| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:LASP1-KMT2A |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: LASP1-KMT2A | FusionPDB ID: 44183 | FusionGDB2.0 ID: 44183 | Hgene | Tgene | Gene symbol | LASP1 | KMT2A | Gene ID | 3927 | 4297 |
| Gene name | LIM and SH3 protein 1 | lysine methyltransferase 2A | |
| Synonyms | Lasp-1|MLN50 | ALL-1|CXXC7|HRX|HTRX1|MLL|MLL1|MLL1A|TRX1|WDSTS | |
| Cytomap | 17q12 | 11q23.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | LIM and SH3 domain protein 1metastatic lymph node gene 50 protein | histone-lysine N-methyltransferase 2ACXXC-type zinc finger protein 7lysine (K)-specific methyltransferase 2Alysine N-methyltransferase 2Amixed lineage leukemia 1myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)trithorax-like | |
| Modification date | 20200313 | 20200319 | |
| UniProtAcc | Q14847 | Q03164 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000318008, ENST00000433206, ENST00000435347, | ENST00000354520, ENST00000389506, ENST00000420751, ENST00000534358, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 18 X 17 X 8=2448 | 31 X 72 X 3=6696 |
| # samples | 21 | 79 | |
| ** MAII score | log2(21/2448*10)=-3.54314232502653 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(79/6696*10)=-3.08337496948588 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: LASP1 [Title/Abstract] AND KMT2A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | LASP1-KMT2A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. LASP1-KMT2A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. LASP1-KMT2A seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor by not retaining the major functional domain in the partially deleted in-frame ORF. LASP1-KMT2A seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor by not retaining the major functional domain in the partially deleted in-frame ORF. LASP1-KMT2A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. LASP1-KMT2A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. LASP1-KMT2A seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target by not retaining the major functional domain in the partially deleted in-frame ORF. LASP1-KMT2A seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target by not retaining the major functional domain in the partially deleted in-frame ORF. LASP1-KMT2A seems lost the major protein functional domain in Hgene partner, which is a transcription factor by not retaining the major functional domain in the partially deleted in-frame ORF. LASP1-KMT2A seems lost the major protein functional domain in Hgene partner, which is a transcription factor by not retaining the major functional domain in the partially deleted in-frame ORF. KMT2A-LASP1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. KMT2A-LASP1 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. KMT2A-LASP1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. KMT2A-LASP1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. KMT2A-LASP1 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. KMT2A-LASP1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | KMT2A | GO:0044648 | histone H3-K4 dimethylation | 25561738 |
| Tgene | KMT2A | GO:0045944 | positive regulation of transcription by RNA polymerase II | 20861184 |
| Tgene | KMT2A | GO:0051568 | histone H3-K4 methylation | 19556245 |
| Tgene | KMT2A | GO:0065003 | protein-containing complex assembly | 15199122 |
| Tgene | KMT2A | GO:0080182 | histone H3-K4 trimethylation | 20861184 |
| Tgene | KMT2A | GO:0097692 | histone H3-K4 monomethylation | 25561738|26324722 |
 Fusion gene breakpoints across LASP1 (5'-gene) Fusion gene breakpoints across LASP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across KMT2A (3'-gene) Fusion gene breakpoints across KMT2A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | LASP1 | chr17 | 37071399 | + | KMT2A | chr11 | 118355576 | |
| ChimerKB3 | . | . | LASP1 | chr17 | 37074857 | + | KMT2A | chr11 | 118359328 | + |
| ChimerKB3 | . | . | LASP1 | chr17 | 37078023 | + | KMT2A | chr11 | 118355690 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000318008 | LASP1 | chr17 | 37071399 | + | ENST00000534358 | KMT2A | chr11 | 118355576 | 13296 | 943 | 331 | 8643 | 2770 | |
| ENST00000318008 | LASP1 | chr17 | 37071399 | + | ENST00000389506 | KMT2A | chr11 | 118355576 | 10380 | 943 | 331 | 8634 | 2767 | |
| ENST00000433206 | LASP1 | chr17 | 37071399 | + | ENST00000534358 | KMT2A | chr11 | 118355576 | 13039 | 686 | 242 | 8386 | 2714 | |
| ENST00000433206 | LASP1 | chr17 | 37071399 | + | ENST00000389506 | KMT2A | chr11 | 118355576 | 10123 | 686 | 242 | 8377 | 2711 | |
| ENST00000435347 | LASP1 | chr17 | 37071399 | + | ENST00000534358 | KMT2A | chr11 | 118355576 | 13012 | 659 | 47 | 8359 | 2770 | |
| ENST00000435347 | LASP1 | chr17 | 37071399 | + | ENST00000389506 | KMT2A | chr11 | 118355576 | 10096 | 659 | 47 | 8350 | 2767 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >44183_44183_1_LASP1-KMT2A_LASP1_chr17_37071399_ENST00000318008_KMT2A_chr11_118355576_ENST00000389506_length(amino acids)=2767AA_BP=204 MNPNCARCGKIVYPTEKVNCLDKFWHKACFHCETCKMTLNMKNYKGYEKKPYCNAHYPKQSFTMVADTPENLRLKQQSELQSQVRYKEEF EKNKGKGFSVVADTPELQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDSQDGSSYRRPLEQQQPHHIPTSAPVYQQPQQQPVA QSYGGYKEPAAPVSIQRSAPGGGGEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLE NWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKL FAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALL NSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDG GQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGP GEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVH MAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLN GLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSF TESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGS RRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDG SSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHL RGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKP EFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIES TSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGE EDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTA TTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLD TYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVS SSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDAD HISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQ TLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPS GLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLG SLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHV TLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQL QHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKR FQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEP KTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGI LHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSM DLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAA -------------------------------------------------------------- >44183_44183_2_LASP1-KMT2A_LASP1_chr17_37071399_ENST00000318008_KMT2A_chr11_118355576_ENST00000534358_length(amino acids)=2770AA_BP=204 MNPNCARCGKIVYPTEKVNCLDKFWHKACFHCETCKMTLNMKNYKGYEKKPYCNAHYPKQSFTMVADTPENLRLKQQSELQSQVRYKEEF EKNKGKGFSVVADTPELQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDSQDGSSYRRPLEQQQPHHIPTSAPVYQQPQQQPVA QSYGGYKEPAAPVSIQRSAPGGGGEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLE NWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKL FAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLT ALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAIN SDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKP KGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLK NVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRK FLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSP TSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRS IGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSH LDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPH LHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGN LKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQ IESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRS YGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTS VTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKN LLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDS SVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHM DADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLY VLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISN PPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLL DLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVP GHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEH YQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPK TKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQES AEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRM LGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRA TSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHG -------------------------------------------------------------- >44183_44183_3_LASP1-KMT2A_LASP1_chr17_37071399_ENST00000433206_KMT2A_chr11_118355576_ENST00000389506_length(amino acids)=2711AA_BP=148 MLPLRDLQDDTEHEELQGLREEALLQRVRYKEEFEKNKGKGFSVVADTPELQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDS QDGSSYRRPLEQQQPHHIPTSAPVYQQPQQQPVAQSYGGYKEPAAPVSIQRSAPGGGGEDCEAENVWEMGGLGILTSVPITPRVVCFLCA SSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICT KCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVA YTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDL EGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSL DHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAG RLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHR DLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYT CKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPG CRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLG PLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNT TSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQK GKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKR TVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQE DGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLA SHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSR VKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLG EGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVA SSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQ ISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPS ASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSS TPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTS GSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTA AITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQAS PTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQ SSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIED AWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMF NFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQT DKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCN -------------------------------------------------------------- >44183_44183_4_LASP1-KMT2A_LASP1_chr17_37071399_ENST00000433206_KMT2A_chr11_118355576_ENST00000534358_length(amino acids)=2714AA_BP=148 MLPLRDLQDDTEHEELQGLREEALLQRVRYKEEFEKNKGKGFSVVADTPELQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDS QDGSSYRRPLEQQQPHHIPTSAPVYQQPQQQPVAQSYGGYKEPAAPVSIQRSAPGGGGEDCEAENVWEMGGLGILTSVPITPRVVCFLCA SSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICT KCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPE SVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQP LDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLP PSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSAN DAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQ RHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRC VYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQR SPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDH VLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQV HNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDR RQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAP RKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPK PQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEE RLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHS VSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELL NLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEK SVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPG PSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSL FPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLA PSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSH LTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTP STAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAV QASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTAS VEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAES IEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAF DMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRS IQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKL -------------------------------------------------------------- >44183_44183_5_LASP1-KMT2A_LASP1_chr17_37071399_ENST00000435347_KMT2A_chr11_118355576_ENST00000389506_length(amino acids)=2767AA_BP=204 MNPNCARCGKIVYPTEKVNCLDKFWHKACFHCETCKMTLNMKNYKGYEKKPYCNAHYPKQSFTMVADTPENLRLKQQSELQSQVRYKEEF EKNKGKGFSVVADTPELQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDSQDGSSYRRPLEQQQPHHIPTSAPVYQQPQQQPVA QSYGGYKEPAAPVSIQRSAPGGGGEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLE NWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKL FAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALL NSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDG GQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGP GEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVH MAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLN GLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSF TESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGS RRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDG SSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHL RGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKP EFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIES TSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGE EDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTA TTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLD TYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVS SSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDAD HISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQ TLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPS GLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLG SLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHV TLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQL QHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKR FQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEP KTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGI LHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSM DLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAA -------------------------------------------------------------- >44183_44183_6_LASP1-KMT2A_LASP1_chr17_37071399_ENST00000435347_KMT2A_chr11_118355576_ENST00000534358_length(amino acids)=2770AA_BP=204 MNPNCARCGKIVYPTEKVNCLDKFWHKACFHCETCKMTLNMKNYKGYEKKPYCNAHYPKQSFTMVADTPENLRLKQQSELQSQVRYKEEF EKNKGKGFSVVADTPELQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDSQDGSSYRRPLEQQQPHHIPTSAPVYQQPQQQPVA QSYGGYKEPAAPVSIQRSAPGGGGEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKFCLEENERPLEDQLE NWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKL FAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLT ALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAIN SDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKP KGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLK NVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRK FLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSP TSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRS IGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSH LDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPH LHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGN LKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQ IESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRS YGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTS VTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKN LLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDS SVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHM DADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLY VLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISN PPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLL DLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVP GHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEH YQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPK TKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQES AEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRM LGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRA TSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHG -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:/chr11:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LASP1 | KMT2A |
| FUNCTION: Plays an important role in the regulation of dynamic actin-based, cytoskeletal activities. Agonist-dependent changes in LASP1 phosphorylation may also serve to regulate actin-associated ion transport activities, not only in the parietal cell but also in certain other F-actin-rich secretory epithelial cell types (By similarity). {ECO:0000250}. | FUNCTION: Histone methyltransferase that plays an essential role in early development and hematopoiesis (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalytic subunit of the MLL1/MLL complex, a multiprotein complex that mediates both methylation of 'Lys-4' of histone H3 (H3K4me) complex and acetylation of 'Lys-16' of histone H4 (H4K16ac) (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:24235145, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalyzes methyl group transfer from S-adenosyl-L-methionine to the epsilon-amino group of 'Lys-4' of histone H3 (H3K4) via a non-processive mechanism. Part of chromatin remodeling machinery predominantly forms H3K4me1 and H3K4me2 methylation marks at active chromatin sites where transcription and DNA repair take place (PubMed:25561738, PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Has weak methyltransferase activity by itself, and requires other component of the MLL1/MLL complex to obtain full methyltransferase activity (PubMed:19187761, PubMed:26886794). Has no activity toward histone H3 phosphorylated on 'Thr-3', less activity toward H3 dimethylated on 'Arg-8' or 'Lys-9', while it has higher activity toward H3 acetylated on 'Lys-9' (PubMed:19187761). Binds to unmethylated CpG elements in the promoter of target genes and helps maintain them in the nonmethylated state (PubMed:20010842). Required for transcriptional activation of HOXA9 (PubMed:12453419, PubMed:20677832, PubMed:20010842). Promotes PPP1R15A-induced apoptosis (PubMed:10490642). Plays a critical role in the control of circadian gene expression and is essential for the transcriptional activation mediated by the CLOCK-ARNTL/BMAL1 heterodimer (By similarity). Establishes a permissive chromatin state for circadian transcription by mediating a rhythmic methylation of 'Lys-4' of histone H3 (H3K4me) and this histone modification directs the circadian acetylation at H3K9 and H3K14 allowing the recruitment of CLOCK-ARNTL/BMAL1 to chromatin (By similarity). Also has auto-methylation activity on Cys-3882 in absence of histone H3 substrate (PubMed:24235145). {ECO:0000250|UniProtKB:P55200, ECO:0000269|PubMed:10490642, ECO:0000269|PubMed:12453419, ECO:0000269|PubMed:15960975, ECO:0000269|PubMed:19187761, ECO:0000269|PubMed:19556245, ECO:0000269|PubMed:20010842, ECO:0000269|PubMed:21220120, ECO:0000269|PubMed:24235145, ECO:0000269|PubMed:26886794, ECO:0000305|PubMed:20677832}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000318008 | 6 | 7 | 116_151 | 204.0 | 262.0 | Compositional bias | Basic and acidic residues | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000318008 | 6 | 7 | 152_182 | 204.0 | 262.0 | Compositional bias | Polar residues | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000435347 | 6 | 8 | 116_151 | 204.0 | 708.3333333333334 | Compositional bias | Basic and acidic residues | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000435347 | 6 | 8 | 152_182 | 204.0 | 708.3333333333334 | Compositional bias | Polar residues | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000318008 | 6 | 7 | 5_56 | 204.0 | 262.0 | Domain | LIM zinc-binding | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000435347 | 6 | 8 | 5_56 | 204.0 | 708.3333333333334 | Domain | LIM zinc-binding | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000318008 | 6 | 7 | 111_186 | 204.0 | 262.0 | Region | Disordered | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000435347 | 6 | 8 | 111_186 | 204.0 | 708.3333333333334 | Region | Disordered | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000318008 | 6 | 7 | 61_95 | 204.0 | 262.0 | Repeat | Note=Nebulin 1 | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000318008 | 6 | 7 | 97_131 | 204.0 | 262.0 | Repeat | Note=Nebulin 2 | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000435347 | 6 | 8 | 61_95 | 204.0 | 708.3333333333334 | Repeat | Note=Nebulin 1 | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000435347 | 6 | 8 | 97_131 | 204.0 | 708.3333333333334 | Repeat | Note=Nebulin 2 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1046_1061 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1249_1272 | 0.