| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:KLK2-FGFR2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: KLK2-FGFR2 | FusionPDB ID: 43056 | FusionGDB2.0 ID: 43056 | Hgene | Tgene | Gene symbol | KLK2 | FGFR2 | Gene ID | 3817 | 2263 |
| Gene name | kallikrein related peptidase 2 | fibroblast growth factor receptor 2 | |
| Synonyms | KLK2A2|hGK-1|hK2 | BBDS|BEK|BFR-1|CD332|CEK3|CFD1|ECT1|JWS|K-SAM|KGFR|TK14|TK25 | |
| Cytomap | 19q13.33 | 10q26.13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | kallikrein-2glandular kallikrein 2glandular kallikrein-1kallikrein 2, prostatictissue kallikrein-2 | fibroblast growth factor receptor 2BEK fibroblast growth factor receptorbacteria-expressed kinasekeratinocyte growth factor receptorprotein tyrosine kinase, receptor like 14 | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | P20151 | P21802 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000325321, ENST00000358049, ENST00000391810, ENST00000597509, | ENST00000359354, ENST00000490349, ENST00000478859, ENST00000346997, ENST00000351936, ENST00000356226, ENST00000357555, ENST00000358487, ENST00000360144, ENST00000369056, ENST00000369059, ENST00000369060, ENST00000369061, ENST00000457416, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 21 X 19 X 4=1596 | 11 X 11 X 8=968 |
| # samples | 30 | 16 | |
| ** MAII score | log2(30/1596*10)=-2.41142624572647 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/968*10)=-2.59693514238723 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: KLK2 [Title/Abstract] AND FGFR2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | KLK2(51376775)-FGFR2(123310973), # samples:9 | ||
| Anticipated loss of major functional domain due to fusion event. | KLK2-FGFR2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KLK2-FGFR2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KLK2-FGFR2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. KLK2-FGFR2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. KLK2-FGFR2 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target by not retaining the major functional domain in the partially deleted in-frame ORF. KLK2-FGFR2 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target by not retaining the major functional domain in the partially deleted in-frame ORF. KLK2-FGFR2 seems lost the major protein functional domain in Hgene partner, which is a kinase by not retaining the major functional domain in the partially deleted in-frame ORF. KLK2-FGFR2 seems lost the major protein functional domain in Hgene partner, which is a kinase by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | FGFR2 | GO:0008284 | positive regulation of cell proliferation | 8663044 |
| Tgene | FGFR2 | GO:0008543 | fibroblast growth factor receptor signaling pathway | 8663044|15629145 |
| Tgene | FGFR2 | GO:0018108 | peptidyl-tyrosine phosphorylation | 15629145|16844695 |
| Tgene | FGFR2 | GO:0046777 | protein autophosphorylation | 15629145 |
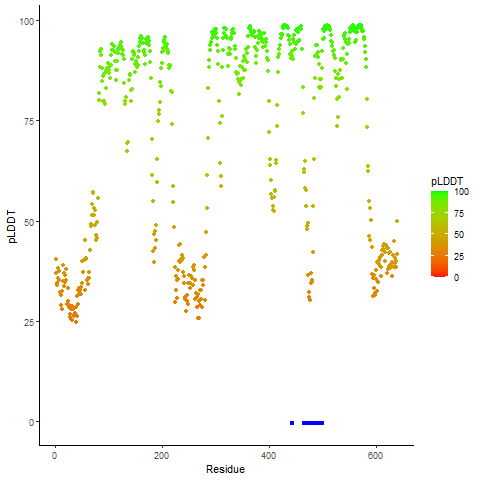
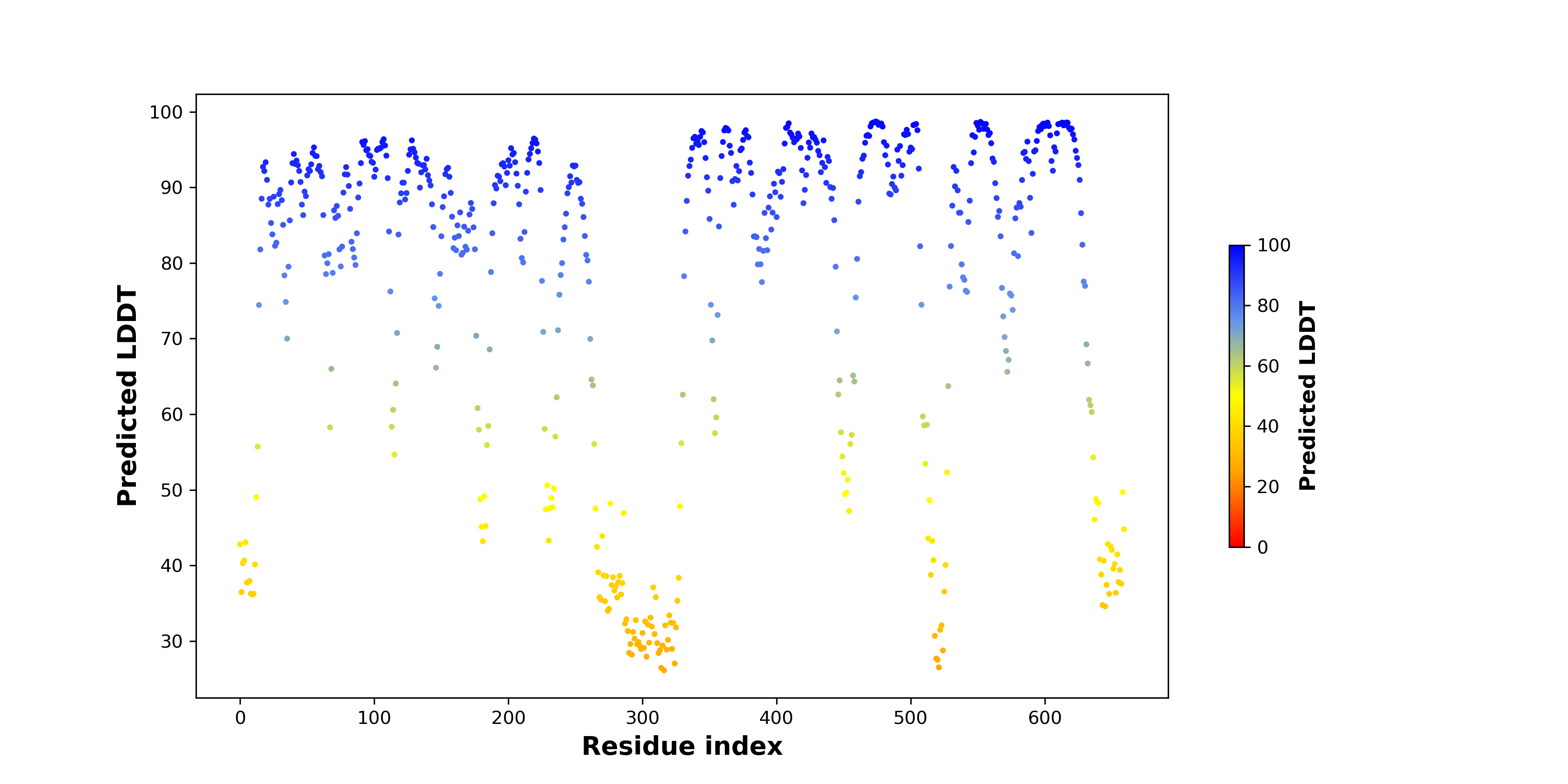
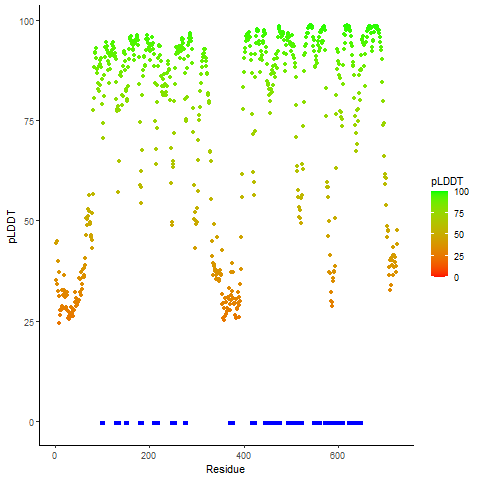
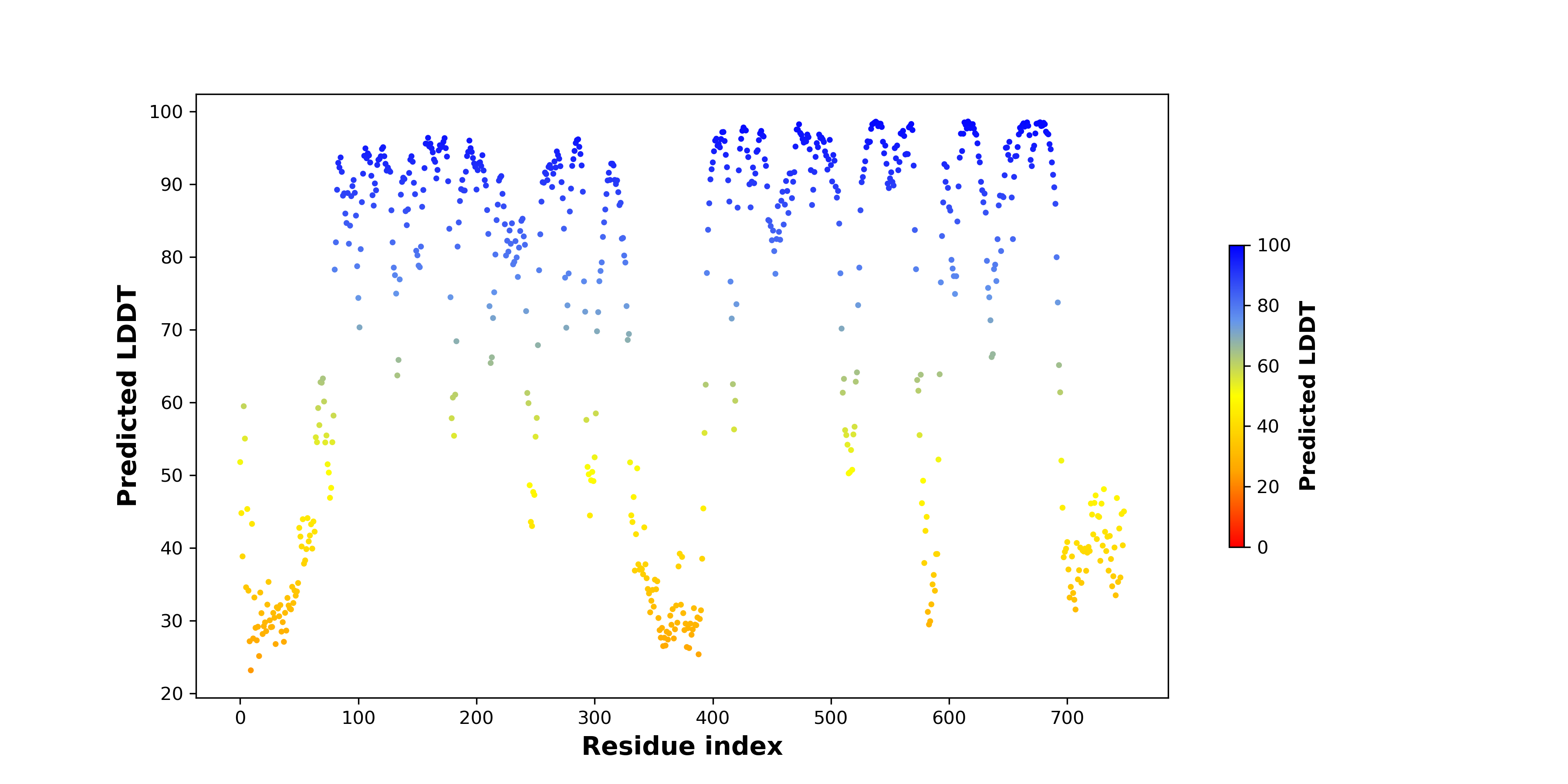
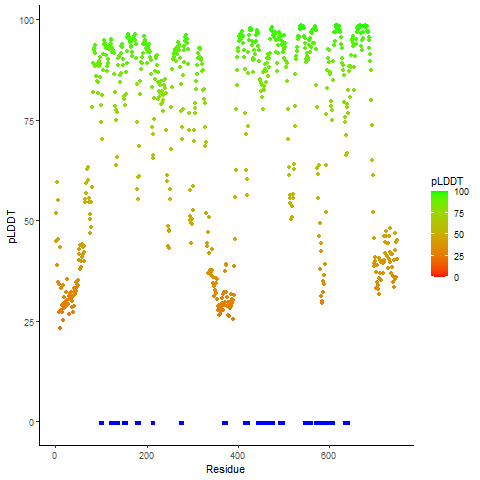
 Fusion gene breakpoints across KLK2 (5'-gene) Fusion gene breakpoints across KLK2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
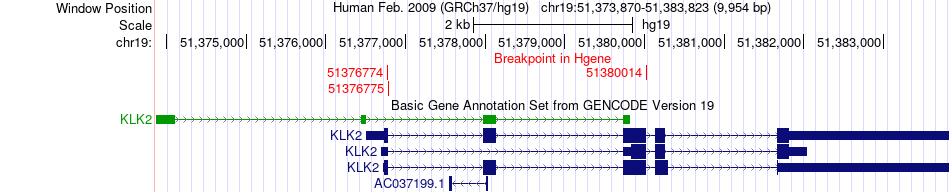 |
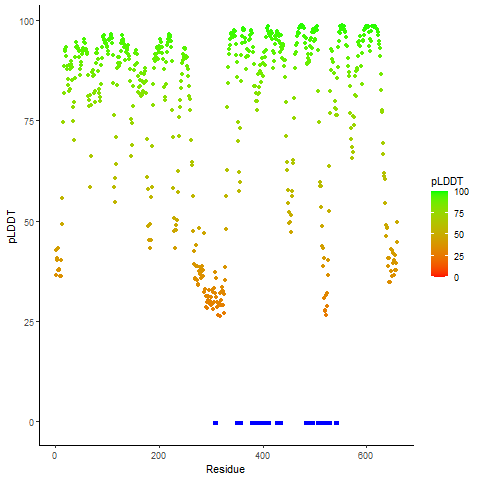
 Fusion gene breakpoints across FGFR2 (3'-gene) Fusion gene breakpoints across FGFR2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
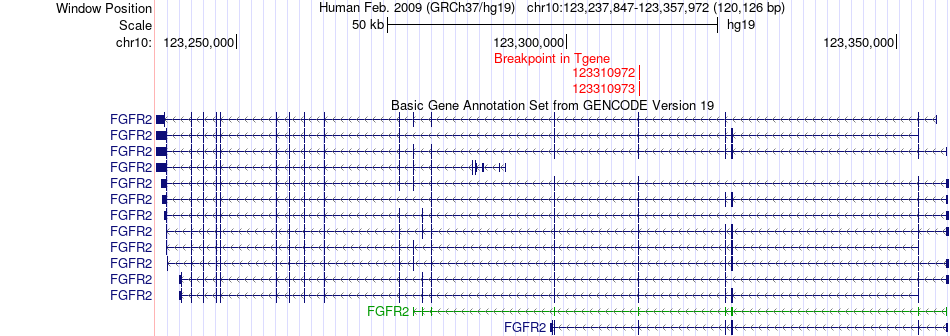 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-V1-A9O5-01A | KLK2 | chr19 | 51376775 | - | FGFR2 | chr10 | 123310973 | - |
| ChimerDB4 | PRAD | TCGA-V1-A9O5-06A | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - |
| ChimerDB4 | PRAD | TCGA-V1-A9O5-06A | KLK2 | chr19 | 51376775 | - | FGFR2 | chr10 | 123310973 | - |
| ChimerDB4 | PRAD | TCGA-V1-A9O5 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - |
| ChimerDB4 | PRAD | TCGA-V1-A9O5 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - |
| ChimerDB4 | UVM | TCGA-V4-A9EA-01A | KLK2 | chr19 | 51376775 | - | FGFR2 | chr10 | 123310973 | - |
| ChimerDB4 | UVM | TCGA-V4-A9EA | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - |
| ChimerKB4 | . | . | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000369061 | FGFR2 | chr10 | 123310972 | 3470 | 271 | 27 | 1946 | 639 | ||
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000357555 | FGFR2 | chr10 | 123310972 | 3455 | 271 | 27 | 2207 | 726 | ||
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000358487 | FGFR2 | chr10 | 123310972 | 3799 | 271 | 27 | 2282 | 751 | ||
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000356226 | FGFR2 | chr10 | 123310972 | 3062 | 271 | 27 | 2276 | 749 | ||
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000369060 | FGFR2 | chr10 | 123310972 | 2574 | 271 | 27 | 1934 | 635 | ||
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000369059 | FGFR2 | chr10 | 123310972 | 2524 | 271 | 27 | 2285 | 752 | ||
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000346997 | FGFR2 | chr10 | 123310972 | 2277 | 271 | 27 | 2276 | 749 | ||
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000457416 | FGFR2 | chr10 | 123310972 | 2286 | 271 | 27 | 2285 | 752 | ||
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000351936 | FGFR2 | chr10 | 123310972 | 2175 | 271 | 27 | 2174 | 715 | ||
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000360144 | FGFR2 | chr10 | 123310972 | 2464 | 271 | 27 | 2126 | 699 | ||
| ENST00000325321 | KLK2 | chr19 | 51376774 | ENST00000369056 | FGFR2 | chr10 | 123310972 | 2443 | 271 | 27 | 2126 | 699 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000369061 | FGFR2 | chr10 | 123310972 | 3261 | 62 | 16 | 1737 | 573 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000357555 | FGFR2 | chr10 | 123310972 | 3246 | 62 | 16 | 1998 | 660 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000358487 | FGFR2 | chr10 | 123310972 | 3590 | 62 | 16 | 2073 | 685 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000356226 | FGFR2 | chr10 | 123310972 | 2853 | 62 | 16 | 2067 | 683 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000369060 | FGFR2 | chr10 | 123310972 | 2365 | 62 | 16 | 1725 | 569 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000369059 | FGFR2 | chr10 | 123310972 | 2315 | 62 | 16 | 2076 | 686 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000346997 | FGFR2 | chr10 | 123310972 | 2068 | 62 | 16 | 2067 | 684 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000457416 | FGFR2 | chr10 | 123310972 | 2077 | 62 | 16 | 2076 | 687 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000351936 | FGFR2 | chr10 | 123310972 | 1966 | 62 | 16 | 1965 | 650 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000360144 | FGFR2 | chr10 | 123310972 | 2255 | 62 | 16 | 1917 | 633 | ||
| ENST00000358049 | KLK2 | chr19 | 51376774 | ENST00000369056 | FGFR2 | chr10 | 123310972 | 2234 | 62 | 16 | 1917 | 633 | ||
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000369061 | FGFR2 | chr10 | 123310973 | - | 3470 | 271 | 27 | 1946 | 639 |
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000357555 | FGFR2 | chr10 | 123310973 | - | 3455 | 271 | 27 | 2207 | 726 |
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000358487 | FGFR2 | chr10 | 123310973 | - | 3799 | 271 | 27 | 2282 | 751 |
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000356226 | FGFR2 | chr10 | 123310973 | - | 3062 | 271 | 27 | 2276 | 749 |
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000369060 | FGFR2 | chr10 | 123310973 | - | 2574 | 271 | 27 | 1934 | 635 |
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000369059 | FGFR2 | chr10 | 123310973 | - | 2524 | 271 | 27 | 2285 | 752 |
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000346997 | FGFR2 | chr10 | 123310973 | - | 2277 | 271 | 27 | 2276 | 749 |
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000457416 | FGFR2 | chr10 | 123310973 | - | 2286 | 271 | 27 | 2285 | 752 |
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000351936 | FGFR2 | chr10 | 123310973 | - | 2175 | 271 | 27 | 2174 | 715 |
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000360144 | FGFR2 | chr10 | 123310973 | - | 2464 | 271 | 27 | 2126 | 699 |
| ENST00000325321 | KLK2 | chr19 | 51376775 | + | ENST00000369056 | FGFR2 | chr10 | 123310973 | - | 2443 | 271 | 27 | 2126 | 699 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000369061 | FGFR2 | chr10 | 123310973 | - | 3261 | 62 | 16 | 1737 | 573 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000357555 | FGFR2 | chr10 | 123310973 | - | 3246 | 62 | 16 | 1998 | 660 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000358487 | FGFR2 | chr10 | 123310973 | - | 3590 | 62 | 16 | 2073 | 685 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000356226 | FGFR2 | chr10 | 123310973 | - | 2853 | 62 | 16 | 2067 | 683 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000369060 | FGFR2 | chr10 | 123310973 | - | 2365 | 62 | 16 | 1725 | 569 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000369059 | FGFR2 | chr10 | 123310973 | - | 2315 | 62 | 16 | 2076 | 686 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000346997 | FGFR2 | chr10 | 123310973 | - | 2068 | 62 | 16 | 2067 | 684 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000457416 | FGFR2 | chr10 | 123310973 | - | 2077 | 62 | 16 | 2076 | 687 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000351936 | FGFR2 | chr10 | 123310973 | - | 1966 | 62 | 16 | 1965 | 650 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000360144 | FGFR2 | chr10 | 123310973 | - | 2255 | 62 | 16 | 1917 | 633 |
| ENST00000358049 | KLK2 | chr19 | 51376775 | + | ENST00000369056 | FGFR2 | chr10 | 123310973 | - | 2234 | 62 | 16 | 1917 | 633 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000369061 | FGFR2 | chr10 | 123310973 | - | 3917 | 718 | 747 | 2393 | 548 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000357555 | FGFR2 | chr10 | 123310973 | - | 3902 | 718 | 747 | 2654 | 635 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000358487 | FGFR2 | chr10 | 123310973 | - | 4246 | 718 | 747 | 2729 | 660 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000356226 | FGFR2 | chr10 | 123310973 | - | 3509 | 718 | 747 | 2723 | 658 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000369060 | FGFR2 | chr10 | 123310973 | - | 3021 | 718 | 747 | 2381 | 544 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000369059 | FGFR2 | chr10 | 123310973 | - | 2971 | 718 | 747 | 2732 | 661 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000346997 | FGFR2 | chr10 | 123310973 | - | 2724 | 718 | 747 | 2723 | 658 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000457416 | FGFR2 | chr10 | 123310973 | - | 2733 | 718 | 747 | 2732 | 661 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000351936 | FGFR2 | chr10 | 123310973 | - | 2622 | 718 | 747 | 2621 | 624 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000360144 | FGFR2 | chr10 | 123310973 | - | 2911 | 718 | 747 | 2573 | 608 |
| ENST00000325321 | KLK2 | chr19 | 51380014 | + | ENST00000369056 | FGFR2 | chr10 | 123310973 | - | 2890 | 718 | 747 | 2573 | 608 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000369061 | FGFR2 | chr10 | 123310973 | - | 3567 | 368 | 397 | 2043 | 548 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000357555 | FGFR2 | chr10 | 123310973 | - | 3552 | 368 | 397 | 2304 | 635 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000358487 | FGFR2 | chr10 | 123310973 | - | 3896 | 368 | 397 | 2379 | 660 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000356226 | FGFR2 | chr10 | 123310973 | - | 3159 | 368 | 397 | 2373 | 658 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000369060 | FGFR2 | chr10 | 123310973 | - | 2671 | 368 | 397 | 2031 | 544 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000369059 | FGFR2 | chr10 | 123310973 | - | 2621 | 368 | 397 | 2382 | 661 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000346997 | FGFR2 | chr10 | 123310973 | - | 2374 | 368 | 397 | 2373 | 659 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000457416 | FGFR2 | chr10 | 123310973 | - | 2383 | 368 | 397 | 2382 | 662 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000351936 | FGFR2 | chr10 | 123310973 | - | 2272 | 368 | 397 | 2271 | 625 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000360144 | FGFR2 | chr10 | 123310973 | - | 2561 | 368 | 397 | 2223 | 608 |
| ENST00000391810 | KLK2 | chr19 | 51380014 | + | ENST00000369056 | FGFR2 | chr10 | 123310973 | - | 2540 | 368 | 397 | 2223 | 608 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000369061 | FGFR2 | chr10 | 123310973 | - | 3708 | 509 | 538 | 2184 | 548 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000357555 | FGFR2 | chr10 | 123310973 | - | 3693 | 509 | 538 | 2445 | 635 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000358487 | FGFR2 | chr10 | 123310973 | - | 4037 | 509 | 538 | 2520 | 660 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000356226 | FGFR2 | chr10 | 123310973 | - | 3300 | 509 | 538 | 2514 | 658 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000369060 | FGFR2 | chr10 | 123310973 | - | 2812 | 509 | 538 | 2172 | 544 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000369059 | FGFR2 | chr10 | 123310973 | - | 2762 | 509 | 538 | 2523 | 661 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000346997 | FGFR2 | chr10 | 123310973 | - | 2515 | 509 | 538 | 2514 | 659 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000457416 | FGFR2 | chr10 | 123310973 | - | 2524 | 509 | 538 | 2523 | 662 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000351936 | FGFR2 | chr10 | 123310973 | - | 2413 | 509 | 538 | 2412 | 625 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000360144 | FGFR2 | chr10 | 123310973 | - | 2702 | 509 | 538 | 2364 | 608 |
| ENST00000358049 | KLK2 | chr19 | 51380014 | + | ENST00000369056 | FGFR2 | chr10 | 123310973 | - | 2681 | 509 | 538 | 2364 | 608 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000369061 | FGFR2 | chr10 | 123310972 | - | 3470 | 271 | 27 | 1946 | 639 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000357555 | FGFR2 | chr10 | 123310972 | - | 3455 | 271 | 27 | 2207 | 726 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000358487 | FGFR2 | chr10 | 123310972 | - | 3799 | 271 | 27 | 2282 | 751 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000356226 | FGFR2 | chr10 | 123310972 | - | 3062 | 271 | 27 | 2276 | 749 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000369060 | FGFR2 | chr10 | 123310972 | - | 2574 | 271 | 27 | 1934 | 635 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000369059 | FGFR2 | chr10 | 123310972 | - | 2524 | 271 | 27 | 2285 | 752 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000346997 | FGFR2 | chr10 | 123310972 | - | 2277 | 271 | 27 | 2276 | 749 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000457416 | FGFR2 | chr10 | 123310972 | - | 2286 | 271 | 27 | 2285 | 752 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000351936 | FGFR2 | chr10 | 123310972 | - | 2175 | 271 | 27 | 2174 | 715 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000360144 | FGFR2 | chr10 | 123310972 | - | 2464 | 271 | 27 | 2126 | 699 |
| ENST00000325321 | KLK2 | chr19 | 51376774 | + | ENST00000369056 | FGFR2 | chr10 | 123310972 | - | 2443 | 271 | 27 | 2126 | 699 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000369061 | FGFR2 | chr10 | 123310972 | - | 3261 | 62 | 16 | 1737 | 573 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000357555 | FGFR2 | chr10 | 123310972 | - | 3246 | 62 | 16 | 1998 | 660 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000358487 | FGFR2 | chr10 | 123310972 | - | 3590 | 62 | 16 | 2073 | 685 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000356226 | FGFR2 | chr10 | 123310972 | - | 2853 | 62 | 16 | 2067 | 683 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000369060 | FGFR2 | chr10 | 123310972 | - | 2365 | 62 | 16 | 1725 | 569 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000369059 | FGFR2 | chr10 | 123310972 | - | 2315 | 62 | 16 | 2076 | 686 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000346997 | FGFR2 | chr10 | 123310972 | - | 2068 | 62 | 16 | 2067 | 684 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000457416 | FGFR2 | chr10 | 123310972 | - | 2077 | 62 | 16 | 2076 | 687 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000351936 | FGFR2 | chr10 | 123310972 | - | 1966 | 62 | 16 | 1965 | 650 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000360144 | FGFR2 | chr10 | 123310972 | - | 2255 | 62 | 16 | 1917 | 633 |
