| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:KRT17-CUL3 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: KRT17-CUL3 | FusionPDB ID: 43530 | FusionGDB2.0 ID: 43530 | Hgene | Tgene | Gene symbol | KRT17 | CUL3 | Gene ID | 3872 | 8452 |
| Gene name | keratin 17 | cullin 3 | |
| Synonyms | 39.1|CK-17|K17|PC|PC2|PCHC1 | CUL-3|PHA2E | |
| Cytomap | 17q21.2 | 2q36.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | keratin, type I cytoskeletal 17cytokeratin-17keratin 17, type I | cullin-3 | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | Q04695 | Q13618 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000311208, | ENST00000432260, ENST00000264414, ENST00000344951, ENST00000409096, ENST00000409777, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 15 X 17 X 5=1275 | 8 X 8 X 5=320 |
| # samples | 18 | 9 | |
| ** MAII score | log2(18/1275*10)=-2.82442843541655 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/320*10)=-1.83007499855769 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: KRT17 [Title/Abstract] AND CUL3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | KRT17(39777218)-CUL3(225346688), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | KRT17-CUL3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KRT17-CUL3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KRT17-CUL3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. KRT17-CUL3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CUL3 | GO:0000209 | protein polyubiquitination | 19261606 |
| Tgene | CUL3 | GO:0006511 | ubiquitin-dependent protein catabolic process | 25401743|27561354 |
| Tgene | CUL3 | GO:0006513 | protein monoubiquitination | 22358839 |
| Tgene | CUL3 | GO:0006888 | ER to Golgi vesicle-mediated transport | 22358839 |
| Tgene | CUL3 | GO:0016567 | protein ubiquitination | 17543862|19782033|19995937|20389280|23213400 |
| Tgene | CUL3 | GO:0031145 | anaphase-promoting complex-dependent catabolic process | 10500095 |
| Tgene | CUL3 | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | 19261606|19782033|20389280 |
| Tgene | CUL3 | GO:0071630 | nuclear protein quality control by the ubiquitin-proteasome system | 27561354 |
 Fusion gene breakpoints across KRT17 (5'-gene) Fusion gene breakpoints across KRT17 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
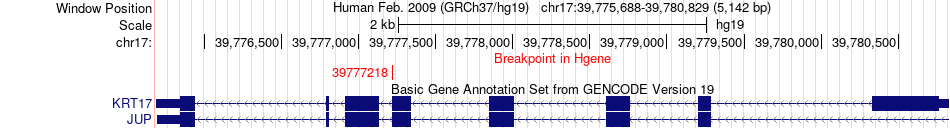 |
 Fusion gene breakpoints across CUL3 (3'-gene) Fusion gene breakpoints across CUL3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
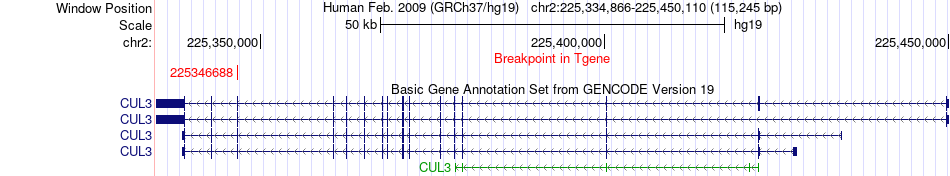 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | BE717377 | KRT17 | chr17 | 39777218 | - | CUL3 | chr2 | 225346688 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000311208 | KRT17 | chr17 | 39777218 | - | ENST00000264414 | CUL3 | chr2 | 225346688 | - | 5481 | 1028 | 47 | 1159 | 370 |
| ENST00000311208 | KRT17 | chr17 | 39777218 | - | ENST00000344951 | CUL3 | chr2 | 225346688 | - | 5479 | 1028 | 47 | 1159 | 370 |
| ENST00000311208 | KRT17 | chr17 | 39777218 | - | ENST00000409096 | CUL3 | chr2 | 225346688 | - | 1709 | 1028 | 47 | 1159 | 370 |
| ENST00000311208 | KRT17 | chr17 | 39777218 | - | ENST00000409777 | CUL3 | chr2 | 225346688 | - | 1692 | 1028 | 47 | 1159 | 370 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000311208 | ENST00000264414 | KRT17 | chr17 | 39777218 | - | CUL3 | chr2 | 225346688 | - | 0.000185253 | 0.9998148 |
| ENST00000311208 | ENST00000344951 | KRT17 | chr17 | 39777218 | - | CUL3 | chr2 | 225346688 | - | 0.000184974 | 0.99981505 |