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1278_1302 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1303_1317 | 0.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1822_1847 | 0.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 184_203 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 204_226 | 0.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2096_2118 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2146_2174 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2189_2232 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2275_2320 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 236_253 | 0.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2402_2418 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2419_2445 | 0.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2526_2594 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2721_2746 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2747_2784 | 0.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2785_2821 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3011_3064 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3166_3183 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3195_3222 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3230_3244 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 325_351 | 0.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3464_3530 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3563_3606 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 37_58 | 0.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 445_495 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 535_556 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 557_576 | 0.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 713_731 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 747_780 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 798_845 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 81_103 | 0.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 846_898 | 0.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1822_1847 | 1406.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2096_2118 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2146_2174 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2189_2232 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2275_2320 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2402_2418 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2419_2445 | 1406.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2526_2594 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2721_2746 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2747_2784 | 1406.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2785_2821 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3011_3064 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3166_3183 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3195_3222 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3230_3244 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3464_3530 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3563_3606 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1822_1847 | 1406.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2096_2118 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2146_2174 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2189_2232 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2275_2320 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2402_2418 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2419_2445 | 1406.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2526_2594 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2721_2746 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2747_2784 | 1406.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2785_2821 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3011_3064 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3166_3183 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3195_3222 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3230_3244 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3464_3530 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3563_3606 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 169_180 | 0.0 | 3932.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 217_227 | 0.0 | 3932.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 301_309 | 0.0 | 3932.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1703_1748 | 0.0 | 3932.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2018_2074 | 0.0 | 3932.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3666_3747 | 0.0 | 3932.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3829_3945 | 0.0 | 3932.0 | Domain | SET | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3953_3969 | 0.0 | 3932.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1703_1748 | 1406.0 | 3970.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2018_2074 | 1406.0 | 3970.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3666_3747 | 1406.0 | 3970.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3829_3945 | 1406.0 | 3970.0 | Domain | SET | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3953_3969 | 1406.0 | 3970.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1703_1748 | 1406.0 | 3973.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2018_2074 | 1406.0 | 3973.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3666_3747 | 1406.0 | 3973.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3829_3945 | 1406.0 | 3973.0 | Domain | SET | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3953_3969 | 1406.0 | 3973.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 123_134 | 0.0 | 3932.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 147_152 | 0.0 | 3932.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 6_25 | 0.0 | 3932.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1038_1066 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1106_1166 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1200_1375 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 132_253 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1663_1713 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1806_1869 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1_108 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2081_2133 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2145_2232 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2275_2333 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2373_2460 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2475_2618 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2647_2675 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2713_2821 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 2961_3064 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 301_352 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3166_3244 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3464_3608 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3620_3643 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3785_3808 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 3906_3907 | 0.0 | 3932.