP-D-glycero-beta-D-manno-heptose binding |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000177648 | - | 15 | 16 | 236_237 | 1242.3333333333333 | 1245.0 | Region | ADP-D-glycero-beta-D-manno-heptose binding |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000458497 | - | 15 | 16 | 150_153 | 1242.3333333333333 | 1245.0 | Region | ADP-D-glycero-beta-D-manno-heptose binding |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000458497 | - | 15 | 16 | 236_237 | 1242.3333333333333 | 1245.0 | Region | ADP-D-glycero-beta-D-manno-heptose binding |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000504176 | - | 14 | 15 | 150_153 | 1164.3333333333333 | 1167.0 | Region | ADP-D-glycero-beta-D-manno-heptose binding |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000504176 | - | 14 | 15 | 236_237 | 1164.3333333333333 | 1167.0 | Region | ADP-D-glycero-beta-D-manno-heptose binding |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 629_632 | 584.3333333333334 | 867.0 | Compositional bias | Note=Poly-Asp | |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 612_629 | 584.3333333333334 | 867.0 | Motif | Bipartite nuclear localization signal | |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 672_705 | 584.3333333333334 | 867.0 | Repeat | Note=TPR 8 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ALPK1 | chr4:113362261 | chr4:140291365 | ENST00000504176 | - | 14 | 15 | 1017_1237 | 1164.3333333333333 | 1167.0 | Domain | Alpha-type protein kinase |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 148_184 | 584.3333333333334 | 867.0 | Repeat | Note=TPR 3 | |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 224_257 | 584.3333333333334 | 867.0 | Repeat | Note=TPR 4 | |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 374_407 | 584.3333333333334 | 867.0 | Repeat | Note=TPR 5 | |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 409_441 | 584.3333333333334 | 867.0 | Repeat | Note=TPR 6 | |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 46_79 | 584.3333333333334 | 867.0 | Repeat | Note=TPR 1 | |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 485_518 | 584.3333333333334 | 867.0 | Repeat | Note=TPR 7 | |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 80_113 | 584.3333333333334 | 867.0 | Repeat | Note=TPR 2 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ALPK1 | |
| NAA15 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | NAA15 | chr4:113362261 | chr4:140291365 | ENST00000296543 | 13 | 20 | 500_866 | 584.3333333333334 | 867.0 | HYPK |
Top |
Related Drugs to ALPK1-NAA15 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ALPK1-NAA15 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