| ENST00000358049 | KLK2 | chr19 | 51376774 | + | ENST00000369056 | FGFR2 | chr10 | 123310972 | - | 2234 | 62 | 16 | 1917 | 633 |
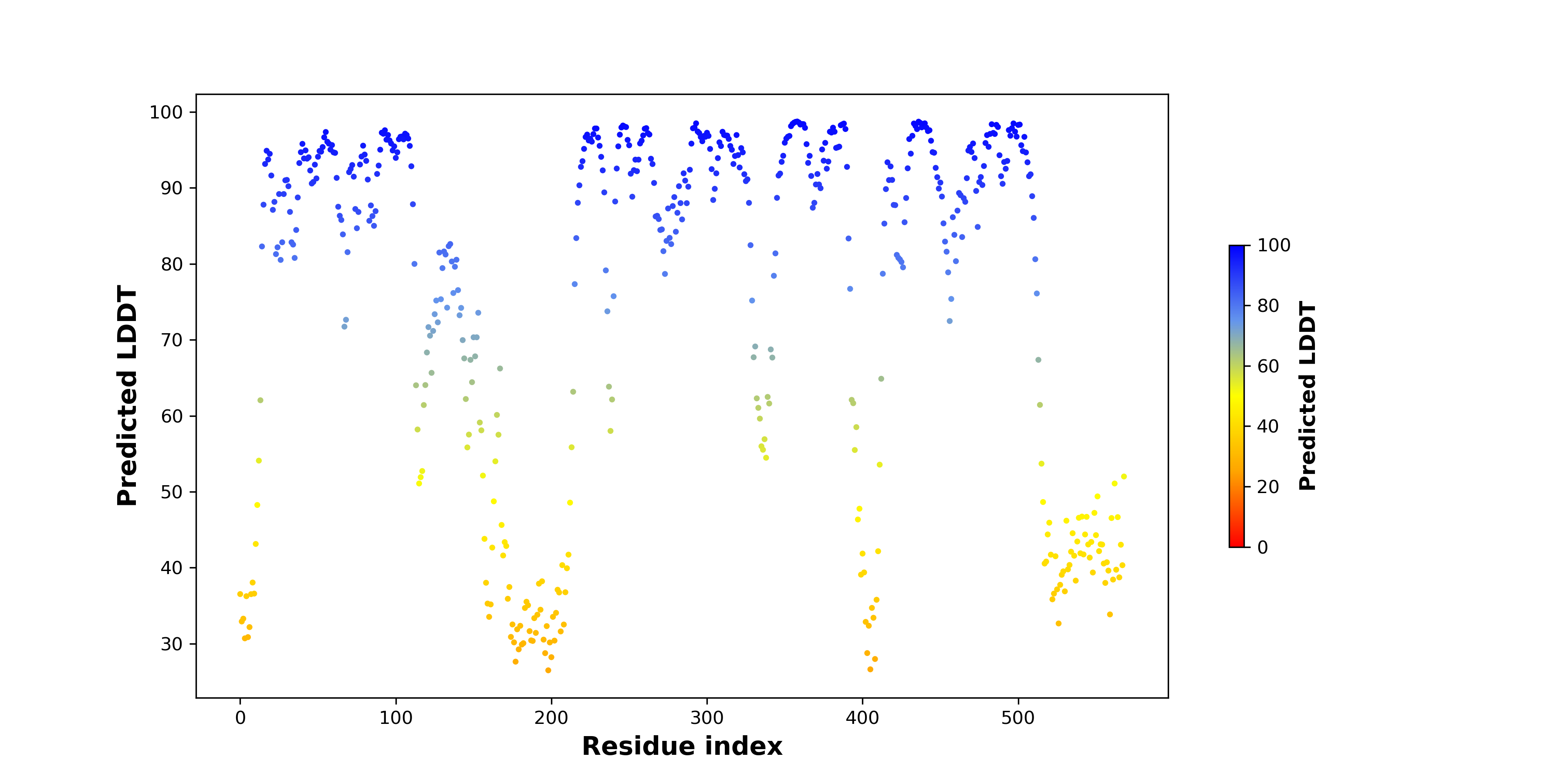
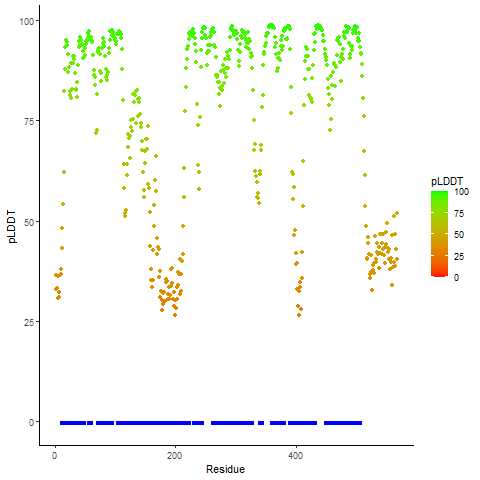
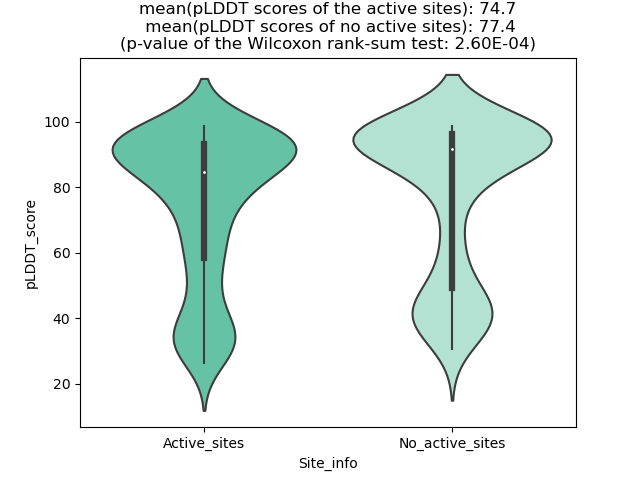
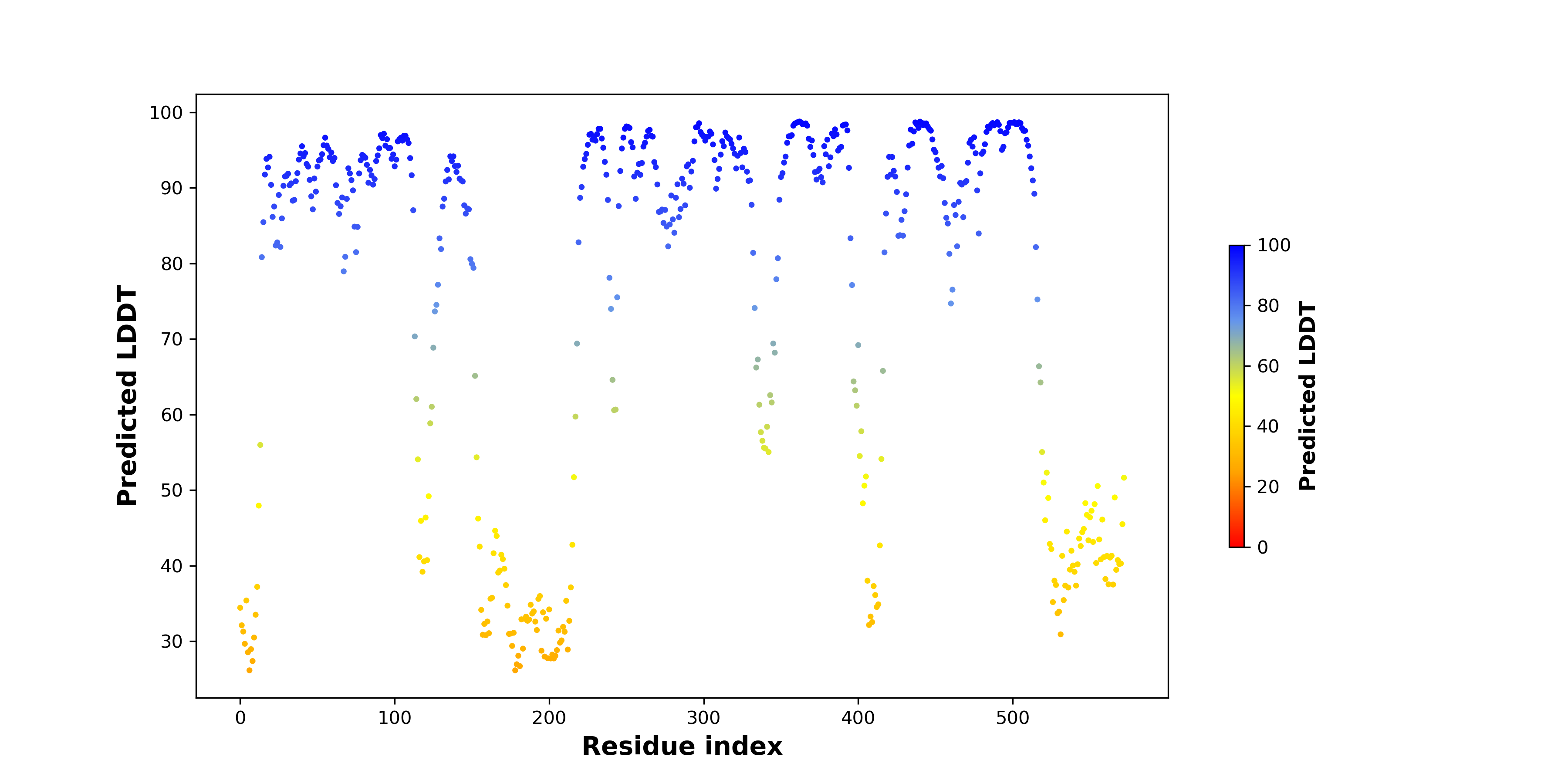
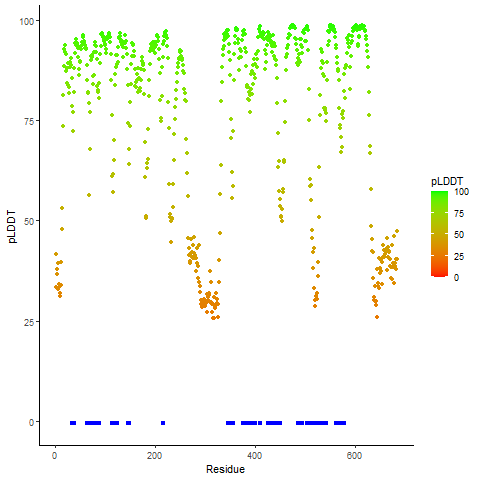
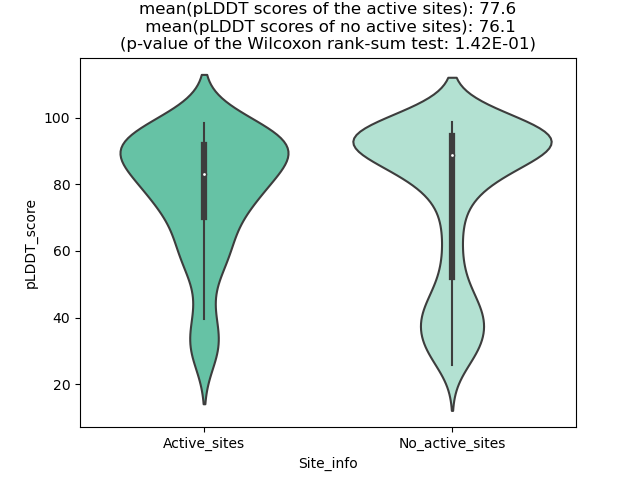
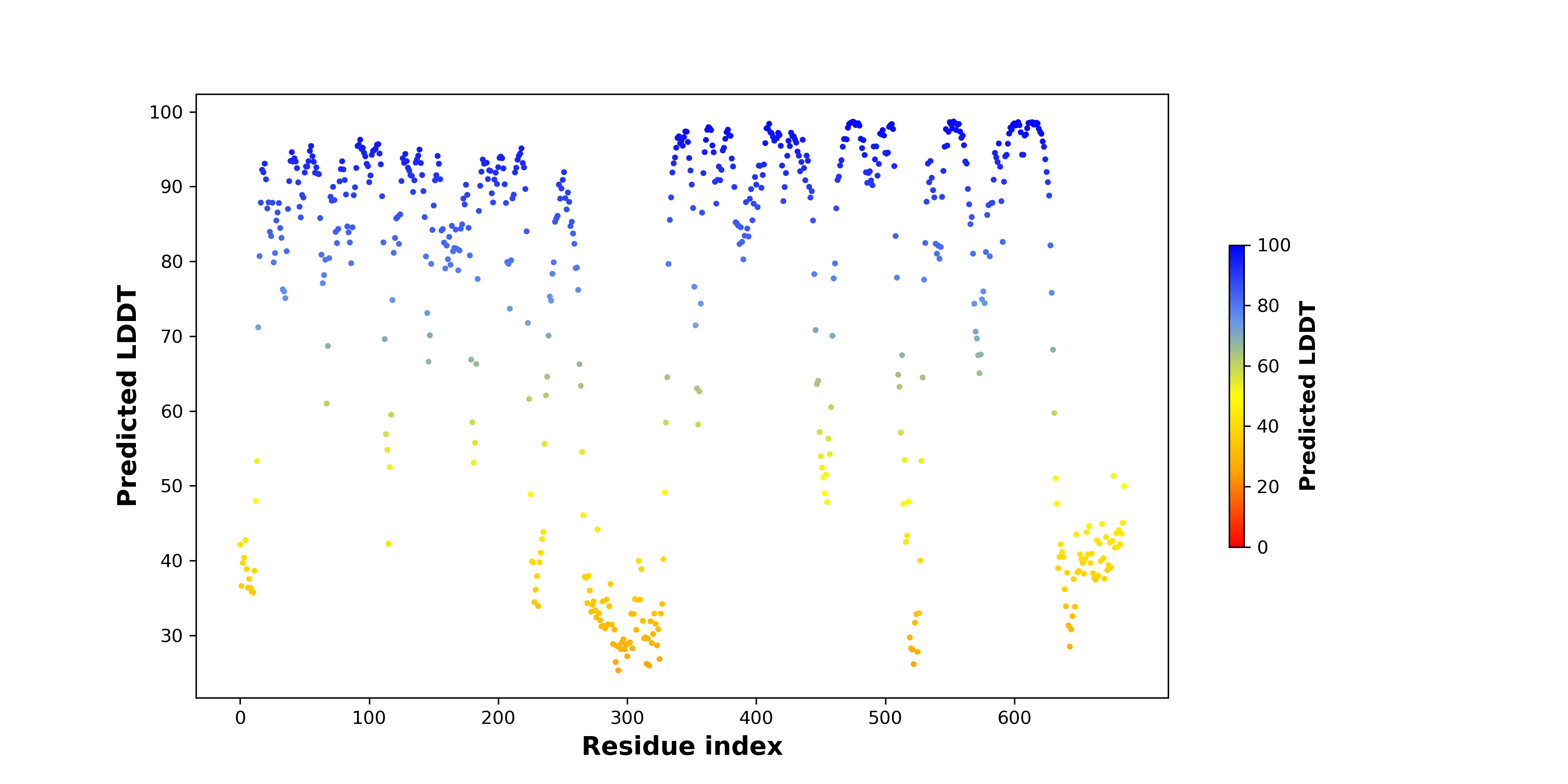
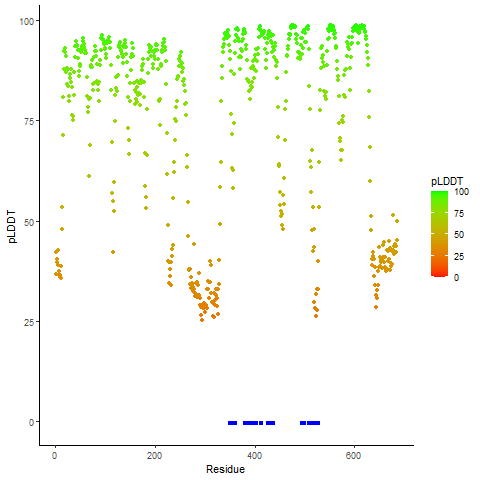
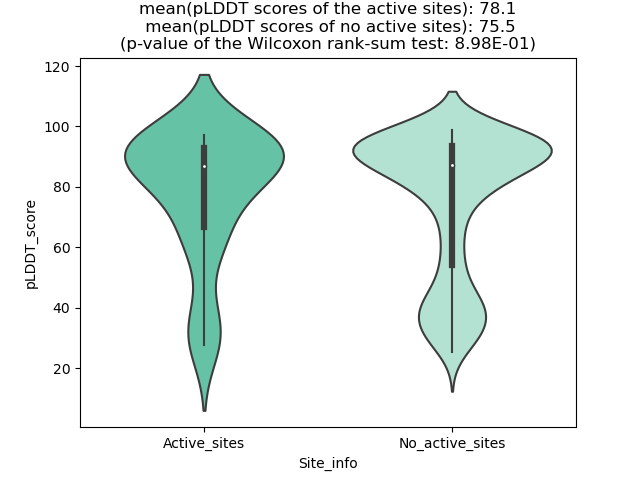
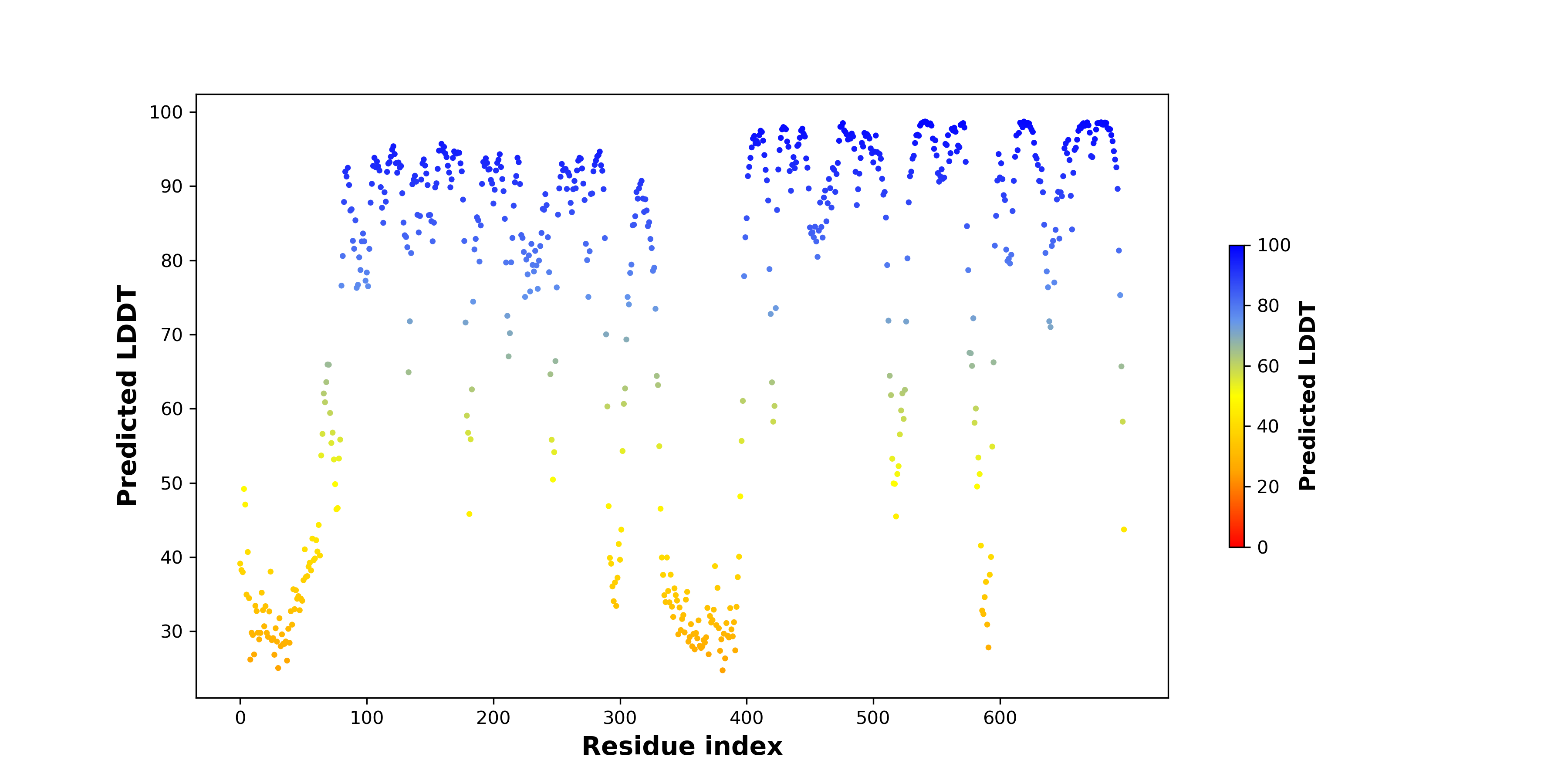
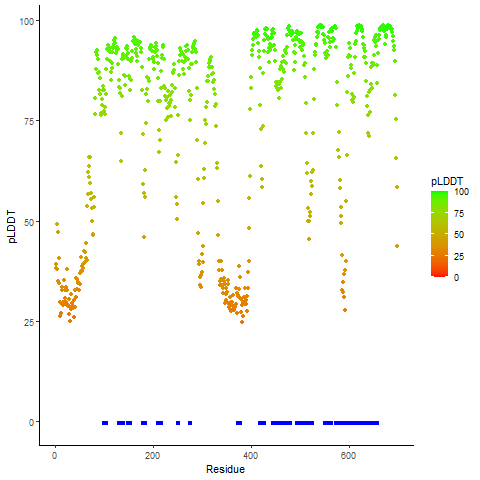
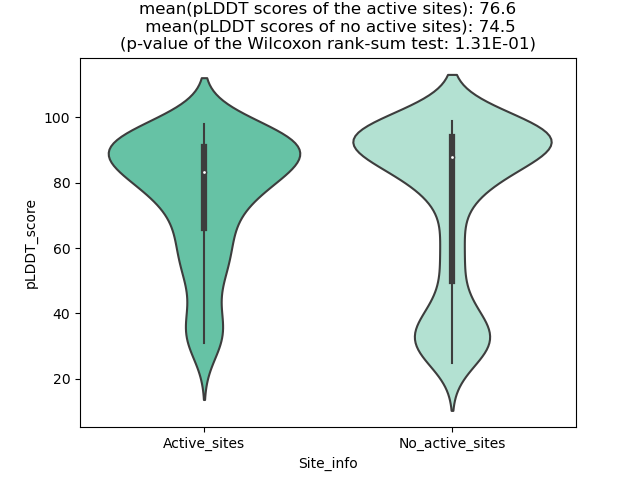
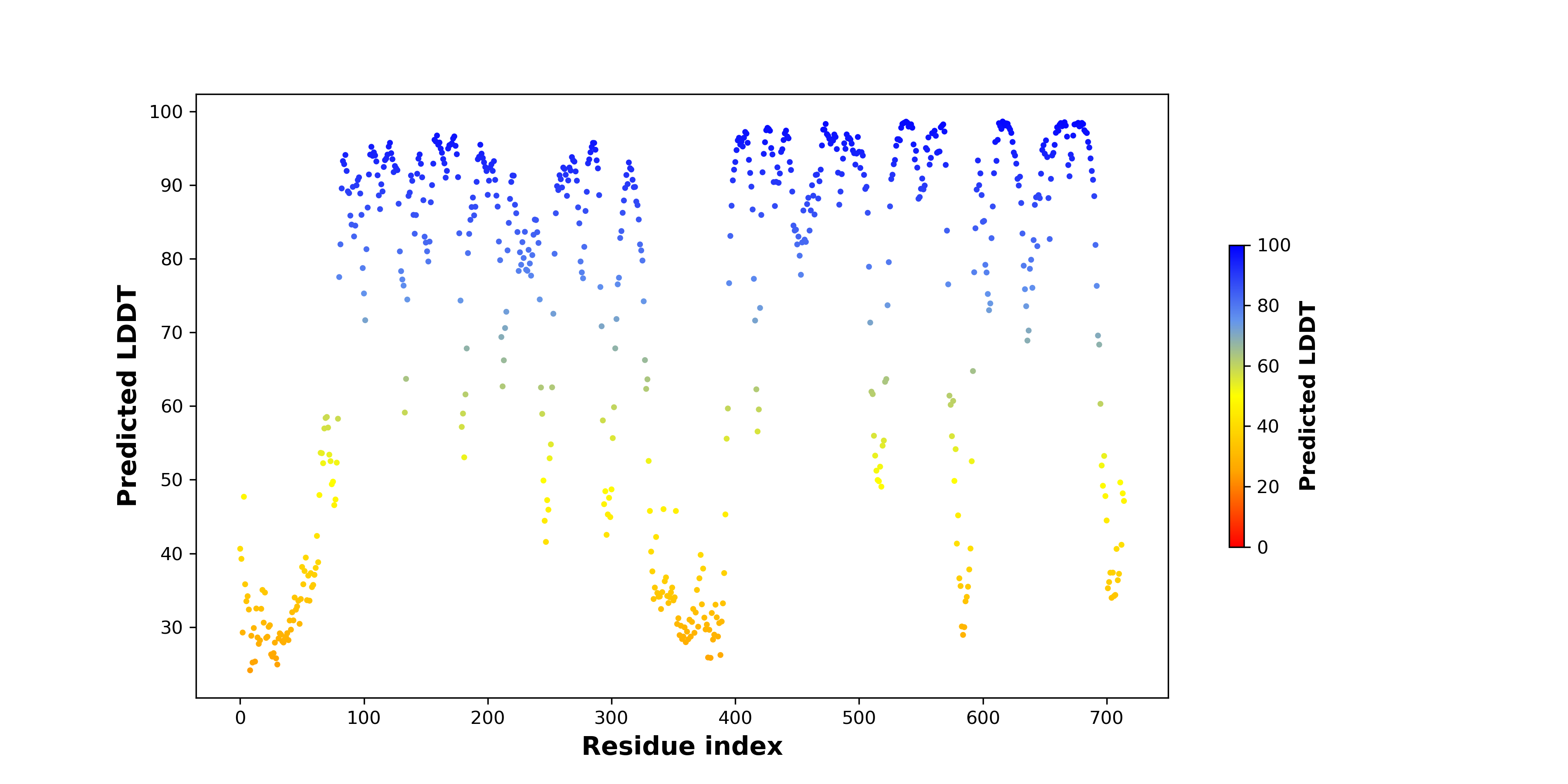
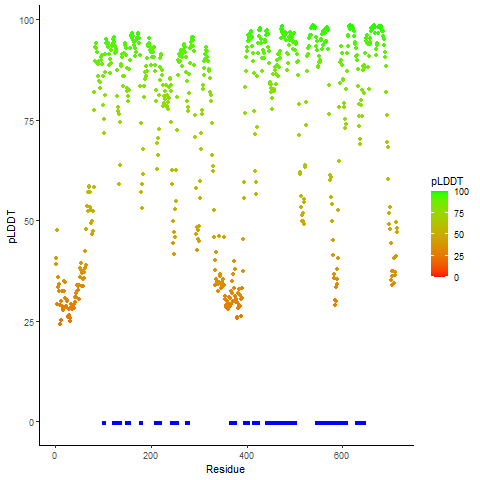
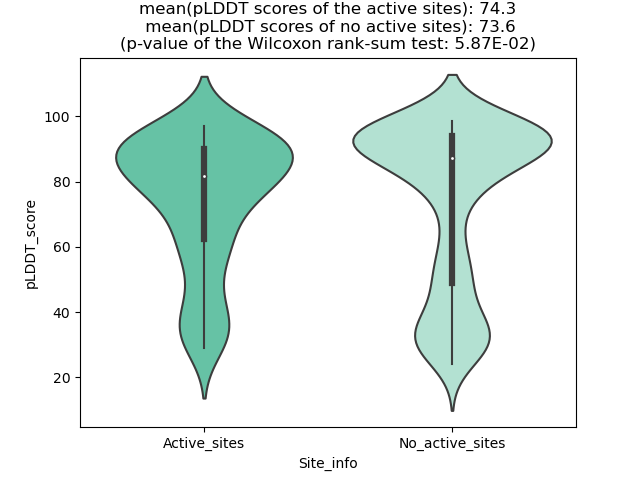
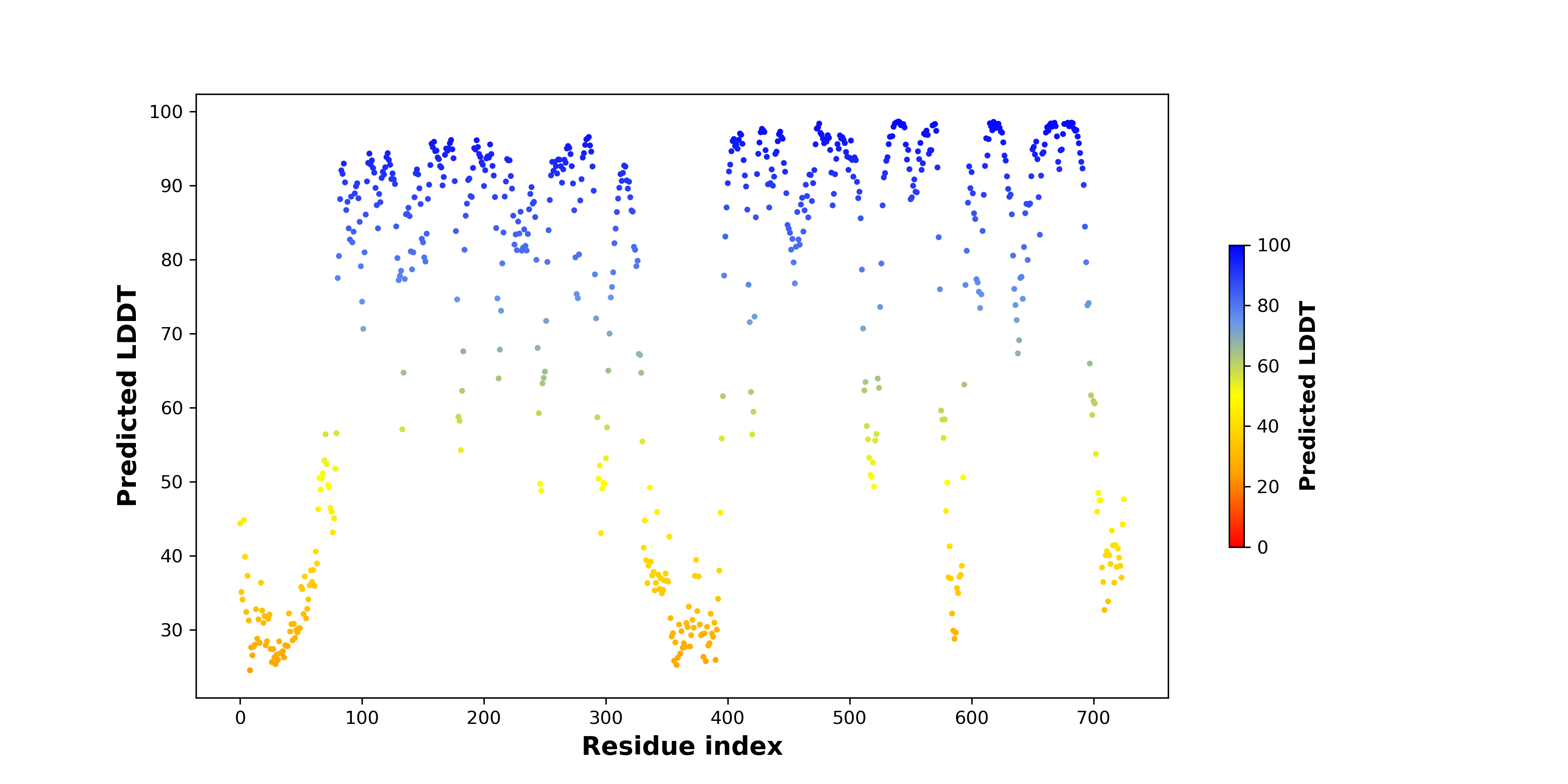
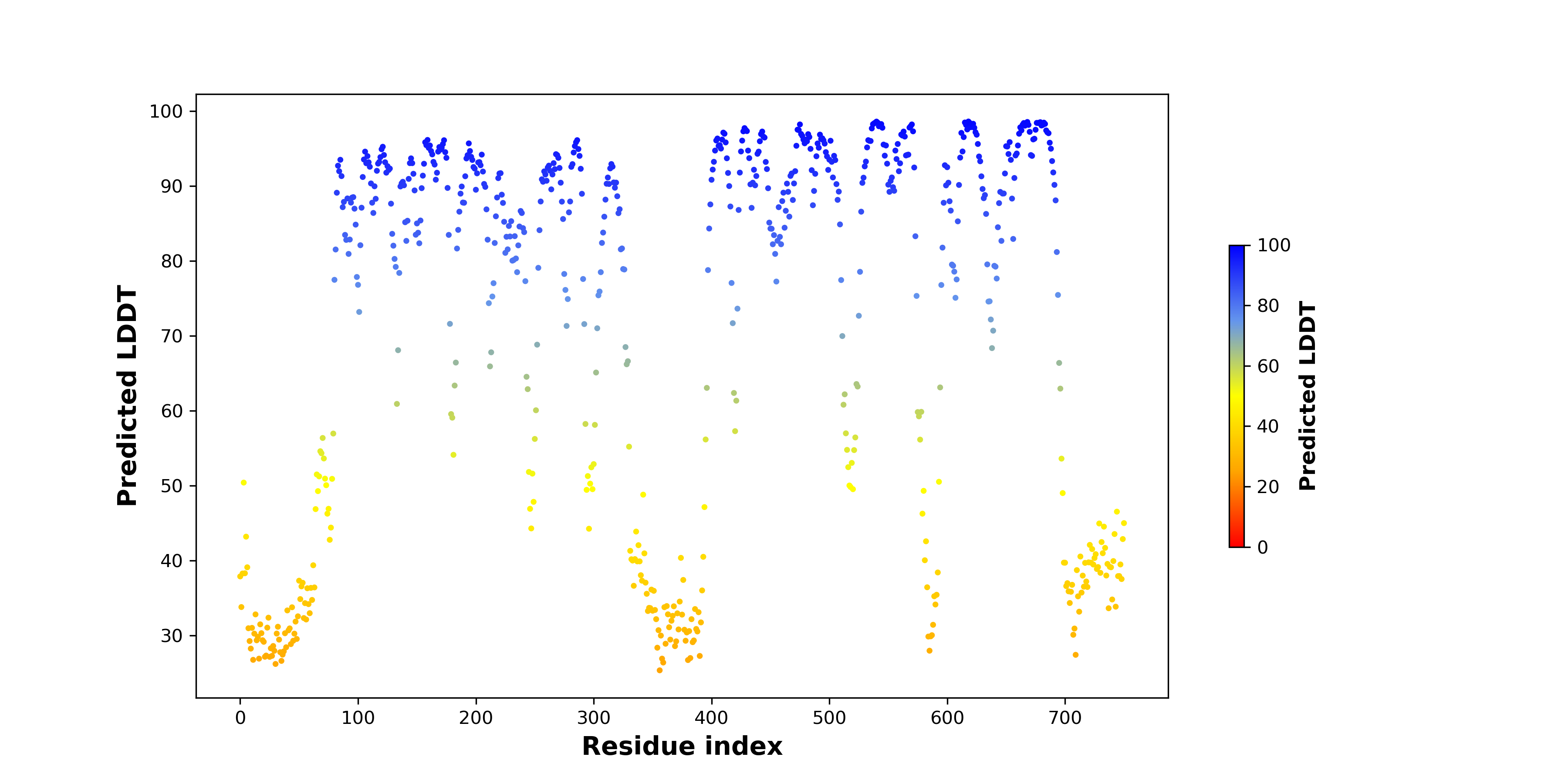
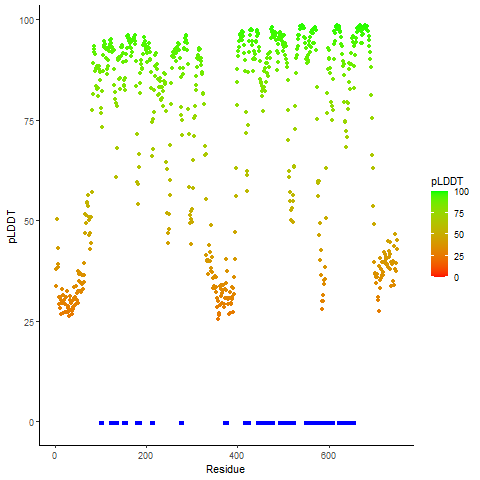
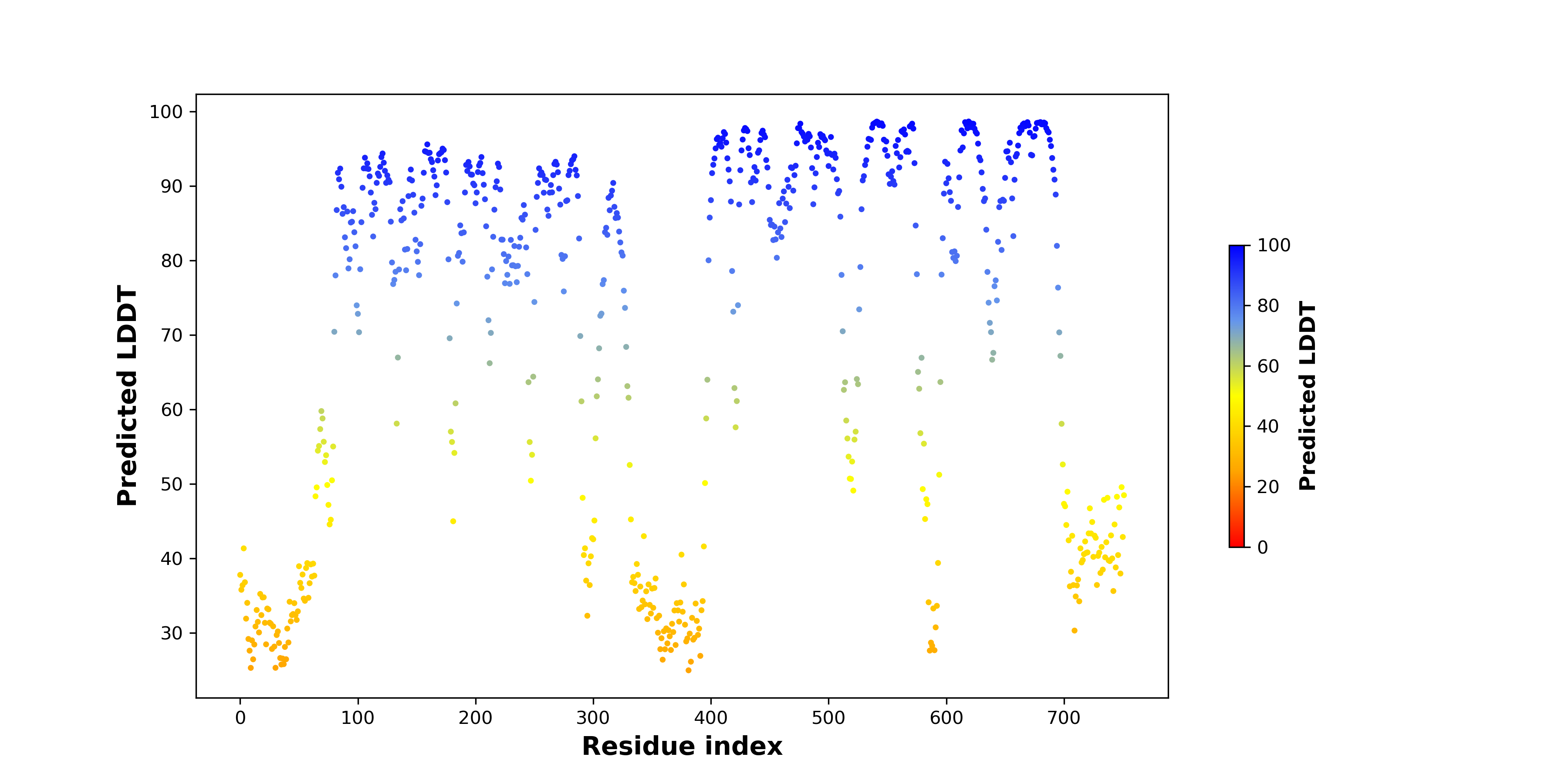
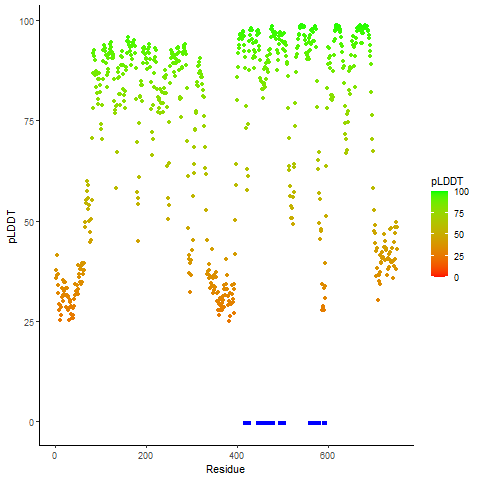
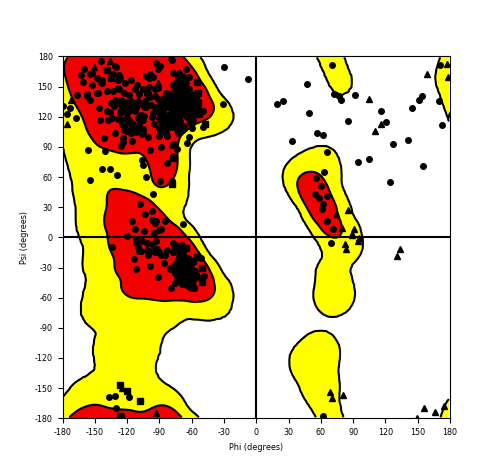
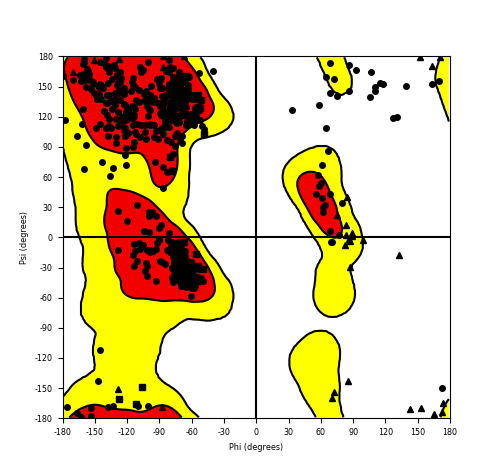
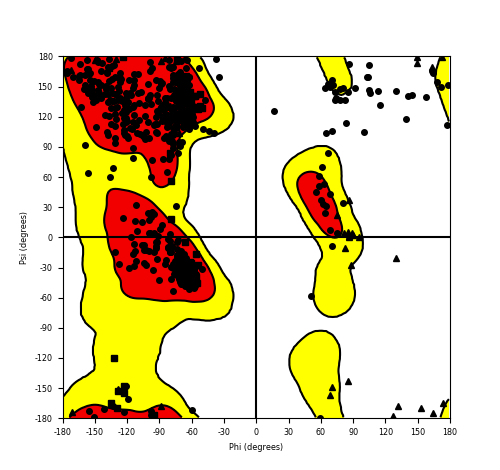
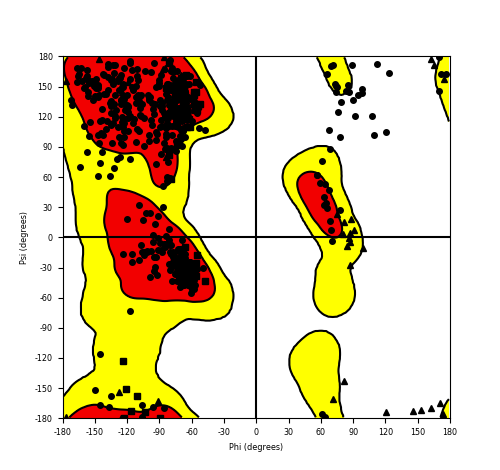
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000325321 | ENST00000369061 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.000416372 | 0.99958366 |
| ENST00000325321 | ENST00000357555 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.000333672 | 0.99966633 |
| ENST00000325321 | ENST00000358487 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.000336792 | 0.99966323 |
| ENST00000325321 | ENST00000356226 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.000759771 | 0.9992403 |
| ENST00000325321 | ENST00000369060 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.000560225 | 0.9994398 |
| ENST00000325321 | ENST00000369059 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.001491766 | 0.99850816 |
| ENST00000325321 | ENST00000346997 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.002005886 | 0.9979942 |
| ENST00000325321 | ENST00000457416 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.001818574 | 0.99818146 |
| ENST00000325321 | ENST00000351936 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.002095243 | 0.9979048 |
| ENST00000325321 | ENST00000360144 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.001740112 | 0.99825984 |
| ENST00000325321 | ENST00000369056 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.002086493 | 0.99791354 |
| ENST00000358049 | ENST00000369061 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.000380013 | 0.99961996 |
| ENST00000358049 | ENST00000357555 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.000301072 | 0.999699 |
| ENST00000358049 | ENST00000358487 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.00030815 | 0.99969184 |
| ENST00000358049 | ENST00000356226 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.000654728 | 0.99934524 |
| ENST00000358049 | ENST00000369060 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.000455312 | 0.9995447 |
| ENST00000358049 | ENST00000369059 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.001289997 | 0.99871004 |
| ENST00000358049 | ENST00000346997 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.001688006 | 0.998312 |
| ENST00000358049 | ENST00000457416 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.00157876 | 0.9984212 |
| ENST00000358049 | ENST00000351936 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.001776542 | 0.9982235 |
| ENST00000358049 | ENST00000360144 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.001523459 | 0.99847656 |
| ENST00000358049 | ENST00000369056 | KLK2 | chr19 | 51376775 | + | FGFR2 | chr10 | 123310973 | - | 0.001874698 | 0.99812526 |
| ENST00000325321 | ENST00000369061 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.000470186 | 0.99952984 |
| ENST00000325321 | ENST00000357555 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.000827256 | 0.99917275 |
| ENST00000325321 | ENST00000358487 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.000863062 | 0.999137 |
| ENST00000325321 | ENST00000356226 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.001291971 | 0.998708 |
| ENST00000325321 | ENST00000369060 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.001469928 | 0.99853003 |
| ENST00000325321 | ENST00000369059 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002309828 | 0.9976902 |
| ENST00000325321 | ENST00000346997 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.003038274 | 0.99696165 |
| ENST00000325321 | ENST00000457416 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002703334 | 0.99729663 |
| ENST00000325321 | ENST00000351936 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.003346135 | 0.9966538 |
| ENST00000325321 | ENST00000360144 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002759313 | 0.99724066 |
| ENST00000325321 | ENST00000369056 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.003169281 | 0.99683076 |
| ENST00000391810 | ENST00000369061 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.000450814 | 0.9995492 |
| ENST00000391810 | ENST00000357555 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.000778994 | 0.99922097 |
| ENST00000391810 | ENST00000358487 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.000825051 | 0.9991749 |
| ENST00000391810 | ENST00000356226 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.001273834 | 0.9987262 |
| ENST00000391810 | ENST00000369060 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.001437859 | 0.99856216 |
| ENST00000391810 | ENST00000369059 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002358725 | 0.9976413 |
| ENST00000391810 | ENST00000346997 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.003135372 | 0.9968646 |
| ENST00000391810 | ENST00000457416 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002762056 | 0.9972379 |
| ENST00000391810 | ENST00000351936 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.00346849 | 0.99653155 |
| ENST00000391810 | ENST00000360144 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002820517 | 0.9971795 |
| ENST00000391810 | ENST00000369056 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.003294881 | 0.9967051 |
| ENST00000358049 | ENST00000369061 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.000427248 | 0.9995727 |
| ENST00000358049 | ENST00000357555 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.000756408 | 0.9992436 |
| ENST00000358049 | ENST00000358487 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.000798171 | 0.99920183 |
| ENST00000358049 | ENST00000356226 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.001151027 | 0.9988489 |
| ENST00000358049 | ENST00000369060 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.001273705 | 0.9987263 |
| ENST00000358049 | ENST00000369059 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002062625 | 0.99793744 |
| ENST00000358049 | ENST00000346997 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.00271355 | 0.99728644 |
| ENST00000358049 | ENST00000457416 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002417671 | 0.9975823 |
| ENST00000358049 | ENST00000351936 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002993604 | 0.99700636 |
| ENST00000358049 | ENST00000360144 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002451569 | 0.9975484 |
| ENST00000358049 | ENST00000369056 | KLK2 | chr19 | 51380014 | + | FGFR2 | chr10 | 123310973 | - | 0.002860428 | 0.9971396 |
| ENST00000325321 | ENST00000369061 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.000416372 | 0.99958366 |
| ENST00000325321 | ENST00000357555 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.000333672 | 0.99966633 |
| ENST00000325321 | ENST00000358487 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.000336792 | 0.99966323 |
| ENST00000325321 | ENST00000356226 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.000759771 | 0.9992403 |
| ENST00000325321 | ENST00000369060 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.000560225 | 0.9994398 |
| ENST00000325321 | ENST00000369059 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.001491766 | 0.99850816 |
| ENST00000325321 | ENST00000346997 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.002005886 | 0.9979942 |
| ENST00000325321 | ENST00000457416 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.001818574 | 0.99818146 |
| ENST00000325321 | ENST00000351936 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.002095243 | 0.9979048 |
| ENST00000325321 | ENST00000360144 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.001740112 | 0.99825984 |
| ENST00000325321 | ENST00000369056 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.002086493 | 0.99791354 |
| ENST00000358049 | ENST00000369061 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.000380013 | 0.99961996 |
| ENST00000358049 | ENST00000357555 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.000301072 | 0.999699 |
| ENST00000358049 | ENST00000358487 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.00030815 | 0.99969184 |
| ENST00000358049 | ENST00000356226 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.000654728 | 0.99934524 |
| ENST00000358049 | ENST00000369060 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.000455312 | 0.9995447 |
| ENST00000358049 | ENST00000369059 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.001289997 | 0.99871004 |
| ENST00000358049 | ENST00000346997 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.001688006 | 0.998312 |
| ENST00000358049 | ENST00000457416 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.00157876 | 0.9984212 |
| ENST00000358049 | ENST00000351936 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.001776542 | 0.9982235 |
| ENST00000358049 | ENST00000360144 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.001523459 | 0.99847656 |
| ENST00000358049 | ENST00000369056 | KLK2 | chr19 | 51376774 | + | FGFR2 | chr10 | 123310972 | - | 0.001874698 | 0.99812526 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >43056_43056_1_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000346997_length(amino acids)=749AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEY TCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSA ESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDD ATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLAR GMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGS PYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSS -------------------------------------------------------------- >43056_43056_2_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000351936_length(amino acids)=715AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEY TCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSA ESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDD ATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLAR GMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGS -------------------------------------------------------------- >43056_43056_3_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000356226_length(amino acids)=749AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEY TCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSA ESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDD ATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLAR GMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGS PYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSS -------------------------------------------------------------- >43056_43056_4_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000357555_length(amino acids)=726AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEY TCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG GSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEEKKVSGAVDCHKPPCNPSHLPC -------------------------------------------------------------- >43056_43056_5_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000358487_length(amino acids)=751AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEY TCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG GSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSC -------------------------------------------------------------- >43056_43056_6_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000360144_length(amino acids)=699AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYIC KVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVT VSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKML KDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQ LARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTL -------------------------------------------------------------- >43056_43056_7_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000369056_length(amino acids)=699AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYIC KVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVT VSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKML KDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQ LARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTL -------------------------------------------------------------- >43056_43056_8_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000369059_length(amino acids)=752AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYIC KVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVT VSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKML KDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQ LARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTL GGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSS -------------------------------------------------------------- >43056_43056_9_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000369060_length(amino acids)=635AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKVSAESSSSMNSNTPLVRITTRLSSTAD TPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKN IINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTE NNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPA NCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQYPHIN -------------------------------------------------------------- >43056_43056_10_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000369061_length(amino acids)=639AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVA PGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLS STADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIG KHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNV LVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRM DKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQY -------------------------------------------------------------- >43056_43056_11_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000325321_FGFR2_chr10_123310972_ENST00000457416_length(amino acids)=752AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYIC KVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVT VSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKML KDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQ LARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTL GGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSS -------------------------------------------------------------- >43056_43056_12_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000346997_length(amino acids)=684AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAG VNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKK PDFSSQPAVHKLTKRIPLRRQVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMA EAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSY DINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDR VYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEE -------------------------------------------------------------- >43056_43056_13_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000351936_length(amino