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 445_585 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 713_780 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 798_949 | 0.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1663_1713 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1806_1869 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2081_2133 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2145_2232 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2275_2333 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2373_2460 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2475_2618 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2647_2675 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2713_2821 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 2961_3064 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3166_3244 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3464_3608 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3620_3643 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3785_3808 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 3906_3907 | 1406.0 | 3970.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1663_1713 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1806_1869 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2081_2133 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2145_2232 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2275_2333 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2373_2460 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2475_2618 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2647_2675 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2713_2821 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 2961_3064 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3166_3244 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3464_3608 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3620_3643 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3785_3808 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 3906_3907 | 1406.0 | 3973.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1147_1195 | 0.0 | 3932.0 | Zinc finger | CXXC-type | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1431_1482 | 0.0 | 3932.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1479_1533 | 0.0 | 3932.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1566_1627 | 0.0 | 3932.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1870_1910 | 0.0 | 3932.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000354520 | 0 | 35 | 1931_1978 | 0.0 | 3932.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1431_1482 | 1406.0 | 3970.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1479_1533 | 1406.0 | 3970.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1566_1627 | 1406.0 | 3970.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1870_1910 | 1406.0 | 3970.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1931_1978 | 1406.0 | 3970.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1431_1482 | 1406.0 | 3973.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1479_1533 | 1406.0 | 3973.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1566_1627 | 1406.0 | 3973.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1870_1910 | 1406.0 | 3973.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1931_1978 | 1406.0 | 3973.0 | Zinc finger | PHD-type 4 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000318008 | 6 | 7 | 202_261 | 204.0 | 262.0 | Domain | SH3 | |
| Hgene | LASP1 | chr17:37071399 | chr11:118355576 | ENST00000435347 | 6 | 8 | 202_261 | 204.0 | 708.3333333333334 | Domain | SH3 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1046_1061 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1249_1272 | 1406.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1278_1302 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1303_1317 | 1406.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 184_203 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 204_226 | 1406.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 236_253 | 1406.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 325_351 | 1406.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 37_58 | 1406.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 445_495 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 535_556 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 557_576 | 1406.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 713_731 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 747_780 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 798_845 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 81_103 | 1406.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 846_898 | 1406.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1046_1061 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1249_1272 | 1406.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1278_1302 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1303_1317 | 1406.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 184_203 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 204_226 | 1406.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 236_253 | 1406.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 325_351 | 1406.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 37_58 | 1406.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 445_495 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 535_556 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 557_576 | 1406.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 713_731 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 747_780 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 798_845 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 81_103 | 1406.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 846_898 | 1406.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 169_180 | 1406.0 | 3970.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 217_227 | 1406.0 | 3970.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 301_309 | 1406.0 | 3970.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 169_180 | 1406.0 | 3973.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 217_227 | 1406.0 | 3973.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 301_309 | 1406.0 | 3973.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 123_134 | 1406.0 | 3970.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 147_152 | 1406.0 | 3970.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 6_25 | 1406.0 | 3970.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 123_134 | 1406.0 | 3973.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 147_152 | 1406.0 | 3973.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 6_25 | 1406.0 | 3973.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1038_1066 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1106_1166 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1200_1375 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 132_253 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1_108 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 301_352 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 445_585 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 713_780 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 798_949 | 1406.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1038_1066 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1106_1166 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1200_1375 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 132_253 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1_108 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 301_352 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 445_585 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 713_780 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 798_949 | 1406.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000389506 | 8 | 36 | 1147_1195 | 1406.0 | 3970.0 | Zinc finger | CXXC-type | |