| ENST00000311208 | ENST00000409096 | KRT17 | chr17 | 39777218 | - | CUL3 | chr2 | 225346688 | - | 0.003448487 | 0.9965515 |
| ENST00000311208 | ENST00000409777 | KRT17 | chr17 | 39777218 | - | CUL3 | chr2 | 225346688 | - | 0.003238743 | 0.99676126 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >43530_43530_1_KRT17-CUL3_KRT17_chr17_39777218_ENST00000311208_CUL3_chr2_225346688_ENST00000264414_length(amino acids)=370AA_BP= MPPAAATMTTSIRQFTSSSSIKGSSGLGGGSSRTSCRLSGGLGAGSCRLGSAGGLGSTLGGSSYSSCYSFGSGGGYGSSFGGVDGLLAGG EKATMQNLNDRLASYLDKVRALEEANTELEVKIRDWYQRQAPGPARDYSQYYRTIEELQNKILTATVDNANILLQIDNARLAADDFRTKF ETEQALRLSVEADINGLRRVLDELTLARADLEMQIENLKEELAYLKKNHEEEMNALRGQVGGEINVEMDAAPGVDLSRILNEMRDQYEKM AEKNRKDAEDWFFSKTEELNREVATNSELVQSGKSEISELRRTMQALEIELQSQLSMVTQQLKARFLPSPVVIKKRIEGLIEREYLARTP -------------------------------------------------------------- >43530_43530_2_KRT17-CUL3_KRT17_chr17_39777218_ENST00000311208_CUL3_chr2_225346688_ENST00000344951_length(amino acids)=370AA_BP= MPPAAATMTTSIRQFTSSSSIKGSSGLGGGSSRTSCRLSGGLGAGSCRLGSAGGLGSTLGGSSYSSCYSFGSGGGYGSSFGGVDGLLAGG EKATMQNLNDRLASYLDKVRALEEANTELEVKIRDWYQRQAPGPARDYSQYYRTIEELQNKILTATVDNANILLQIDNARLAADDFRTKF ETEQALRLSVEADINGLRRVLDELTLARADLEMQIENLKEELAYLKKNHEEEMNALRGQVGGEINVEMDAAPGVDLSRILNEMRDQYEKM AEKNRKDAEDWFFSKTEELNREVATNSELVQSGKSEISELRRTMQALEIELQSQLSMVTQQLKARFLPSPVVIKKRIEGLIEREYLARTP -------------------------------------------------------------- >43530_43530_3_KRT17-CUL3_KRT17_chr17_39777218_ENST00000311208_CUL3_chr2_225346688_ENST00000409096_length(amino acids)=370AA_BP= MPPAAATMTTSIRQFTSSSSIKGSSGLGGGSSRTSCRLSGGLGAGSCRLGSAGGLGSTLGGSSYSSCYSFGSGGGYGSSFGGVDGLLAGG EKATMQNLNDRLASYLDKVRALEEANTELEVKIRDWYQRQAPGPARDYSQYYRTIEELQNKILTATVDNANILLQIDNARLAADDFRTKF ETEQALRLSVEADINGLRRVLDELTLARADLEMQIENLKEELAYLKKNHEEEMNALRGQVGGEINVEMDAAPGVDLSRILNEMRDQYEKM AEKNRKDAEDWFFSKTEELNREVATNSELVQSGKSEISELRRTMQALEIELQSQLSMVTQQLKARFLPSPVVIKKRIEGLIEREYLARTP -------------------------------------------------------------- >43530_43530_4_KRT17-CUL3_KRT17_chr17_39777218_ENST00000311208_CUL3_chr2_225346688_ENST00000409777_length(amino acids)=370AA_BP= MPPAAATMTTSIRQFTSSSSIKGSSGLGGGSSRTSCRLSGGLGAGSCRLGSAGGLGSTLGGSSYSSCYSFGSGGGYGSSFGGVDGLLAGG EKATMQNLNDRLASYLDKVRALEEANTELEVKIRDWYQRQAPGPARDYSQYYRTIEELQNKILTATVDNANILLQIDNARLAADDFRTKF ETEQALRLSVEADINGLRRVLDELTLARADLEMQIENLKEELAYLKKNHEEEMNALRGQVGGEINVEMDAAPGVDLSRILNEMRDQYEKM AEKNRKDAEDWFFSKTEELNREVATNSELVQSGKSEISELRRTMQALEIELQSQLSMVTQQLKARFLPSPVVIKKRIEGLIEREYLARTP -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:39777218/chr2:225346688) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| KRT17 | CUL3 |
| FUNCTION: Type I keratin involved in the formation and maintenance of various skin appendages, specifically in determining shape and orientation of hair (By similarity). Required for the correct growth of hair follicles, in particular for the persistence of the anagen (growth) state (By similarity). Modulates the function of TNF-alpha in the specific context of hair cycling. Regulates protein synthesis and epithelial cell growth through binding to the adapter protein SFN and by stimulating Akt/mTOR pathway (By similarity). Involved in tissue repair. May be a marker of basal cell differentiation in complex epithelia and therefore indicative of a certain type of epithelial 'stem cells'. Acts as a promoter of epithelial proliferation by acting a regulator of immune response in skin: promotes Th1/Th17-dominated immune environment contributing to the development of basaloid skin tumors (By similarity). May act as an autoantigen in the immunopathogenesis of psoriasis, with certain peptide