acids)=650AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAG VNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKK PDFSSQPAVHKLTKRIPLRRQVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMA EAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSY DINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDR VYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEE -------------------------------------------------------------- >43056_43056_14_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000356226_length(amino acids)=683AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAG VNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKK PDFSSQPAVHKLTKRIPLRRQVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMA EAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSY DINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDR VYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEE -------------------------------------------------------------- >43056_43056_15_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000357555_length(amino acids)=660AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAG VNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKK PDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVV MAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEY SYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALF DRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTN -------------------------------------------------------------- >43056_43056_16_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000358487_length(amino acids)=685AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAG VNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKK PDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVV MAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEY SYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALF DRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTN -------------------------------------------------------------- >43056_43056_17_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000360144_length(amino acids)=633AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSG INSSNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTK KPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQV VMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEAL FDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTT -------------------------------------------------------------- >43056_43056_18_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000369056_length(amino acids)=633AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSG INSSNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTK KPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQV VMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEAL FDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTT -------------------------------------------------------------- >43056_43056_19_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000369059_length(amino acids)=686AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSG INSSNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTK KPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQV VMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEAL FDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTT -------------------------------------------------------------- >43056_43056_20_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000369060_length(amino acids)=569AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKVSA ESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDD ATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLAR GMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGS PYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSS -------------------------------------------------------------- >43056_43056_21_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000369061_length(amino acids)=573AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQV TVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKM LKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTY QLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFT LGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRS -------------------------------------------------------------- >43056_43056_22_KLK2-FGFR2_KLK2_chr19_51376774_ENST00000358049_FGFR2_chr10_123310972_ENST00000457416_length(amino acids)=687AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSG INSSNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTK KPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQV VMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEAL FDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTT -------------------------------------------------------------- >43056_43056_23_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000346997_length(amino acids)=749AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEY TCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSA ESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDD ATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLAR GMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGS PYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSS -------------------------------------------------------------- >43056_43056_24_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000351936_length(amino acids)=715AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEY TCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSA ESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDD ATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLAR GMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGS -------------------------------------------------------------- >43056_43056_25_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000356226_length(amino acids)=749AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEY TCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSA ESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDD ATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLAR GMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGS PYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSS -------------------------------------------------------------- >43056_43056_26_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000357555_length(amino acids)=726AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEY TCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG GSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEEKKVSGAVDCHKPPCNPSHLPC -------------------------------------------------------------- >43056_43056_27_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000358487_length(amino acids)=751AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEY TCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG GSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSC -------------------------------------------------------------- >43056_43056_28_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000360144_length(amino acids)=699AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYIC KVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVT VSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKML KDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQ LARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTL -------------------------------------------------------------- >43056_43056_29_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000369056_length(amino acids)=699AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYIC KVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVT VSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKML KDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQ LARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTL -------------------------------------------------------------- >43056_43056_30_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000369059_length(amino acids)=752AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYIC KVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVT VSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKML KDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQ LARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTL GGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSS -------------------------------------------------------------- >43056_43056_31_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000369060_length(amino acids)=635AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKVSAESSSSMNSNTPLVRITTRLSSTAD TPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKN IINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTE NNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPA NCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQYPHIN -------------------------------------------------------------- >43056_43056_32_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000369061_length(amino acids)=639AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVA PGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLS STADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIG KHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNV LVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRM DKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQY -------------------------------------------------------------- >43056_43056_33_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000325321_FGFR2_chr10_123310973_ENST00000457416_length(amino acids)=752AA_BP=80 MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLREAPQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTE KMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVE RSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYIC KVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVT VSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKML KDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQ LARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTL GGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSS -------------------------------------------------------------- >43056_43056_34_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000346997_length(amino acids)=684AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAG VNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKK PDFSSQPAVHKLTKRIPLRRQVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMA EAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSY DINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDR VYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEE -------------------------------------------------------------- >43056_43056_35_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000351936_length(amino acids)=650AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAG VNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKK PDFSSQPAVHKLTKRIPLRRQVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMA EAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSY DINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDR VYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEE -------------------------------------------------------------- >43056_43056_36_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000356226_length(amino acids)=683AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAG VNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKK PDFSSQPAVHKLTKRIPLRRQVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMA EAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSY DINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDR VYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEE -------------------------------------------------------------- >43056_43056_37_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000357555_length(amino acids)=660AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAG VNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKK PDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVV MAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEY SYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALF DRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTN -------------------------------------------------------------- >43056_43056_38_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000358487_length(amino acids)=685AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAG VNTTDKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKK PDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVV MAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEY SYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALF DRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTN -------------------------------------------------------------- >43056_43056_39_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000360144_length(amino acids)=633AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSG INSSNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTK KPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQV VMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEAL FDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTT -------------------------------------------------------------- >43056_43056_40_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000369056_length(amino acids)=633AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSG INSSNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTK KPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQV VMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEAL FDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTT -------------------------------------------------------------- >43056_43056_41_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000369059_length(amino acids)=686AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSG INSSNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTK KPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQV VMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEAL FDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTT -------------------------------------------------------------- >43056_43056_42_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000369060_length(amino acids)=569AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKVSA ESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDD ATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLAR GMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGS PYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSS -------------------------------------------------------------- >43056_43056_43_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000369061_length(amino acids)=573AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQV TVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKM LKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTY QLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFT LGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRS -------------------------------------------------------------- >43056_43056_44_KLK2-FGFR2_KLK2_chr19_51376775_ENST00000358049_FGFR2_chr10_123310973_ENST00000457416_length(amino acids)=687AA_BP=14 MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDK GNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSG INSSNAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTK KPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQV VMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEAL FDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTT -------------------------------------------------------------- >43056_43056_45_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000346997_length(amino acids)=658AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAE SSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDA TEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARG MEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSP YPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSG -------------------------------------------------------------- >43056_43056_46_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000351936_length(amino acids)=624AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAE SSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDA TEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARG MEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSP -------------------------------------------------------------- >43056_43056_47_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000356226_length(amino acids)=658AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAE SSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDA TEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARG MEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSP YPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSG -------------------------------------------------------------- >43056_43056_48_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000357555_length(amino acids)=635AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVS AESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKD DATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLA RGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGG SPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEEKKVSGAVDCHKPPCNPSHLPCV -------------------------------------------------------------- >43056_43056_49_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000358487_length(amino acids)=660AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVS AESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKD DATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLA RGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGG SPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCS -------------------------------------------------------------- >43056_43056_50_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000360144_length(amino acids)=608AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG -------------------------------------------------------------- >43056_43056_51_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000369056_length(amino acids)=608AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG -------------------------------------------------------------- >43056_43056_52_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000369059_length(amino acids)=661AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG GSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSC -------------------------------------------------------------- >43056_43056_53_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000369060_length(amino acids)=544AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKVSAESSSSMNSNTPLVRITTRLSSTADT PMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNI INLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTEN NVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPAN CTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQYPHING -------------------------------------------------------------- >43056_43056_54_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000369061_length(amino acids)=548AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVAP GREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSS TADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGK HKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVL VTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMD KPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQYP -------------------------------------------------------------- >43056_43056_55_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000325321_FGFR2_chr10_123310973_ENST00000457416_length(amino acids)=661AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG GSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSC -------------------------------------------------------------- >43056_43056_56_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000346997_length(amino acids)=659AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAE SSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDA TEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARG MEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSP YPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSG -------------------------------------------------------------- >43056_43056_57_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000351936_length(amino acids)=625AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAE SSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDA TEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARG MEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSP -------------------------------------------------------------- >43056_43056_58_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000356226_length(amino acids)=658AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAE SSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDA TEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARG MEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSP YPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSG -------------------------------------------------------------- >43056_43056_59_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000357555_length(amino acids)=635AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVS AESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKD DATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLA RGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGG SPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEEKKVSGAVDCHKPPCNPSHLPCV -------------------------------------------------------------- >43056_43056_60_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000358487_length(amino acids)=660AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVS AESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKD DATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLA RGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGG SPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCS -------------------------------------------------------------- >43056_43056_61_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000360144_length(amino acids)=608AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG -------------------------------------------------------------- >43056_43056_62_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000369056_length(amino acids)=608AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG -------------------------------------------------------------- >43056_43056_63_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000369059_length(amino acids)=661AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG GSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSC -------------------------------------------------------------- >43056_43056_64_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000369060_length(amino acids)=544AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKVSAESSSSMNSNTPLVRITTRLSSTADT PMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNI INLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTEN NVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPAN CTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQYPHING -------------------------------------------------------------- >43056_43056_65_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000369061_length(amino acids)=548AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVAP GREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSS TADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGK HKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVL VTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMD KPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQYP -------------------------------------------------------------- >43056_43056_66_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000358049_FGFR2_chr10_123310973_ENST00000457416_length(amino acids)=662AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG GSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSC -------------------------------------------------------------- >43056_43056_67_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000346997_length(amino acids)=659AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAE SSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDA TEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARG MEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSP YPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSG -------------------------------------------------------------- >43056_43056_68_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000351936_length(amino acids)=625AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAE SSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDA TEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARG MEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSP -------------------------------------------------------------- >43056_43056_69_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000356226_length(amino acids)=658AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAE SSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDA TEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARG MEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSP YPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSG -------------------------------------------------------------- >43056_43056_70_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000357555_length(amino acids)=635AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVS AESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKD DATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLA RGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGG SPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEEKKVSGAVDCHKPPCNPSHLPCV -------------------------------------------------------------- >43056_43056_71_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000358487_length(amino acids)=660AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFEDAGEYT CLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVS AESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKD DATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLA RGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGG SPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCS -------------------------------------------------------------- >43056_43056_72_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000360144_length(amino acids)=608AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG -------------------------------------------------------------- >43056_43056_73_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000369056_length(amino acids)=608AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG -------------------------------------------------------------- >43056_43056_74_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000369059_length(amino acids)=661AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG GSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSC -------------------------------------------------------------- >43056_43056_75_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000369060_length(amino acids)=544AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKVSAESSSSMNSNTPLVRITTRLSSTADT PMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNI INLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTEN NVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPAN CTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQYPHING -------------------------------------------------------------- >43056_43056_76_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000369061_length(amino acids)=548AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVAP GREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSS TADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGK HKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVL VTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMD KPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQYP -------------------------------------------------------------- >43056_43056_77_KLK2-FGFR2_KLK2_chr19_51380014_ENST00000391810_FGFR2_chr10_123310973_ENST00000457416_length(amino acids)=662AA_BP= MEKRLHAVPAANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENEYGSINHTYHLDVVER SPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAGEYICK VSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTV SAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQL ARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLG GSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSC -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:51376775/chr10:123310973) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KLK2 | FGFR2 |
| FUNCTION: Glandular kallikreins cleave Met-Lys and Arg-Ser bonds in kininogen to release Lys-bradykinin. | FUNCTION: Tyrosine-protein kinase that acts as cell-surface receptor for fibroblast growth factors and plays an essential role in the regulation of cell proliferation, differentiation, migration and apoptosis, and in the regulation of embryonic development. Required for normal embryonic patterning, trophoblast function, limb bud development, lung morphogenesis, osteogenesis and skin development. Plays an essential role in the regulation of osteoblast differentiation, proliferation and apoptosis, and is required for normal skeleton development. Promotes cell proliferation in keratinocytes and immature osteoblasts, but promotes apoptosis in differentiated osteoblasts. Phosphorylates PLCG1, FRS2 and PAK4. Ligand binding leads to the activation of several signaling cascades. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate. Phosphorylation of FRS2 triggers recruitment of GRB2, GAB1, PIK3R1 and SOS1, and mediates activation of RAS, MAPK1/ERK2, MAPK3/ERK1 and the MAP kinase signaling pathway, as well as of the AKT1 signaling pathway. FGFR2 signaling is down-regulated by ubiquitination, internalization and degradation. Mutations that lead to constitutive kinase activation or impair normal FGFR2 maturation, internalization and degradation lead to aberrant signaling. Over-expressed FGFR2 promotes activation of STAT1. {ECO:0000269|PubMed:12529371, ECO:0000269|PubMed:15190072, ECO:0000269|PubMed:15629145, ECO:0000269|PubMed:16384934, ECO:0000269|PubMed:16597617, ECO:0000269|PubMed:17311277, ECO:0000269|PubMed:17623664, ECO:0000269|PubMed:18374639, ECO:0000269|PubMed:19103595, ECO:0000269|PubMed:19387476, ECO:0000269|PubMed:19410646, ECO:0000269|PubMed:21596750, ECO:0000269|PubMed:8663044}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 154_247 | 151.33333333333334 | 820.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 256_358 | 151.33333333333334 | 820.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 481_770 | 151.33333333333334 | 820.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 154_247 | 151.33333333333334 | 786.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 256_358 | 151.33333333333334 | 786.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 481_770 | 151.33333333333334 | 786.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 154_247 | 36.333333333333336 | 705.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 256_358 | 36.333333333333336 | 705.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 481_770 | 36.333333333333336 | 705.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 154_247 | 62.333333333333336 | 708.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 256_358 | 62.333333333333336 | 708.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 481_770 | 62.333333333333336 | 708.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 154_247 | 151.33333333333334 | 822.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 256_358 | 151.33333333333334 | 822.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 481_770 | 151.33333333333334 | 822.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 154_247 | 151.33333333333334 | 255.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 256_358 | 151.33333333333334 | 255.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 481_770 | 151.33333333333334 | 255.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 154_247 | 62.333333333333336 | 681.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 256_358 | 62.333333333333336 | 681.