| Tgene | KMT2A | chr17:37071399 | chr11:118355576 | ENST00000534358 | 8 | 36 | 1147_1195 | 1406.0 | 3973.0 | Zinc finger | CXXC-type |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (1190) | LASP1 | chr17 | 37071399 | + | KMT2A | chr11 | 118355576 | MLPLRDLQDDTEHEELQGLREEALLQRVRYKEEFEKNKGKGFSVVADTPE LQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDSQDGSSYRRPL EQQQPHHIPTSAPVYQQPQQQPVAQSYGGYKEPAAPVSIQRSAPGGGGED CEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKF CLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPE CLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHD CAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYE ILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTS HLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDL EGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFF IRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQER EENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDS PELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSA EVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMC SRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRR KFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWST TDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSK ESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPG CRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLR IMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQ NTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKT KVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVS FSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMS NRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEG NLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGS AKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEP ISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQE DGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGK RSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQ CDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRP EDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSR RVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTP SMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVD SSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEK RVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPP CGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQ ISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSV SSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSH HQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLS TTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFT PSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGG TAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASV PGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQ LGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQL LASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQAS PTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVS HLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPA GQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKE SITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQ LSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPL NPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARR ATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEY AGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHS CEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCN | 2711 | |
| PDB file (1191) | LASP1 | chr17 | 37071399 | + | KMT2A | chr11 | 118355576 | MLPLRDLQDDTEHEELQGLREEALLQRVRYKEEFEKNKGKGFSVVADTPE LQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDSQDGSSYRRPL EQQQPHHIPTSAPVYQQPQQQPVAQSYGGYKEPAAPVSIQRSAPGGGGED CEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCCEPFHKF CLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPE CLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHD CAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDE MYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSR TTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQP LDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVK SFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQW QEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSR EDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCAL WSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYH FMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGIS LRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVY WSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTES SSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQR SPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRS KLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTS LGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKG EKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPS SVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLS GMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTG EEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQV EGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSP TEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPK PQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKK RGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFRE EEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKI DRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLE SSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLK NTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTE PVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDS GEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHIS SPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPG PSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLT SSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLP MSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRT DLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLI NFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQS VGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSS ASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGM FPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHV NQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAV QASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKH KVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECG QPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQK RKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNAR LKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANE PPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKS ARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMV IEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFI NHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKL | 2714 | |