regions being a major target for autoreactive T-cells and hence causing their proliferation. {ECO:0000250|UniProtKB:Q9QWL7, ECO:0000269|PubMed:10844551, ECO:0000269|PubMed:15795121, ECO:0000269|PubMed:16713453}. | FUNCTION: Core component of multiple cullin-RING-based BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complexes which mediate the ubiquitination and subsequent proteasomal degradation of target proteins. BCR complexes and ARIH1 collaborate in tandem to mediate ubiquitination of target proteins (PubMed:27565346). As a scaffold protein may contribute to catalysis through positioning of the substrate and the ubiquitin-conjugating enzyme. The E3 ubiquitin-protein ligase activity of the complex is dependent on the neddylation of the cullin subunit and is inhibited by the association of the deneddylated cullin subunit with TIP120A/CAND1. The functional specificity of the BCR complex depends on the BTB domain-containing protein as the substrate recognition component. BCR(KLHL42) is involved in ubiquitination of KATNA1. BCR(SPOP) is involved in ubiquitination of BMI1/PCGF4, BRMS1, MACROH2A1 and DAXX, GLI2 and GLI3. Can also form a cullin-RING-based BCR (BTB-CUL3-RBX1) E3 ubiquitin-protein ligase complex containing homodimeric SPOPL or the heterodimer formed by SPOP and SPOPL; these complexes have lower ubiquitin ligase activity. BCR(KLHL9-KLHL13) controls the dynamic behavior of AURKB on mitotic chromosomes and thereby coordinates faithful mitotic progression and completion of cytokinesis. BCR(KLHL12) is involved in ER-Golgi transport by regulating the size of COPII coats, thereby playing a key role in collagen export, which is required for embryonic stem (ES) cells division: BCR(KLHL12) acts by mediating monoubiquitination of SEC31 (SEC31A or SEC31B) (PubMed:22358839, PubMed:27716508). BCR(KLHL3) acts as a regulator of ion transport in the distal nephron; by mediating ubiquitination of WNK4 (PubMed:23387299, PubMed:23453970, PubMed:23576762). The BCR(KLHL20) E3 ubiquitin ligase complex is involved in interferon response and anterograde Golgi to endosome transport: it mediates both ubiquitination leading to degradation and 'Lys-33'-linked ubiquitination (PubMed:20389280, PubMed:21840486, PubMed:21670212, PubMed:24768539). The BCR(KLHL21) E3 ubiquitin ligase complex regulates localization of the chromosomal passenger complex (CPC) from chromosomes to the spindle midzone in anaphase and mediates the ubiquitination of AURKB (PubMed:19995937). The BCR(KLHL22) ubiquitin ligase complex mediates monoubiquitination of PLK1, leading to PLK1 dissociation from phosphoreceptor proteins and subsequent removal from kinetochores, allowing silencing of the spindle assembly checkpoint (SAC) and chromosome segregation (PubMed:23455478). The BCR(KLHL22) ubiquitin ligase complex is also responsible for the amino acid-stimulated 'Lys-48' polyubiquitination and proteasomal degradation of DEPDC5. Through the degradation of DEPDC5, releases the GATOR1 complex-mediated inhibition of the TORC1 pathway (PubMed:29769719). The BCR(KLHL25) ubiquitin ligase complex is involved in translational homeostasis by mediating ubiquitination and subsequent degradation of hypophosphorylated EIF4EBP1 (4E-BP1) (PubMed:22578813). The BCR(KBTBD8) complex acts by mediating monoubiquitination of NOLC1 and TCOF1, leading to remodel the translational program of differentiating cells in favor of neural crest specification (PubMed:26399832). Involved in ubiquitination of cyclin E and of cyclin D1 (in vitro) thus involved in regulation of