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 481_770 | 62.333333333333336 | 681.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 154_247 | 151.33333333333334 | 770.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 256_358 | 151.33333333333334 | 770.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 481_770 | 151.33333333333334 | 770.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 154_247 | 151.33333333333334 | 706.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 256_358 | 151.33333333333334 | 706.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 481_770 | 151.33333333333334 | 706.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 154_247 | 151.33333333333334 | 710.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 256_358 | 151.33333333333334 | 710.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 481_770 | 151.33333333333334 | 710.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 154_247 | 151.33333333333334 | 823.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 256_358 | 151.33333333333334 | 823.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 481_770 | 151.33333333333334 | 823.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000346997 | 2 | 17 | 154_247 | 151.33333333333334 | 820.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000346997 | 2 | 17 | 256_358 | 151.33333333333334 | 820.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000346997 | 2 | 17 | 481_770 | 151.33333333333334 | 820.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000351936 | 3 | 18 | 154_247 | 151.33333333333334 | 786.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000351936 | 3 | 18 | 256_358 | 151.33333333333334 | 786.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000351936 | 3 | 18 | 481_770 | 151.33333333333334 | 786.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000356226 | 1 | 16 | 154_247 | 36.333333333333336 | 705.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000356226 | 1 | 16 | 256_358 | 36.333333333333336 | 705.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000356226 | 1 | 16 | 481_770 | 36.333333333333336 | 705.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000357555 | 2 | 17 | 154_247 | 62.333333333333336 | 708.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000357555 | 2 | 17 | 256_358 | 62.333333333333336 | 708.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000357555 | 2 | 17 | 481_770 | 62.333333333333336 | 708.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000358487 | 3 | 18 | 154_247 | 151.33333333333334 | 822.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000358487 | 3 | 18 | 256_358 | 151.33333333333334 | 822.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000358487 | 3 | 18 | 481_770 | 151.33333333333334 | 822.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000359354 | 3 | 7 | 154_247 | 151.33333333333334 | 255.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000359354 | 3 | 7 | 256_358 | 151.33333333333334 | 255.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000359354 | 3 | 7 | 481_770 | 151.33333333333334 | 255.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000360144 | 2 | 17 | 154_247 | 62.333333333333336 | 681.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000360144 | 2 | 17 | 256_358 | 62.333333333333336 | 681.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000360144 | 2 | 17 | 481_770 | 62.333333333333336 | 681.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369056 | 2 | 17 | 154_247 | 151.33333333333334 | 770.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369056 | 2 | 17 | 256_358 | 151.33333333333334 | 770.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369056 | 2 | 17 | 481_770 | 151.33333333333334 | 770.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369060 | 3 | 16 | 154_247 | 151.33333333333334 | 706.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369060 | 3 | 16 | 256_358 | 151.33333333333334 | 706.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369060 | 3 | 16 | 481_770 | 151.33333333333334 | 706.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369061 | 2 | 15 | 154_247 | 151.33333333333334 | 710.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369061 | 2 | 15 | 256_358 | 151.33333333333334 | 710.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369061 | 2 | 15 | 481_770 | 151.33333333333334 | 710.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000457416 | 3 | 18 | 154_247 | 151.33333333333334 | 823.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000457416 | 3 | 18 | 256_358 | 151.33333333333334 | 823.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000457416 | 3 | 18 | 481_770 | 151.33333333333334 | 823.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000346997 | 2 | 17 | 154_247 | 151.33333333333334 | 820.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000346997 | 2 | 17 | 256_358 | 151.33333333333334 | 820.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000346997 | 2 | 17 | 481_770 | 151.33333333333334 | 820.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000351936 | 3 | 18 | 154_247 | 151.33333333333334 | 786.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000351936 | 3 | 18 | 256_358 | 151.33333333333334 | 786.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000351936 | 3 | 18 | 481_770 | 151.33333333333334 | 786.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000356226 | 1 | 16 | 154_247 | 36.333333333333336 | 705.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000356226 | 1 | 16 | 256_358 | 36.333333333333336 | 705.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000356226 | 1 | 16 | 481_770 | 36.333333333333336 | 705.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000357555 | 2 | 17 | 154_247 | 62.333333333333336 | 708.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000357555 | 2 | 17 | 256_358 | 62.333333333333336 | 708.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000357555 | 2 | 17 | 481_770 | 62.333333333333336 | 708.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000358487 | 3 | 18 | 154_247 | 151.33333333333334 | 822.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000358487 | 3 | 18 | 256_358 | 151.33333333333334 | 822.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000358487 | 3 | 18 | 481_770 | 151.33333333333334 | 822.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000359354 | 3 | 7 | 154_247 | 151.33333333333334 | 255.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000359354 | 3 | 7 | 256_358 | 151.33333333333334 | 255.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000359354 | 3 | 7 | 481_770 | 151.33333333333334 | 255.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000360144 | 2 | 17 | 154_247 | 62.333333333333336 | 681.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000360144 | 2 | 17 | 256_358 | 62.333333333333336 | 681.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000360144 | 2 | 17 | 481_770 | 62.333333333333336 | 681.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369056 | 2 | 17 | 154_247 | 151.33333333333334 | 770.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369056 | 2 | 17 | 256_358 | 151.33333333333334 | 770.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369056 | 2 | 17 | 481_770 | 151.33333333333334 | 770.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369060 | 3 | 16 | 154_247 | 151.33333333333334 | 706.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369060 | 3 | 16 | 256_358 | 151.33333333333334 | 706.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369060 | 3 | 16 | 481_770 | 151.33333333333334 | 706.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369061 | 2 | 15 | 154_247 | 151.33333333333334 | 710.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369061 | 2 | 15 | 256_358 | 151.33333333333334 | 710.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369061 | 2 | 15 | 481_770 | 151.33333333333334 | 710.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000457416 | 3 | 18 | 154_247 | 151.33333333333334 | 823.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000457416 | 3 | 18 | 256_358 | 151.33333333333334 | 823.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000457416 | 3 | 18 | 481_770 | 151.33333333333334 | 823.0 | Domain | Protein kinase | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 487_495 | 151.33333333333334 | 820.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 565_567 | 151.33333333333334 | 820.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 487_495 | 151.33333333333334 | 786.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 565_567 | 151.33333333333334 | 786.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 487_495 | 36.333333333333336 | 705.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 565_567 | 36.333333333333336 | 705.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 487_495 | 62.333333333333336 | 708.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 565_567 | 62.333333333333336 | 708.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 487_495 | 151.33333333333334 | 822.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 565_567 | 151.33333333333334 | 822.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 487_495 | 151.33333333333334 | 255.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 565_567 | 151.33333333333334 | 255.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 487_495 | 62.333333333333336 | 681.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 565_567 | 62.333333333333336 | 681.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 487_495 | 151.33333333333334 | 770.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 565_567 | 151.33333333333334 | 770.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 487_495 | 151.33333333333334 | 706.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 565_567 | 151.33333333333334 | 706.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 487_495 | 151.33333333333334 | 710.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 565_567 | 151.33333333333334 | 710.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 487_495 | 151.33333333333334 | 823.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 565_567 | 151.33333333333334 | 823.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000346997 | 2 | 17 | 487_495 | 151.33333333333334 | 820.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000346997 | 2 | 17 | 565_567 | 151.33333333333334 | 820.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000351936 | 3 | 18 | 487_495 | 151.33333333333334 | 786.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000351936 | 3 | 18 | 565_567 | 151.33333333333334 | 786.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000356226 | 1 | 16 | 487_495 | 36.333333333333336 | 705.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000356226 | 1 | 16 | 565_567 | 36.333333333333336 | 705.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000357555 | 2 | 17 | 487_495 | 62.333333333333336 | 708.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000357555 | 2 | 17 | 565_567 | 62.333333333333336 | 708.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000358487 | 3 | 18 | 487_495 | 151.33333333333334 | 822.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000358487 | 3 | 18 | 565_567 | 151.33333333333334 | 822.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000359354 | 3 | 7 | 487_495 | 151.33333333333334 | 255.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000359354 | 3 | 7 | 565_567 | 151.33333333333334 | 255.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000360144 | 2 | 17 | 487_495 | 62.333333333333336 | 681.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000360144 | 2 | 17 | 565_567 | 62.333333333333336 | 681.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369056 | 2 | 17 | 487_495 | 151.33333333333334 | 770.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369056 | 2 | 17 | 565_567 | 151.33333333333334 | 770.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369060 | 3 | 16 | 487_495 | 151.33333333333334 | 706.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369060 | 3 | 16 | 565_567 | 151.33333333333334 | 706.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369061 | 2 | 15 | 487_495 | 151.33333333333334 | 710.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369061 | 2 | 15 | 565_567 | 151.33333333333334 | 710.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000457416 | 3 | 18 | 487_495 | 151.33333333333334 | 823.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000457416 | 3 | 18 | 565_567 | 151.33333333333334 | 823.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000346997 | 2 | 17 | 487_495 | 151.33333333333334 | 820.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000346997 | 2 | 17 | 565_567 | 151.33333333333334 | 820.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000351936 | 3 | 18 | 487_495 | 151.33333333333334 | 786.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000351936 | 3 | 18 | 565_567 | 151.33333333333334 | 786.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000356226 | 1 | 16 | 487_495 | 36.333333333333336 | 705.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000356226 | 1 | 16 | 565_567 | 36.333333333333336 | 705.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000357555 | 2 | 17 | 487_495 | 62.333333333333336 | 708.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000357555 | 2 | 17 | 565_567 | 62.333333333333336 | 708.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000358487 | 3 | 18 | 487_495 | 151.33333333333334 | 822.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000358487 | 3 | 18 | 565_567 | 151.33333333333334 | 822.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000359354 | 3 | 7 | 487_495 | 151.33333333333334 | 255.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000359354 | 3 | 7 | 565_567 | 151.33333333333334 | 255.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000360144 | 2 | 17 | 487_495 | 62.333333333333336 | 681.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000360144 | 2 | 17 | 565_567 | 62.333333333333336 | 681.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369056 | 2 | 17 | 487_495 | 151.33333333333334 | 770.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369056 | 2 | 17 | 565_567 | 151.33333333333334 | 770.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369060 | 3 | 16 | 487_495 | 151.33333333333334 | 706.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369060 | 3 | 16 | 565_567 | 151.33333333333334 | 706.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369061 | 2 | 15 | 487_495 | 151.33333333333334 | 710.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369061 | 2 | 15 | 565_567 | 151.33333333333334 | 710.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000457416 | 3 | 18 | 487_495 | 151.33333333333334 | 823.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000457416 | 3 | 18 | 565_567 | 151.33333333333334 | 823.0 | Nucleotide binding | ATP | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 161_178 | 151.33333333333334 | 820.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 161_178 | 151.33333333333334 | 786.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 131_151 | 36.333333333333336 | 705.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 161_178 | 36.333333333333336 | 705.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 131_151 | 62.333333333333336 | 708.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 161_178 | 62.333333333333336 | 708.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 161_178 | 151.33333333333334 | 822.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 161_178 | 151.33333333333334 | 255.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 131_151 | 62.333333333333336 | 681.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 161_178 | 62.333333333333336 | 681.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 161_178 | 151.33333333333334 | 770.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 161_178 | 151.33333333333334 | 706.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 161_178 | 151.33333333333334 | 710.