| PDB file (1198) | LASP1 | chr17 | 37071399 | + | KMT2A | chr11 | 118355576 | MNPNCARCGKIVYPTEKVNCLDKFWHKACFHCETCKMTLNMKNYKGYEKK PYCNAHYPKQSFTMVADTPENLRLKQQSELQSQVRYKEEFEKNKGKGFSV VADTPELQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDSQDGS SYRRPLEQQQPHHIPTSAPVYQQPQQQPVAQSYGGYKEPAAPVSIQRSAP GGGGEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCC EPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCR NSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHD FSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENL SDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALL NSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDD QQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANS MVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNY AQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTD RSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVN CALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTS NYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFE GISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCS RVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSF TESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSP TQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSP QRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNS STSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSS LKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFA EPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTL KLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQV TTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPP MQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIES TSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPD GPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSST GKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNL FREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTEN LKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLS SLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDF VLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLP TTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVI SDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDAD HISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTD SPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKI QLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKG LLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESS QRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNM TLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLL PQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTP TSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPS SGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQL QHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALS SAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNG KKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQK ECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQ EQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARS NARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEE ANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQ LKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAG EMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAA RFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDAS | 2767 | |
| PDB file (1200) >>>1200.pdbFusion protein BP residue: 204 CIF file (1200) >>>1200.cif | LASP1 | chr17 | 37071399 | + | KMT2A | chr11 | 118355576 | MNPNCARCGKIVYPTEKVNCLDKFWHKACFHCETCKMTLNMKNYKGYEKK PYCNAHYPKQSFTMVADTPENLRLKQQSELQSQVRYKEEFEKNKGKGFSV VADTPELQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDSQDGS SYRRPLEQQQPHHIPTSAPVYQQPQQQPVAQSYGGYKEPAAPVSIQRSAP GGGGEDCEAENVWEMGGLGILTSVPITPRVVCFLCASSGHVEFVYCQVCC EPFHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCR NSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHD FSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENL SGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLT ALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSK QDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKK ANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLD HNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPIL STDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWT HVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTS CTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFV DFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGY QCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSP TSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPS YSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSS LSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGP LNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSK SSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIG SFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEV KTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEP GQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPK APPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQ IESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLM LPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYS SSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLAS HNLFREEEQCDLPKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENG TENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEA QLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDI MDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQ QLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRL AVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHM DADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPN STDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVT QKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSA SKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVS ESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVV SNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPA PLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTT TTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVAL PPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEH YQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNK ALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDK GNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQS SQKECGQPAGQVAVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLW LQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQE ARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHK PEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEE EVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNI DAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHG NAARFINHSCEPNCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIE | 2770 | |
| 3D view using mol* of 1200 (AA BP:204) | ||||||||||
 | ||||||||||
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
LASP1_pLDDT.png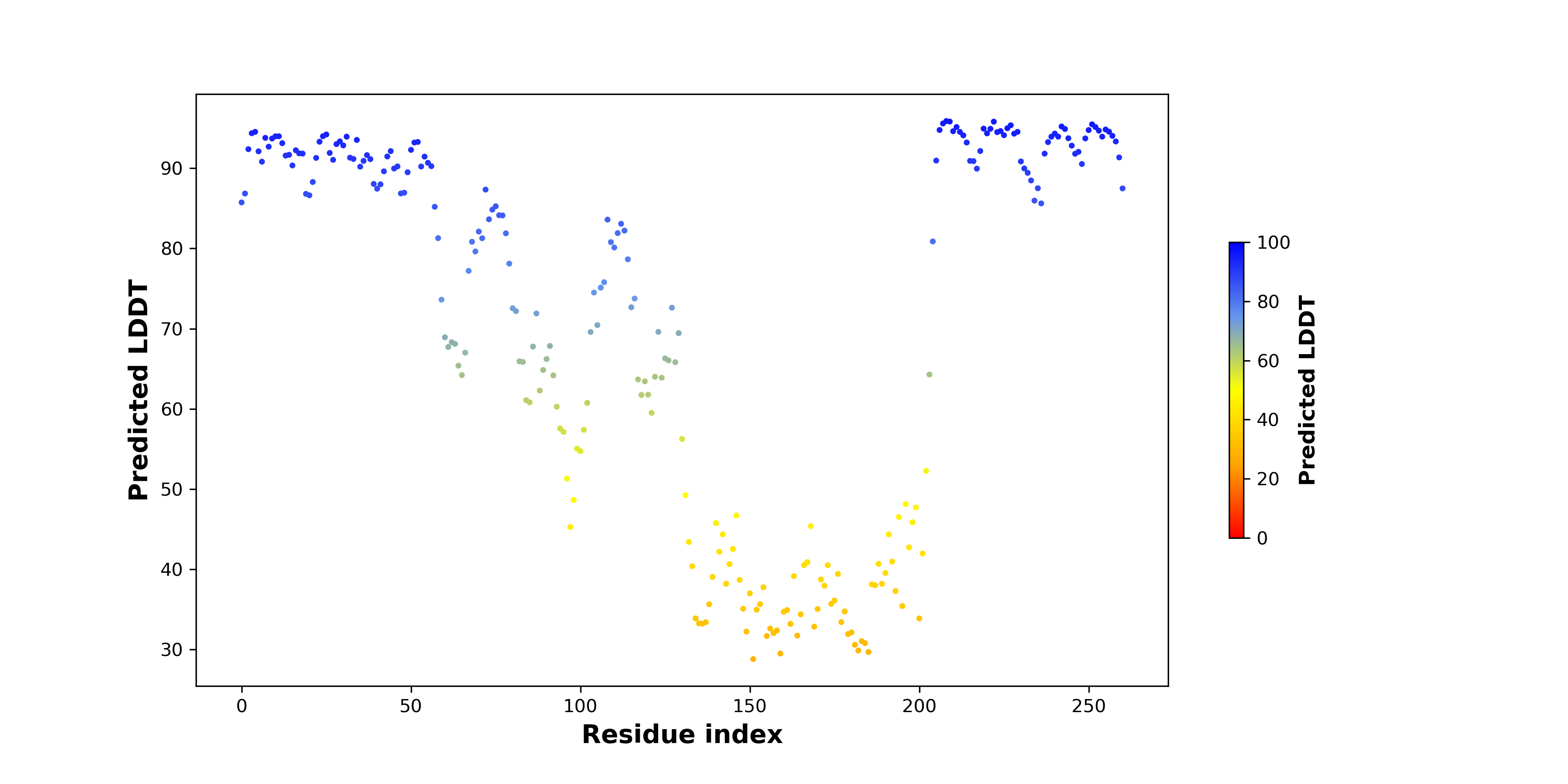 |
KMT2A_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
LASP1_KMT2A_1200_pLDDT.png (AA BP:204)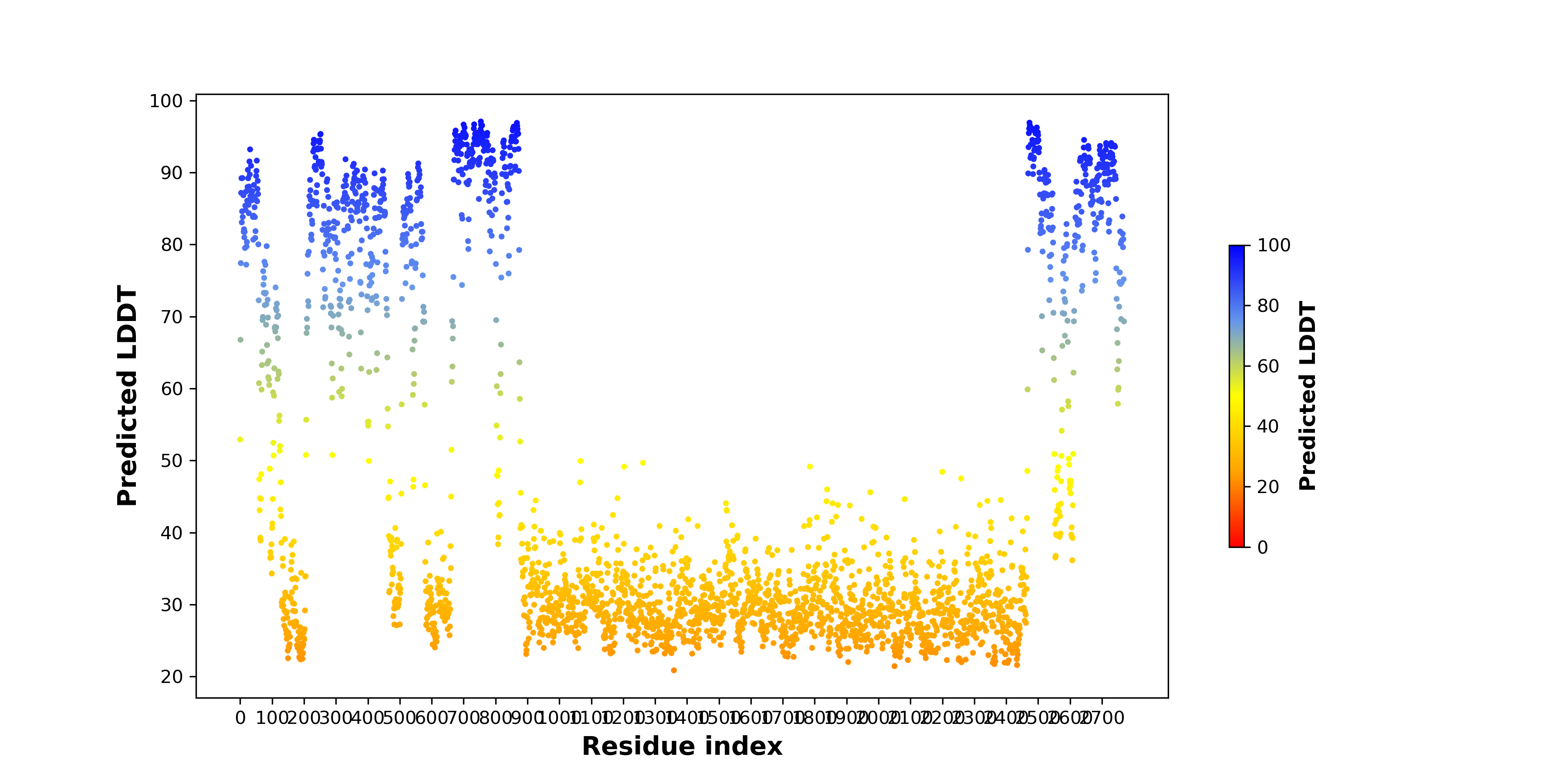 |
LASP1_KMT2A_1200_pLDDT_and_active_sites.png (AA BP:204)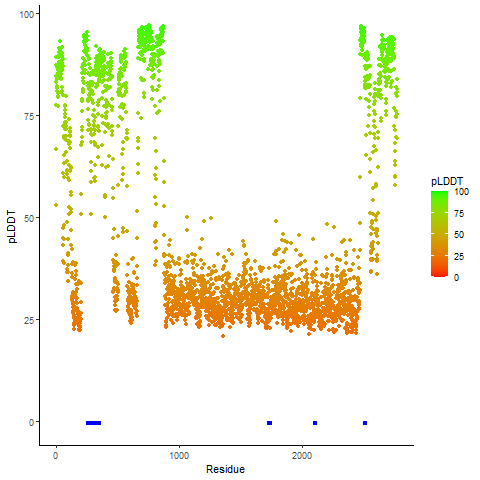 |