G1/S transition. Involved in the ubiquitination of KEAP1, ENC1 and KLHL41 (PubMed:15983046). In concert with ATF2 and RBX1, promotes degradation of KAT5 thereby attenuating its ability to acetylate and activate ATM. The BCR(KCTD17) E3 ubiquitin ligase complex mediates ubiquitination and degradation of TCHP, a down-regulator of cilium assembly, thereby inducing ciliogenesis (PubMed:25270598). The BCR(KLHL24) E3 ubiquitin ligase complex mediates ubiquitination of KRT14, controls KRT14 levels during keratinocytes differentiation, and is essential for skin integrity (PubMed:27798626). The BCR(KLHL18) E3 ubiquitin ligase complex mediates the ubiquitination of AURKA leading to its activation at the centrosome which is required for initiating mitotic entry (PubMed:23213400). The BCR(KEAP1) E3 ubiquitin ligase complex acts as a key sensor of oxidative and electrophilic stress by mediating ubiquitination and degradation of NFE2L2/NRF2, a transcription factor regulating expression of many cytoprotective genes (PubMed:15601839, PubMed:16006525). {ECO:0000269|PubMed:10500095, ECO:0000269|PubMed:11311237, ECO:0000269|PubMed:15601839, ECO:0000269|PubMed:15897469, ECO:0000269|PubMed:15983046, ECO:0000269|PubMed:16006525, ECO:0000269|PubMed:16524876, ECO:0000269|PubMed:17543862, ECO:0000269|PubMed:18397884, ECO:0000269|PubMed:19261606, ECO:0000269|PubMed:19995937, ECO:0000269|PubMed:20389280, ECO:0000269|PubMed:21670212, ECO:0000269|PubMed:21840486, ECO:0000269|PubMed:22085717, ECO:0000269|PubMed:22358839, ECO:0000269|PubMed:22578813, ECO:0000269|PubMed:22632832, ECO:0000269|PubMed:23213400, ECO:0000269|PubMed:23387299, ECO:0000269|PubMed:23453970, ECO:0000269|PubMed:23455478, ECO:0000269|PubMed:23576762, ECO:0000269|PubMed:24768539, ECO:0000269|PubMed:25270598, ECO:0000269|PubMed:26399832, ECO:0000269|PubMed:27565346, ECO:0000269|PubMed:27716508, ECO:0000269|PubMed:27798626, ECO:0000269|PubMed:29769719}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 102_116 | 320.0 | 433.0 | Region | Note=Peptide epitope S1%3B induces T-cell and keratinocyte proliferation and IFN-gamma production |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 121_138 | 320.0 | 433.0 | Region | Note=Linker 1 |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 139_230 | 320.0 | 433.0 | Region | Note=Coil 1B |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 153_167 | 320.0 | 433.0 | Region | Note=Peptide epitope S2%3B induces T-cell proliferation and IFN-gamma production |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 1_83 | 320.0 | 433.0 | Region | Note=Head |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 231_250 | 320.0 | 433.0 | Region | Note=Linker 12 |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 84_120 | 320.0 | 433.0 | Region | Note=Coil 1A |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 84_395 | 320.0 | 433.0 | Domain | IF rod |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 251_392 | 320.0 | 433.0 | Region | Note=Coil 2 |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 332_346 | 320.0 | 433.0 | Region | Note=Peptide epitope S4%3B induces T-cell and keratinocyte proliferation and IFN-gamma production |
| Hgene | KRT17 | chr17:39777218 | chr2:225346688 | ENST00000311208 | - | 5 | 8 | 393_432 | 320.0 | 433.0 | Region | Note=Tail |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| KRT17 | |
| CUL3 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to KRT17-CUL3 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to KRT17-CUL3 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