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 161_178 | 151.33333333333334 | 823.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000346997 | 2 | 17 | 161_178 | 151.33333333333334 | 820.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000351936 | 3 | 18 | 161_178 | 151.33333333333334 | 786.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000356226 | 1 | 16 | 161_178 | 36.333333333333336 | 705.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000357555 | 2 | 17 | 161_178 | 62.333333333333336 | 708.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000358487 | 3 | 18 | 161_178 | 151.33333333333334 | 822.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000359354 | 3 | 7 | 161_178 | 151.33333333333334 | 255.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000360144 | 2 | 17 | 161_178 | 62.333333333333336 | 681.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369056 | 2 | 17 | 161_178 | 151.33333333333334 | 770.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369060 | 3 | 16 | 161_178 | 151.33333333333334 | 706.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369061 | 2 | 15 | 161_178 | 151.33333333333334 | 710.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000457416 | 3 | 18 | 161_178 | 151.33333333333334 | 823.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000346997 | 2 | 17 | 161_178 | 151.33333333333334 | 820.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000351936 | 3 | 18 | 161_178 | 151.33333333333334 | 786.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000356226 | 1 | 16 | 161_178 | 36.333333333333336 | 705.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000357555 | 2 | 17 | 161_178 | 62.333333333333336 | 708.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000358487 | 3 | 18 | 161_178 | 151.33333333333334 | 822.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000359354 | 3 | 7 | 161_178 | 151.33333333333334 | 255.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000360144 | 2 | 17 | 161_178 | 62.333333333333336 | 681.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369056 | 2 | 17 | 161_178 | 151.33333333333334 | 770.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369060 | 3 | 16 | 161_178 | 151.33333333333334 | 706.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369061 | 2 | 15 | 161_178 | 151.33333333333334 | 710.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000457416 | 3 | 18 | 161_178 | 151.33333333333334 | 823.0 | Region | Note=Heparin-binding | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 399_821 | 151.33333333333334 | 820.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 399_821 | 151.33333333333334 | 786.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 399_821 | 36.333333333333336 | 705.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 399_821 | 62.333333333333336 | 708.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 399_821 | 151.33333333333334 | 822.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 399_821 | 151.33333333333334 | 255.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 399_821 | 62.333333333333336 | 681.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 399_821 | 151.33333333333334 | 770.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 399_821 | 151.33333333333334 | 706.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 399_821 | 151.33333333333334 | 710.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 399_821 | 151.33333333333334 | 823.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000346997 | 2 | 17 | 399_821 | 151.33333333333334 | 820.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000351936 | 3 | 18 | 399_821 | 151.33333333333334 | 786.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000356226 | 1 | 16 | 399_821 | 36.333333333333336 | 705.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000357555 | 2 | 17 | 399_821 | 62.333333333333336 | 708.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000358487 | 3 | 18 | 399_821 | 151.33333333333334 | 822.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000359354 | 3 | 7 | 399_821 | 151.33333333333334 | 255.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000360144 | 2 | 17 | 399_821 | 62.333333333333336 | 681.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369056 | 2 | 17 | 399_821 | 151.33333333333334 | 770.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369060 | 3 | 16 | 399_821 | 151.33333333333334 | 706.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369061 | 2 | 15 | 399_821 | 151.33333333333334 | 710.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000457416 | 3 | 18 | 399_821 | 151.33333333333334 | 823.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000346997 | 2 | 17 | 399_821 | 151.33333333333334 | 820.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000351936 | 3 | 18 | 399_821 | 151.33333333333334 | 786.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000356226 | 1 | 16 | 399_821 | 36.333333333333336 | 705.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000357555 | 2 | 17 | 399_821 | 62.333333333333336 | 708.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000358487 | 3 | 18 | 399_821 | 151.33333333333334 | 822.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000359354 | 3 | 7 | 399_821 | 151.33333333333334 | 255.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000360144 | 2 | 17 | 399_821 | 62.333333333333336 | 681.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369056 | 2 | 17 | 399_821 | 151.33333333333334 | 770.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369060 | 3 | 16 | 399_821 | 151.33333333333334 | 706.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369061 | 2 | 15 | 399_821 | 151.33333333333334 | 710.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000457416 | 3 | 18 | 399_821 | 151.33333333333334 | 823.0 | Topological domain | Cytoplasmic | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 378_398 | 151.33333333333334 | 820.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 378_398 | 151.33333333333334 | 786.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 378_398 | 36.333333333333336 | 705.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 378_398 | 62.333333333333336 | 708.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 378_398 | 151.33333333333334 | 822.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 378_398 | 151.33333333333334 | 255.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 378_398 | 62.333333333333336 | 681.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 378_398 | 151.33333333333334 | 770.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 378_398 | 151.33333333333334 | 706.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 378_398 | 151.33333333333334 | 710.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 378_398 | 151.33333333333334 | 823.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000346997 | 2 | 17 | 378_398 | 151.33333333333334 | 820.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000351936 | 3 | 18 | 378_398 | 151.33333333333334 | 786.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000356226 | 1 | 16 | 378_398 | 36.333333333333336 | 705.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000357555 | 2 | 17 | 378_398 | 62.333333333333336 | 708.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000358487 | 3 | 18 | 378_398 | 151.33333333333334 | 822.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000359354 | 3 | 7 | 378_398 | 151.33333333333334 | 255.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000360144 | 2 | 17 | 378_398 | 62.333333333333336 | 681.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369056 | 2 | 17 | 378_398 | 151.33333333333334 | 770.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369060 | 3 | 16 | 378_398 | 151.33333333333334 | 706.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369061 | 2 | 15 | 378_398 | 151.33333333333334 | 710.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000457416 | 3 | 18 | 378_398 | 151.33333333333334 | 823.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000346997 | 2 | 17 | 378_398 | 151.33333333333334 | 820.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000351936 | 3 | 18 | 378_398 | 151.33333333333334 | 786.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000356226 | 1 | 16 | 378_398 | 36.333333333333336 | 705.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000357555 | 2 | 17 | 378_398 | 62.333333333333336 | 708.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000358487 | 3 | 18 | 378_398 | 151.33333333333334 | 822.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000359354 | 3 | 7 | 378_398 | 151.33333333333334 | 255.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000360144 | 2 | 17 | 378_398 | 62.333333333333336 | 681.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369056 | 2 | 17 | 378_398 | 151.33333333333334 | 770.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369060 | 3 | 16 | 378_398 | 151.33333333333334 | 706.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369061 | 2 | 15 | 378_398 | 151.33333333333334 | 710.0 | Transmembrane | Helical | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000457416 | 3 | 18 | 378_398 | 151.33333333333334 | 823.0 | Transmembrane | Helical |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KLK2 | chr19:51376774 | chr10:123310972 | ENST00000325321 | 1 | 5 | 25_258 | 15.333333333333334 | 262.0 | Domain | Peptidase S1 | |
| Hgene | KLK2 | chr19:51376774 | chr10:123310972 | ENST00000325321 | + | 1 | 5 | 25_258 | 15.333333333333334 | 262.0 | Domain | Peptidase S1 |
| Hgene | KLK2 | chr19:51376774 | chr10:123310972 | ENST00000358049 | 1 | 5 | 25_258 | 15.333333333333334 | 224.0 | Domain | Peptidase S1 | |
| Hgene | KLK2 | chr19:51376774 | chr10:123310972 | ENST00000358049 | + | 1 | 5 | 25_258 | 15.333333333333334 | 224.0 | Domain | Peptidase S1 |
| Hgene | KLK2 | chr19:51376774 | chr10:123310972 | ENST00000391810 | 1 | 4 | 25_258 | 0 | 160.0 | Domain | Peptidase S1 | |
| Hgene | KLK2 | chr19:51376774 | chr10:123310972 | ENST00000391810 | + | 1 | 4 | 25_258 | 0 | 160.0 | Domain | Peptidase S1 |
| Hgene | KLK2 | chr19:51376775 | chr10:123310973 | ENST00000325321 | + | 1 | 5 | 25_258 | 15.333333333333334 | 262.0 | Domain | Peptidase S1 |
| Hgene | KLK2 | chr19:51376775 | chr10:123310973 | ENST00000358049 | + | 1 | 5 | 25_258 | 15.333333333333334 | 224.0 | Domain | Peptidase S1 |
| Hgene | KLK2 | chr19:51376775 | chr10:123310973 | ENST00000391810 | + | 1 | 4 | 25_258 | 0 | 160.0 | Domain | Peptidase S1 |
| Hgene | KLK2 | chr19:51380014 | chr10:123310973 | ENST00000325321 | + | 3 | 5 | 25_258 | 164.33333333333334 | 262.0 | Domain | Peptidase S1 |
| Hgene | KLK2 | chr19:51380014 | chr10:123310973 | ENST00000358049 | + | 3 | 5 | 25_258 | 164.33333333333334 | 224.0 | Domain | Peptidase S1 |
| Hgene | KLK2 | chr19:51380014 | chr10:123310973 | ENST00000391810 | + | 2 | 4 | 25_258 | 62.333333333333336 | 160.0 | Domain | Peptidase S1 |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 25_125 | 151.33333333333334 | 820.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 25_125 | 151.33333333333334 | 786.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 25_125 | 36.333333333333336 | 705.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 25_125 | 62.333333333333336 | 708.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 25_125 | 151.33333333333334 | 822.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 25_125 | 151.33333333333334 | 255.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 25_125 | 62.333333333333336 | 681.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 25_125 | 151.33333333333334 | 770.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 25_125 | 151.33333333333334 | 706.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 25_125 | 151.33333333333334 | 710.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 25_125 | 151.33333333333334 | 823.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000346997 | 2 | 17 | 25_125 | 151.33333333333334 | 820.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000351936 | 3 | 18 | 25_125 | 151.33333333333334 | 786.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000356226 | 1 | 16 | 25_125 | 36.333333333333336 | 705.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000357555 | 2 | 17 | 25_125 | 62.333333333333336 | 708.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000358487 | 3 | 18 | 25_125 | 151.33333333333334 | 822.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000359354 | 3 | 7 | 25_125 | 151.33333333333334 | 255.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000360144 | 2 | 17 | 25_125 | 62.333333333333336 | 681.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369056 | 2 | 17 | 25_125 | 151.33333333333334 | 770.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369060 | 3 | 16 | 25_125 | 151.33333333333334 | 706.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369061 | 2 | 15 | 25_125 | 151.33333333333334 | 710.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000457416 | 3 | 18 | 25_125 | 151.33333333333334 | 823.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000346997 | 2 | 17 | 25_125 | 151.33333333333334 | 820.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000351936 | 3 | 18 | 25_125 | 151.33333333333334 | 786.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000356226 | 1 | 16 | 25_125 | 36.333333333333336 | 705.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000357555 | 2 | 17 | 25_125 | 62.333333333333336 | 708.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000358487 | 3 | 18 | 25_125 | 151.33333333333334 | 822.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000359354 | 3 | 7 | 25_125 | 151.33333333333334 | 255.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000360144 | 2 | 17 | 25_125 | 62.333333333333336 | 681.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369056 | 2 | 17 | 25_125 | 151.33333333333334 | 770.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369060 | 3 | 16 | 25_125 | 151.33333333333334 | 706.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369061 | 2 | 15 | 25_125 | 151.33333333333334 | 710.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000457416 | 3 | 18 | 25_125 | 151.33333333333334 | 823.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 131_151 | 151.33333333333334 | 820.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 131_151 | 151.33333333333334 | 786.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 131_151 | 151.33333333333334 | 822.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 131_151 | 151.33333333333334 | 255.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 131_151 | 151.33333333333334 | 770.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 131_151 | 151.33333333333334 | 706.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 131_151 | 151.33333333333334 | 710.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 131_151 | 151.33333333333334 | 823.0 | Region | Disordered | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000346997 | 2 | 17 | 22_377 | 151.33333333333334 | 820.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000351936 | 3 | 18 | 22_377 | 151.33333333333334 | 786.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000356226 | 1 | 16 | 22_377 | 36.333333333333336 | 705.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000357555 | 2 | 17 | 22_377 | 62.333333333333336 | 708.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000358487 | 3 | 18 | 22_377 | 151.33333333333334 | 822.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000359354 | 3 | 7 | 22_377 | 151.33333333333334 | 255.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000360144 | 2 | 17 | 22_377 | 62.333333333333336 | 681.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369056 | 2 | 17 | 22_377 | 151.33333333333334 | 770.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369060 | 3 | 16 | 22_377 | 151.33333333333334 | 706.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000369061 | 2 | 15 | 22_377 | 151.33333333333334 | 710.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376774 | chr10:123310972 | ENST00000457416 | 3 | 18 | 22_377 | 151.33333333333334 | 823.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000346997 | 2 | 17 | 22_377 | 151.33333333333334 | 820.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000351936 | 3 | 18 | 22_377 | 151.33333333333334 | 786.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000356226 | 1 | 16 | 22_377 | 36.333333333333336 | 705.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000357555 | 2 | 17 | 22_377 | 62.333333333333336 | 708.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000358487 | 3 | 18 | 22_377 | 151.33333333333334 | 822.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000359354 | 3 | 7 | 22_377 | 151.33333333333334 | 255.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000360144 | 2 | 17 | 22_377 | 62.333333333333336 | 681.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369056 | 2 | 17 | 22_377 | 151.33333333333334 | 770.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369060 | 3 | 16 | 22_377 | 151.33333333333334 | 706.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000369061 | 2 | 15 | 22_377 | 151.33333333333334 | 710.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51376775 | chr10:123310973 | ENST00000457416 | 3 | 18 | 22_377 | 151.33333333333334 | 823.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000346997 | 2 | 17 | 22_377 | 151.33333333333334 | 820.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000351936 | 3 | 18 | 22_377 | 151.33333333333334 | 786.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000356226 | 1 | 16 | 22_377 | 36.333333333333336 | 705.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000357555 | 2 | 17 | 22_377 | 62.333333333333336 | 708.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000358487 | 3 | 18 | 22_377 | 151.33333333333334 | 822.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000359354 | 3 | 7 | 22_377 | 151.33333333333334 | 255.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000360144 | 2 | 17 | 22_377 | 62.333333333333336 | 681.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369056 | 2 | 17 | 22_377 | 151.33333333333334 | 770.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369060 | 3 | 16 | 22_377 | 151.33333333333334 | 706.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000369061 | 2 | 15 | 22_377 | 151.33333333333334 | 710.0 | Topological domain | Extracellular | |