LASP1_KMT2A_1200_violinplot.png (AA BP:204)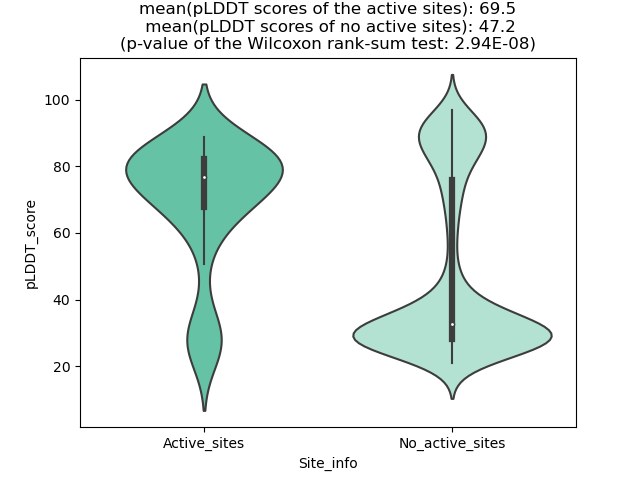 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| LASP1_KMT2A_1200.png |
 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 1200 | 1.0051 | 416 | 1.0481 | 1214.906 | 0.6539 | 0.6603 | 0.8209 | 0.4865 | 0.8837 | 0.5505 | 1.0979 | Chain A: 259,261,262,263,264,265,268,271,272,273,2 75,276,278,279,281,287,290,291,292,293,294,295,297 ,298,299,300,301,302,305,306,308,309,310,311,312,3 13,314,315,316,317,318,319,321,322,323,326,329,337 ,338,339,340,342,343,344,345,346,347,348,349,1734, 1735,1736,1737,1738,1739,1740,2101,2102,2103,2104, 2508,2509,2510,2511 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| KMT2A | PPIE, PPP1R15A, KMT2A, ASH2L, HCFC1, HCFC2, MEN1, RBBP5, WDR5, AVP, INS, OXT, MAP3K5, HDAC1, CTBP1, CBX4, BMI1, CREBBP, SMARCB1, CXXC1, MYB, CTNNB1, SNW1, E2F2, E2F4, E2F6, PSIP1, MLLT4, POLR2A, KAT8, RNF2, TP53, tat, SBF1, MTM1, SET, HIST1H3A, HIST1H4A, KAT6A, ELL, AFF1, AFF4, CDK9, CCNT1, CTR9, LEO1, PAF1, CDC73, WDR61, MLLT3, DOT1L, SKP2, HIST3H3, SVIL, HIST2H3C, SIN3A, MLLT1, RUNX1, CBFB, H3F3A, SIRT7, ASB2, TCEB1, TCEB2, CBX8, TOP1, TAF6, NCL, HECW2, LGR4, CSNK2A2, SENP3, SYMPK, PKN1, PIH1D1, KRAS, TAF1, CHD3, SMARCA2, SMARCC2, SMARCC1, HDAC2, RBBP4, RBBP7, TBP, MBD3, SAP30, RAN, TAF9, TASP1, HIST1H2BG, EWSR1, DYNLT1, KIF11, ING4, Cbx1, Set, ZNF131, ASB7, AR, CSNK2A1, OGT, BRD4, KDM5B, KDM5D, KDM6B, CKS1B, CKS2, CHD4, ESR2, AGR2, EZH2, FBXW7, SOX2, MED17, WDR82, ACTR8, GEMIN5, RUVBL1, MCM2, RTF1, MED15, MED8, MRGBP, MED14, HIST1H2BB, HIST1H2AB, KIAA1429, SP1, MYC, NR2C2, IRF2, PLEKHA4, DPY30, PTEN, NPM1, RPL13, CCT6A, TMCO1, ITGB1, ESR1, MYCN, SUPT5H, CIT, Pik3r2, FASN, LDLR, CDC27, KMT2B, SETD1A, KDM6A, KANSL2, BUB3, SETD1B, KANSL1, RERE, MCRS1, KMT2C, ZZZ3, MBIP, YEATS2, PAXIP1, KANSL3, KMT2D, BOD1L1, CSRP2BP, ZNF608, ATN1, PHF20L1, PHF20, NCOA6, TADA3, APEX1, ASF1A, CBX3, CD3EAP, CENPA, NUP50, POLR1E, TERF2IP, ZNF330, NAA40, PRKRA, SRSF4, KRR1, RPL13A, ABT1, CSNK2B, OASL, SRSF5, SRSF6, RPS9, FGFBP1, RPLP0, RPL36, KAL1, RPL3, PML, ELF1, ELF2, FEV, NHLH1, EN1, KLF12, KLF3, NFIX, NFYC, YY1, BRD3, |
| LASP1 | TRIP13, PLSCR1, FHL3, MDFI, ZYX, Fancc, TERF1, TINF2, ACD, POT1, ATXN1, FN1, DAZAP2, Prkg1, Prkaca, CD81, GOLGA2, PSMA3, REL, TRIM27, TCF4, FXR2, SPRY2, ARHGEF15, ZC2HC1A, THAP1, SEPT3, LZTS2, RHOXF2, KRTAP4-2, ZBTB9, ZDHHC17, CCDC8, ILK, HAX1, DNAAF2, THAP8, FYTTD1, NFYA, OR10H3, RDH12, SLC25A32, SLC25A44, C11orf65, TAS2R41, AHNAK, ARFGAP1, DDB1, EIF4B, HSPB1, HSPE1, LGALS1, MAP4, NXF1, CRK, STAT3, SH2D2A, CEP170, FBF1, SCLT1, DCTN1, Prkcz, U2AF2, CDH1, ZNF764, SNRNP27, PGK1, EZR, SPTAN1, NCL, UHRF1, NAT10, PDIA5, API5, KRT18, KRT2, G3BP2, AP3M1, NCOR2, CDK12, MYO1C, ACTR3, CLTC, DNMT1, EHMT2, SNAI1, HIST1H3A, TRIM25, TES, PPP6C, EFTUD2, TNIP2, RBPMS, DPF2, COPS5, SFN, MYC, CDK9, CANX, ATG16L1, DCAF15, BICD1, BICD2, BMH1, BMH2, TRIM28, FAM168A, POLD1, DMRT3, POU1F1, ANKS1A, PITX1, CAMK2A, RBFOX1, CHERP, DTX2, NUTF2, VAC14, MED25, HOXA1, CRYBA1, TENC1, TFG, OTX1, GCM2, TEKT3, CPSF3L, ARHGEF16, BHLHE40, ZC3H10, VEZF1, RNF38, SPAG8, GUCD1, BAHD1, GAS8, LMO4, YTHDF1, FAM168B, TEKT5, BAG4, FOXH1, PRR35, PPP1R32, OXER1, TEKT4, CRYBA2, TLX3, RBPMS2, CERCAM, LENG8, MGAT5B, VPS37C, HNRNPF, EVX2, CATSPER1, YES1, UFSP1, CSTF2T, MKRN3, C14orf119, CTNNA3, HGS, WWOX, KRTAP19-6, KRTAP6-2, KRTAP19-4, KRTAP10-8, UBQLN2, C19orf54, NOXA1, ARID5A, CYSRT1, KRTAP26-1, QRICH1, PRR20B, PRR20A, DOK6, VGLL3, KRTAP3-3, KRTAP3-2, KRTAP12-4, PRR20D, PRR20C, PRR20E, KRTAP19-3, FBXO17, PLEKHA4, HCVgp1, PRNP, NUPR1, BRD4, USP15, RIN3, ACTB, PFN1, PRPH, RDX, TJP2, VASP, VIM, NAA40, FAM24B, RPL35A, HSPA1A, BTF3, EP300, STAT1, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| LASP1 |  |
| KMT2A |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to LASP1-KMT2A |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to LASP1-KMT2A |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | LASP1 | C0004352 | Autistic Disorder | 1 | CTD_human |
| Hgene | LASP1 | C0014170 | Endometrial Neoplasms | 1 | CTD_human |
| Hgene | LASP1 | C0036341 | Schizophrenia | 1 | PSYGENET |
| Hgene | LASP1 | C0476089 | Endometrial Carcinoma | 1 | CTD_human |
| Tgene | KMT2A | C2826025 | Mixed phenotype acute leukemia | 3 | ORPHANET |
| Tgene | KMT2A | C0023418 | leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023452 | Childhood Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023453 | L2 Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023466 | Leukemia, Monocytic, Chronic | 2 | CTD_human |
| Tgene | KMT2A | C0023467 | Leukemia, Myelocytic, Acute | 2 | CTD_human |
| Tgene | KMT2A | C0023470 | Myeloid Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0026998 | Acute Myeloid Leukemia, M1 | 2 | CTD_human |
| Tgene | KMT2A | C1854630 | Growth Deficiency and Mental Retardation with Facial Dysmorphism | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | KMT2A | C1879321 | Acute Myeloid Leukemia (AML-M2) | 2 | CTD_human |
| Tgene | KMT2A | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 2 | CTD_human |
| Tgene | KMT2A | C0001418 | Adenocarcinoma | 1 | CTD_human |
| Tgene | KMT2A | C0004403 | Autosome Abnormalities | 1 | CTD_human |
| Tgene | KMT2A | C0005684 | Malignant neoplasm of urinary bladder | 1 | CTD_human |
| Tgene | KMT2A | C0005695 | Bladder Neoplasm | 1 | CTD_human |
| Tgene | KMT2A | C0007138 | Carcinoma, Transitional Cell | 1 | CTD_human |
| Tgene | KMT2A | C0008625 | Chromosome Aberrations | 1 | CTD_human |
| Tgene | KMT2A | C0023448 | Lymphoid leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0023465 | Acute monocytic leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0023479 | Acute myelomonocytic leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human |
| Tgene | KMT2A | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Tgene | KMT2A | C0036341 | Schizophrenia | 1 | PSYGENET |
| Tgene | KMT2A | C0038356 | Stomach Neoplasms | 1 | CTD_human |
| Tgene | KMT2A | C0149925 | Small cell carcinoma of lung | 1 | CTD_human |
| Tgene | KMT2A | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| Tgene | KMT2A | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| Tgene | KMT2A | C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| Tgene | KMT2A | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| Tgene | KMT2A | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| Tgene | KMT2A | C0270972 | Cornelia De Lange Syndrome | 1 | ORPHANET |
| Tgene | KMT2A | C0280141 | Acute Undifferentiated Leukemia | 1 | ORPHANET |
| Tgene | KMT2A | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| Tgene | KMT2A | C0856823 | Undifferentiated type acute leukemia | 1 | ORPHANET |
| Tgene | KMT2A | C1535926 | Neurodevelopmental Disorders | 1 | CTD_human |
| Tgene | KMT2A | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human |
| Tgene | KMT2A | C2239176 | Liver carcinoma | 1 | CTD_human |
| Tgene | KMT2A | C2930974 | Acute erythroleukemia | 1 | CTD_human |
| Tgene | KMT2A | C2930975 | Acute erythroleukemia - M6a subtype | 1 | CTD_human |
| Tgene | KMT2A | C2930976 | Acute myeloid leukemia FAB-M6 | 1 | CTD_human |
| Tgene | KMT2A | C2930977 | Acute erythroleukemia - M6b subtype | 1 | CTD_human |