| Tgene | FGFR2 | chr19:51380014 | chr10:123310973 | ENST00000457416 | 3 | 18 | 22_377 | 151.33333333333334 | 823.0 | Topological domain | Extracellular |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (423) >>>423.pdbFusion protein BP residue: 14 CIF file (423) >>>423.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPM PTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQP HIQWIKHVEKNGSKYGPDGLPYLKVLKVSAESSSSMNSNTPLVRITTRLS STADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVG IDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNIINLLG ACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQMTFKD LVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLARDIN NIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGS PYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQ LVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDP | 569 | ||
| 3D view using mol* of 423 (AA BP:14) | ||||||||||
 | ||||||||||
| PDB file (434) >>>434.pdbFusion protein BP residue: 14 CIF file (434) >>>434.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPM PTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVAPGREKEITASPDYLEIAIYCIGVFLIACMVVTVILC RMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSSSMNSNTPLVRIT TRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLGEGCFGQVVMA EAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMMKMIGKHKNII NLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSYDINRVPEEQM TFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENNVMKIADFGLA RDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWSFGVLMWEIFT LGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRDCWHAVPSQRP TFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRSSCSSGDDSVF | 573 | ||
| 3D view using mol* of 434 (AA BP:14) | ||||||||||
 | ||||||||||
| PDB file (505) >>>505.pdbFusion protein BP residue: 14 CIF file (505) >>>505.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPM PTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQP HIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAG EYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGV FLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESS SSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGK PLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEM EMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVT ENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSD VWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMM | 633 | ||
| 3D view using mol* of 505 (AA BP:14) | ||||||||||
 | ||||||||||
| PDB file (507) >>>507.pdbFusion protein BP residue: 80 CIF file (507) >>>507.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLRE APQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVP AANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMES VVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKVSAESSS SMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKP LGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEME MMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEY SYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTE NNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDV WSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMM RDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDT | 635 | ||
| 3D view using mol* of 507 (AA BP:80) | ||||||||||
 | ||||||||||
| PDB file (511) >>>511.pdbFusion protein BP residue: 80 CIF file (511) >>>511.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLRE APQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVP AANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMES VVPSDKGNYTCVVENEYGSINHTYHLDVVAPGREKEITASPDYLEIAIYC IGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSA ESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLT LGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLV SEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPP GMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNV LVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTH QSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNEL YMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPS | 639 | ||
| 3D view using mol* of 511 (AA BP:80) | ||||||||||
 | ||||||||||
| PDB file (525) | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPM PTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQP HIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFED AGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVF LIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAESSSSM NSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLG EGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMM KMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSY DINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENN VMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWS FGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRD CWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEPYSPCYPDPRX | 650 | ||
| PDB file (538) >>>538.pdbFusion protein BP residue: 14 CIF file (538) >>>538.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPM PTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQP HIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFED AGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVF LIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSS SMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKP LGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEME MMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEY SYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTE NNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDV WSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMM RDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEEKKVSGAVDCHKPPCNPS | 660 | ||
| 3D view using mol* of 538 (AA BP:14) | ||||||||||
 | ||||||||||
| PDB file (555) >>>555.pdbFusion protein BP residue: 14 CIF file (555) >>>555.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPM PTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQP HIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFED AGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVF LIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAESSSSM NSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLG EGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMM KMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSY DINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENN VMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWS FGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRD CWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRS | 683 | ||
| 3D view using mol* of 555 (AA BP:14) | ||||||||||
 | ||||||||||
| PDB file (557) | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPM PTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQP HIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFED AGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVF LIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVSAESSSSM NSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKPLG EGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEMEMM KMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEYSY DINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTENN VMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDVWS FGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMMRD CWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDTRS | 684 | ||
| PDB file (558) >>>558.pdbFusion protein BP residue: 14 CIF file (558) >>>558.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPM PTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQP HIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTTDKEIEVLYIRNVTFED AGEYTCLAGNSIGISFHSAWLTVLPAPGREKEITASPDYLEIAIYCIGVF LIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESSS SMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGKP LGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEME MMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGMEY SYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVTE NNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSDV WSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMMM RDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPDT | 685 | ||
| 3D view using mol* of 558 (AA BP:14) | ||||||||||
 | ||||||||||
| PDB file (560) >>>560.pdbFusion protein BP residue: 14 CIF file (560) >>>560.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPM PTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQP HIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAG EYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGV FLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESS SSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGK PLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEM EMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVT ENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSD VWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMM MRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPD | 686 | ||
| 3D view using mol* of 560 (AA BP:14) | ||||||||||
 | ||||||||||
| PDB file (561) | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVPAANTVKFRCPAGGNPM PTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMESVVPSDKGNYTCVVENE YGSINHTYHLDVVERSPHRPILQAGLPANASTVVGGDVEFVCKVYSDAQP HIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSSNAEVLALFNVTEADAG EYICKVSNYIGQANQSAWLTVLPKQQAPGREKEITASPDYLEIAIYCIGV FLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTKRIPLRRQVTVSAESS SSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDPKWEFPRDKLTLGK PLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDDATEKDLSDLVSEM EMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNLREYLRARRPPGME YSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKCIHRDLAARNVLVT ENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAPEALFDRVYTHQSD VWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRMDKPANCTNELYMM MRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDLSQPLEQYSPSYPD | 687 | ||
| PDB file (570) >>>570.pdbFusion protein BP residue: 80 CIF file (570) >>>570.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLRE APQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVP AANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMES VVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSS NAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPKQQAPGREKEI TASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLT KRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELP EDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKML KDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASK GNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLAS QKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKW MAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEG | 699 | ||
| 3D view using mol* of 570 (AA BP:80) | ||||||||||
 | ||||||||||
| PDB file (587) >>>587.pdbFusion protein BP residue: 80 CIF file (587) >>>587.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLRE APQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVP AANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMES VVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTT DKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEIT ASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTK RIPLRRQVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDP KWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDD ATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNL REYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKC IHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAP EALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRM DKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDL | 715 | ||
| 3D view using mol* of 587 (AA BP:80) | ||||||||||
 | ||||||||||
| PDB file (594) >>>594.pdbFusion protein BP residue: 80 CIF file (594) >>>594.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLRE APQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVP AANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMES VVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTT DKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEIT ASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTK RIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPE DPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKG NLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQ KCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWM APEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGH RMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEEK | 726 | ||
| 3D view using mol* of 594 (AA BP:80) | ||||||||||
 | ||||||||||
| PDB file (613) >>>613.pdbFusion protein BP residue: 80 CIF file (613) >>>613.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLRE APQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVP AANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMES VVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTT DKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEIT ASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTK RIPLRRQVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPEDP KWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLKDD ATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKGNL REYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQKC IHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWMAP EALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGHRM DKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYLDL | 749 | ||
| 3D view using mol* of 613 (AA BP:80) | ||||||||||
 | ||||||||||
| PDB file (615) >>>615.pdbFusion protein BP residue: 80 CIF file (615) >>>615.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLRE APQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVP AANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMES VVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKAAGVNTT DKEIEVLYIRNVTFEDAGEYTCLAGNSIGISFHSAWLTVLPAPGREKEIT ASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLTK RIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELPE DPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKMLK DDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASKG NLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLASQ KCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKWM APEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEGH RMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEYL DLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQYPHINGSVK | 751 | ||
| 3D view using mol* of 615 (AA BP:80) | ||||||||||
 | ||||||||||
| PDB file (617) >>>617.pdbFusion protein BP residue: 80 CIF file (617) >>>617.cif | KLK2 | chr19 | 51376774 | FGFR2 | chr10 | 123310972 | MALPPLPQLWVWEGVVQPPAAWGGPWSASRCQQGKGGVLENEGFIGLLRE APQPQTHHLAVDTCVSMWDLVLSIALSVGCTGAPYWTNTEKMEKRLHAVP AANTVKFRCPAGGNPMPTMRWLKNGKEFKQEHRIGGYKVRNQHWSLIMES VVPSDKGNYTCVVENEYGSINHTYHLDVVERSPHRPILQAGLPANASTVV GGDVEFVCKVYSDAQPHIQWIKHVEKNGSKYGPDGLPYLKVLKHSGINSS NAEVLALFNVTEADAGEYICKVSNYIGQANQSAWLTVLPKQQAPGREKEI TASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTKKPDFSSQPAVHKLT KRIPLRRQVTVSAESSSSMNSNTPLVRITTRLSSTADTPMLAGVSEYELP EDPKWEFPRDKLTLGKPLGEGCFGQVVMAEAVGIDKDKPKEAVTVAVKML KDDATEKDLSDLVSEMEMMKMIGKHKNIINLLGACTQDGPLYVIVEYASK GNLREYLRARRPPGMEYSYDINRVPEEQMTFKDLVSCTYQLARGMEYLAS QKCIHRDLAARNVLVTENNVMKIADFGLARDINNIDYYKKTTNGRLPVKW MAPEALFDRVYTHQSDVWSFGVLMWEIFTLGGSPYPGIPVEELFKLLKEG HRMDKPANCTNELYMMMRDCWHAVPSQRPTFKQLVEDLDRILTLTTNEEY LDLSQPLEQYSPSYPDTRSSCSSGDDSVFSPDPMPYEPCLPQYPHINGSV | 752 | ||
| 3D view using mol* of 617 (AA BP:80) | ||||||||||
 | ||||||||||
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
KLK2_pLDDT.png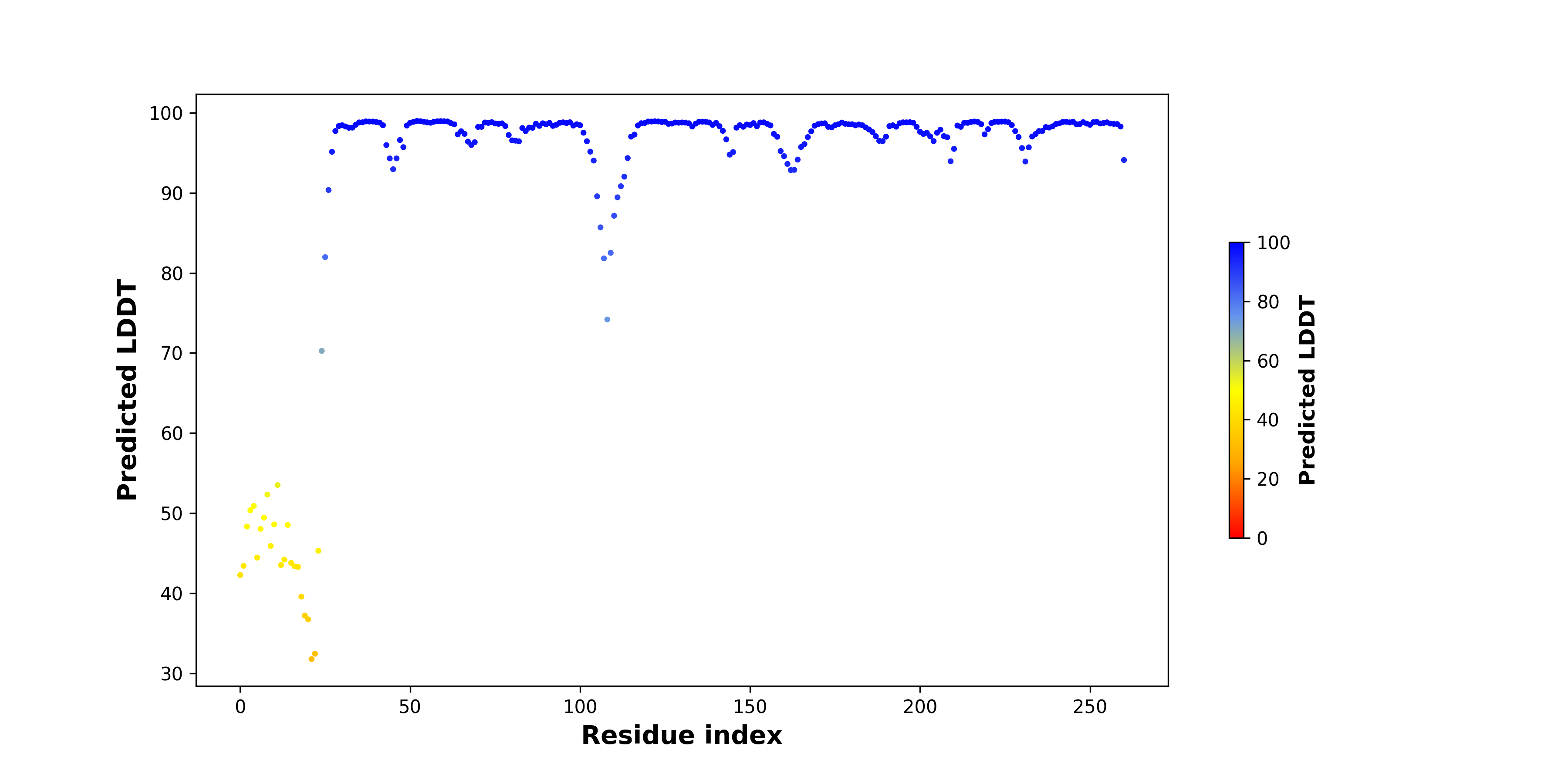 |
FGFR2_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 423 | 0.931 | 6372 | 1.04 | 2945.341 | 0.679 | 0.439 | 0.393 | 0.239 | 0.5 | 0.479 | 0.904 | Chain A: 12,13,14,16,19,20,21,22,23,24,25,26,27,28 ,29,30,31,32,33,34,36,37,38,39,40,41,42,44,45,46,4 7,48,56,57,58,59,72,74,76,77,78,79,81,83,84,92,93, 94,104,105,106,107,108,109,113,114,115,116,117,118 ,119,120,121,122,123,124,125,126,127,128,129,130,1 31,132,133,134,135,136,138,139,140,141,142,143,144 ,145,147,148,149,150,151,152,153,154,155,156,157,1 58,159,160,161,162,163,164,165,171,172,173,174,175 ,176,177,178,179,180,181,182,183,184,185,186,187,1 88,189,190,191,192,193,194,195,196,197,198,199,200 ,201,202,203,204,205,206,207,208,209,210,211,212,2 13,214,215,216,217,218,219,221,222,223,231,235,236 ,237,238,239,240,241,242,243,244,263,265,266,267,2 68,269,270,271,272,273,274,275,276,277,278,279,280 ,281,282,283,284,285,286,287,288,289,290,292,296,3 03,304,305,306,307,308,309,312,313,314,315,316,317 ,318,319,321,322,323,326,327,340,341,342,343,344,3 60,363,364,366,367,368,369,370,372,373,374,378,379 ,381,391,392,393,394,395,396,397,398,399,400,401,4 02,403,404,405,406,407,408,409,410,411,412,413,414 ,415,416,417,418,420,421,422,423,424,425,426,427,4 28,429,430,431,432,450,451,457,461,464,465,466,467 ,468,469,470,471,472,478,479,481,482,483,485,486,4 88,489,490,491,492,493,494,495,496,497,498,499,500 ,501,502,503,504,506,507 |
| 434 | 1.059 | 326 | 1.065 | 818.741 | 0.455 | 0.787 | 1.032 | 0.988 | 1.064 | 0.928 | 0.679 | Chain A: 239,240,242,243,244,245,246,247,267,269,2 70,271,278,279,282,283,285,286,300,316,317,318,319 ,320,322,323,376,377,378,382,383,385,395,396,397,3 98,399,400,401,405,406,407,408,409,410,411,412,413 ,415,416,417,418,419,422,426,427,428,429,430,431,4 32 |
| 505 | 1.03 | 617 | 1.048 | 1777.426 | 0.517 | 0.739 | 0.95 | 0.623 | 1.029 | 0.606 | 0.754 | Chain A: 36,57,62,65,66,67,68,69,70,83,84,86,88,89 ,93,308,309,310,311,312,352,353,355,356,357,358,35 9,360,380,382,383,384,391,392,394,395,396,397,398, 399,401,402,403,405,406,413,429,431,432,435,436,43 8,485,487,490,491,495,496,498,508,509,510,511,512, 513,514,515,518,519,520,521,522,523,524,525,526,52 7,528,529,530,531,532,533,534,535,539,540,541,543, 544,545,567,568,570,571,573,574,575 |
| 507 | 1.07 | 154 | 1.1 | 555.66 | 0.544 | 0.77 | 0.94 | 1.163 | 0.925 | 1.257 | 0.719 | Chain A: 301,302,304,305,306,307,308,309,329,331,3 32,333,340,341,344,345,347,348,352,362,378,379,380 ,381,384,385,438,440,444,445,447,457,458,459,460,4 61,462,463,473 |
| 511 | 1.076 | 128 | 0.963 | 237.356 | 0.399 | 0.812 | 1.128 | 0.471 | 1.423 | 0.331 | 0.711 | Chain A: 443,465,467,471,472,473,474,475,476,477,4 82,483,484,488,492,493,494,495,496,498 |
| 538 | 1.032 | 391 | 1.05 | 1282.477 | 0.514 | 0.742 | 0.943 | 0.726 | 1.027 | 0.707 | 0.921 | Chain A: 309,311,351,354,355,356,357,358,359,379,3 81,382,383,390,391,393,394,395,396,397,398,400,401 ,402,404,405,412,428,429,430,431,434,435,484,485,4 86,489,490,494,495,497,507,508,509,510,511,513,514 ,517,518,519,520,521,522,523,524,525,526,527,528,5 29,530,541,542,543,544 |
| 557 | 1.049 | 653 | 1.076 | 1689.961 | 0.526 | 0.748 | 0.946 | 0.748 | 0.961 | 0.779 | 0.919 | Chain A: 77,78,79,81,82,106,108,109,110,111,114,11 8,119,120,121,138,141,142,144,145,146,147,148,149, 150,201,202,203,204,205,206,207,208,209,210,211,21 2,379,380,381,382,383,384,385,386,387,389,391,399, 401,404,405,419,423,432,434,445,447,448,449,450,45 1,452,453,454,456,457,461,492,493,494,495,496,498, 499,501,511,512,513,514,515,516,517,519,520,521,52 2,523,524,527,528,529,530,531,532,533,534,537,538, 539,542,545,548,549,550,551,555,556,559 |
| 558 | 1.052 | 748 | 1.001 | 2309.762 | 0.447 | 0.776 | 0.989 | 0.616 | 1.241 | 0.496 | 0.628 | Chain A: 34,35,36,37,38,64,65,67,68,69,70,71,72,74 ,81,83,84,86,87,113,114,115,116,117,118,119,121,12 3,145,146,147,215,347,348,349,350,351,352,353,354, 355,356,357,377,379,380,381,388,389,392,393,396,40 0,410,426,427,428,429,430,431,432,433,435,436,437, 439,442,448,449,450,487,488,492,493,495,505,506,50 7,508,509,511,517,518,519,520,521,522,523,524,525, 526,527,528,529,530,531,537,538,540,542,563,564,56 5,567,568,570,571,572,575,579 |
| 560 | 1.077 | 188 | 1.126 | 636.951 | 0.535 | 0.741 | 0.978 | 1.369 | 0.793 | 1.726 | 0.811 | Chain A: 350,352,353,355,356,357,358,359,360,380,3 82,383,384,389,391,392,395,396,398,399,403,413,427 ,429,430,431,432,435,436,495,496,498,508,509,510,5 11,512,514,523,524,525,526,527,528 |
| 570 | 1.006 | 773 | 1.016 | 2681.917 | 0.568 | 0.708 | 0.922 | 0.518 | 1.07 | 0.484 | 0.763 | Chain A: 100,101,102,103,130,131,133,134,135,136,1 37,138,149,150,151,152,153,180,181,182,183,184,211 ,212,213,214,215,216,251,275,276,373,374,375,376,3 77,378,418,419,420,421,422,423,424,425,426,446,448 ,449,450,457,458,460,461,462,463,464,465,467,468,4 69,471,472,479,495,496,497,498,499,500,501,502,505 ,506,508,509,510,511,515,516,517,519,520,521,522,5 24,551,552,553,555,556,557,558,561,562,564,574,575 ,576,577,578,579,580,581,584,585,586,587,588,589,5 90,591,592,593,594,595,596,597,598,599,600,601,605 ,606,607,609,611,619,625,628,629,632,633,634,635,6 36,637,638,639,640,641,644,651,653,654,656,657,658 |
| 587 | 1.014 | 692 | 1.032 | 2557.065 | 0.582 | 0.719 | 0.889 | 0.492 | 1.041 | 0.472 | 0.939 | Chain A: 101,102,123,126,127,128,132,133,134,135,1 50,151,152,153,154,155,180,210,211,212,213,214,215 ,216,217,218,245,247,249,250,252,253,255,276,277,3 68,369,370,371,372,373,374,375,396,397,398,399,402 ,415,416,418,419,420,421,422,423,443,445,446,447,4 54,455,457,458,459,460,461,462,463,464,465,466,467 ,468,469,470,476,481,483,490,492,493,494,495,496,4 98,499,501,548,549,550,553,554,558,559,561,571,572 ,573,574,575,577,578,581,582,583,584,585,586,587,5 88,589,590,591,592,593,594,595,596,597,598,602,603 ,604,606,607,608,630,631,637,638,641,645 |
| 594 | 1.021 | 695 | 1.034 | 2835.238 | 0.581 | 0.73 | 0.931 | 0.576 | 1.055 | 0.546 | 0.914 | Chain A: 100,101,102,130,135,136,150,151,152,153,1 79,180,181,183,184,210,211,212,213,214,215,216,217 ,218,247,249,250,251,255,275,276,277,278,371,373,3 74,375,376,377,417,418,420,421,422,423,424,425,445 ,447,448,449,456,457,459,460,461,462,463,464,466,4 67,468,470,471,478,494,496,497,500,501,503,504,507 ,508,510,512,516,517,518,519,522,524,550,551,552,5 54,555,556,560,561,563,573,574,575,576,577,579,580 ,583,584,585,586,588,589,590,591,592,593,594,595,5 96,597,598,599,600,604,605,606,608,610,624,628,630 ,631,632,633,635,636,637,639,640,643,644,647,648 |
| 615 | 1.007 | 920 | 1.025 | 2430.155 | 0.517 | 0.709 | 0.906 | 0.511 | 1.044 | 0.489 | 0.815 | Chain A: 100,101,102,103,123,124,126,127,128,131,1 32,133,134,135,152,153,154,155,180,181,182,183,184 ,185,213,214,215,276,277,278,373,374,375,376,377,4 17,418,420,421,422,423,424,425,445,447,448,449,456 ,457,459,460,461,462,463,464,466,467,468,470,471,4 78,494,495,496,497,498,500,501,507,508,510,511,512 ,514,515,516,517,518,519,522,523,524,525,550,551,5 52,553,555,556,560,561,563,573,574,575,576,577,578 ,579,580,581,583,584,585,586,587,588,589,590,591,5 92,593,594,595,596,597,598,599,600,604,605,606,608 ,609,610,624,628,630,631,632,634,635,636,637,638,6 39,640,643,650,652,653,654,655,656 |
| 617 | 1.103 | 154 | 1.132 | 537.481 | 0.502 | 0.813 | 1.025 | 1.336 | 0.907 | 1.474 | 0.878 | Chain A: 418,419,421,422,423,424,425,426,446,448,4 49,450,457,458,461,462,465,469,479,495,496,497,498 ,501,502,561,562,564,574,575,576,577,578,580,590,5 94 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| KLK2 | SERPINF2, SERPINA3, SERPINA5, SERPINB6, APP, SUZ12, RNF2, ADAMTS1, SERPINE2, APLP2, GLG1, EXT2, BMP1, PIGT, FGFR4, MAN2B2, SUMF1, LRP6, JAG2, IL17RA, GALNT18, CERCAM, CHID1, PGAM2, ERGIC1, ROR2, PCSK9, B4GALT3, PCSK6, ARSK, MAN2A2, MANBA, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| KLK2 |  |
| FGFR2 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to KLK2-FGFR2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
| KLK2 | FGFR2 | Fgfr Inhibitors | PubMed | 31043681 |
Top |
Related Diseases to KLK2-FGFR2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| KLK2 | FGFR2 | Metastatic Prostate Cancer | PubMed | 31043681 |
| KLK2 | FGFR2 | Cholangiocarcinoma | MyCancerGenome | |
| KLK2 | FGFR2 | Adenocarcinoma Of Unknown Primary | MyCancerGenome | |
| KLK2 | FGFR2 | Adenocarcinoma Of The Gastroesophageal Junction | MyCancerGenome | |
| KLK2 | FGFR2 | Breast Invasive Ductal Carcinoma | MyCancerGenome | |
| KLK2 | FGFR2 | Esophageal Adenocarcinoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | KLK2 | C0033578 | Prostatic Neoplasms | 2 | CTD_human |
| Hgene | KLK2 | C0376358 | Malignant neoplasm of prostate | 2 | CTD_human |
| Tgene | FGFR2 | C2931196 | Craniofacial dysostosis type 1 | 23 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | FGFR2 | C0220658 | Pfeiffer Syndrome | 21 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | FGFR2 | C0001193 | Apert syndrome | 19 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | FGFR2 | C0795998 | JACKSON-WEISS SYNDROME | 10 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | FGFR2 | C0175699 | Saethre-Chotzen Syndrome | 8 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | FGFR2 | C1852406 | Cutis Gyrata Syndrome of Beare And Stevenson | 8 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | FGFR2 | C2936791 | Antley-Bixler Syndrome, Autosomal Dominant | 7 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | FGFR2 | C1510455 | Acrocephalosyndactylia | 6 | CTD_human;ORPHANET |
| Tgene | FGFR2 | C0265269 | Lacrimoauriculodentodigital syndrome | 5 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | FGFR2 | C0010278 | Craniosynostosis | 4 | CTD_human;GENOMICS_ENGLAND |
| Tgene | FGFR2 | C1863389 | Apert-Crouzon Disease | 4 | CTD_human |
| Tgene | FGFR2 | C1865070 | SCAPHOCEPHALY, MAXILLARY RETRUSION, AND MENTAL RETARDATION | 4 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | FGFR2 | C0006142 | Malignant neoplasm of breast | 3 | CTD_human;UNIPROT |
| Tgene | FGFR2 | C0030044 | Acrocephaly | 3 | CTD_human |
| Tgene | FGFR2 | C0036341 | Schizophrenia | 3 | PSYGENET |
| Tgene | FGFR2 | C0221356 | Brachycephaly | 3 | CTD_human |
| Tgene | FGFR2 | C0265534 | Scaphycephaly | 3 | CTD_human |
| Tgene | FGFR2 | C0265535 | Trigonocephaly | 3 | CTD_human |
| Tgene | FGFR2 | C0376634 | Craniofacial Abnormalities | 3 | CTD_human |
| Tgene | FGFR2 | C0678222 | Breast Carcinoma | 3 | CTD_human |
| Tgene | FGFR2 | C1257931 | Mammary Neoplasms, Human | 3 | CTD_human |
| Tgene | FGFR2 | C1458155 | Mammary Neoplasms | 3 | CTD_human |
| Tgene | FGFR2 | C1833340 | Synostotic Posterior Plagiocephaly | 3 | CTD_human |
| Tgene | FGFR2 | C1860819 | Metopic synostosis | 3 | CTD_human |
| Tgene | FGFR2 | C2931150 | Synostotic Anterior Plagiocephaly | 3 | CTD_human |
| Tgene | FGFR2 | C3281247 | BENT BONE DYSPLASIA SYNDROME | 3 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | FGFR2 | C4551902 | Craniosynostosis, Type 1 | 3 | CTD_human |
| Tgene | FGFR2 | C4704874 | Mammary Carcinoma, Human | 3 | CTD_human |
| Tgene | FGFR2 | C0008925 | Cleft Palate | 2 | CTD_human |
| Tgene | FGFR2 | C0011570 | Mental Depression | 2 | PSYGENET |
| Tgene | FGFR2 | C0011581 | Depressive disorder | 2 | PSYGENET |
| Tgene | FGFR2 | C0024623 | Malignant neoplasm of stomach | 2 | CGI;CTD_human |
| Tgene | FGFR2 | C0038356 | Stomach Neoplasms | 2 | CGI;CTD_human |
| Tgene | FGFR2 | C1708349 | Hereditary Diffuse Gastric Cancer | 2 | CTD_human |
| Tgene | FGFR2 | C1837218 | Cleft palate, isolated | 2 | CTD_human |
| Tgene | FGFR2 | C0000772 | Multiple congenital anomalies | 1 | CTD_human |
| Tgene | FGFR2 | C0003090 | Ankylosis | 1 | CTD_human |
| Tgene | FGFR2 | C0005586 | Bipolar Disorder | 1 | PSYGENET |
| Tgene | FGFR2 | C0008924 | Cleft upper lip | 1 | CTD_human |
| Tgene | FGFR2 | C0010273 | Craniofacial Dysostosis | 1 | CTD_human |
| Tgene | FGFR2 | C0011757 | Developmental Coordination Disorder | 1 | CTD_human |
| Tgene | FGFR2 | C0014170 | Endometrial Neoplasms | 1 | CTD_human |
| Tgene | FGFR2 | C0018553 | Hamartoma Syndrome, Multiple | 1 | CTD_human |
| Tgene | FGFR2 | C0020796 | Profound Mental Retardation | 1 | CTD_human |
| Tgene | FGFR2 | C0023890 | Liver Cirrhosis | 1 | CTD_human |
| Tgene | FGFR2 | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Tgene | FGFR2 | C0025363 | Mental Retardation, Psychosocial | 1 | CTD_human |
| Tgene | FGFR2 | C0026613 | Motor Skills Disorders | 1 | CTD_human |
| Tgene | FGFR2 | C0033975 | Psychotic Disorders | 1 | PSYGENET |
| Tgene | FGFR2 | C0037268 | Skin Abnormalities | 1 | CTD_human |
| Tgene | FGFR2 | C0037274 | Dermatologic disorders | 1 | CTD_human |
| Tgene | FGFR2 | C0038219 | Status Dysraphicus | 1 | CTD_human |
| Tgene | FGFR2 | C0040427 | Tooth Abnormalities | 1 | CTD_human |
| Tgene | FGFR2 | C0080178 | Spina Bifida | 1 | CTD_human |
| Tgene | FGFR2 | C0152423 | Congenital small ears | 1 | GENOMICS_ENGLAND |
| Tgene | FGFR2 | C0206698 | Cholangiocarcinoma | 1 | CTD_human |
| Tgene | FGFR2 | C0206762 | Limb Deformities, Congenital | 1 | CTD_human |
| Tgene | FGFR2 | C0239946 | Fibrosis, Liver | 1 | CTD_human |
| Tgene | FGFR2 | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Tgene | FGFR2 | C0265326 | Bannayan-Riley-Ruvalcaba Syndrome | 1 | CTD_human |
| Tgene | FGFR2 | C0266508 | Rachischisis | 1 | CTD_human |
| Tgene | FGFR2 | C0345905 | Intrahepatic Cholangiocarcinoma | 1 | CTD_human |
| Tgene | FGFR2 | C0349204 | Nonorganic psychosis | 1 | PSYGENET |
| Tgene | FGFR2 | C0391826 | Lhermitte-Duclos disease | 1 | CTD_human |
| Tgene | FGFR2 | C0476089 | Endometrial Carcinoma | 1 | CGI;CTD_human |
| Tgene | FGFR2 | C0524730 | Odontome | 1 | CTD_human |
| Tgene | FGFR2 | C0699791 | Stomach Carcinoma | 1 | CGI;GENOMICS_ENGLAND |
| Tgene | FGFR2 | C0917816 | Mental deficiency | 1 | CTD_human |
| Tgene | FGFR2 | C1450010 | Plagiocephaly, Nonsynostotic | 1 | CTD_human |
| Tgene | FGFR2 | C1860042 | Antley-Bixler Syndrome with Disordered Steroidogenesis | 1 | CTD_human |
| Tgene | FGFR2 | C1867564 | SCAPHOCEPHALY AND AXENFELD-RIEGER ANOMALY | 1 | GENOMICS_ENGLAND |
| Tgene | FGFR2 | C1959582 | PTEN Hamartoma Tumor Syndrome | 1 | CTD_human |
| Tgene | FGFR2 | C2350233 | Antley-Bixler Syndrome Phenotype | 1 | CTD_human |
| Tgene | FGFR2 | C3267076 | Familial scaphocephaly syndrome | 1 | GENOMICS_ENGLAND |
| Tgene | FGFR2 | C3714756 | Intellectual Disability | 1 | CTD_human |
| Tgene | FGFR2 | C3805278 | Extrahepatic Cholangiocarcinoma | 1 | CTD_human |