| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:LDLR-ACY1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: LDLR-ACY1 | FusionPDB ID: 44351 | FusionGDB2.0 ID: 44351 | Hgene | Tgene | Gene symbol | LDLR | ACY1 | Gene ID | 3949 | 95 |
| Gene name | low density lipoprotein receptor | aminoacylase 1 | |
| Synonyms | FH|FHC|FHCL1|LDLCQ2 | ACY-1|ACY1D|HEL-S-5 | |
| Cytomap | 19p13.2 | 3p21.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | low-density lipoprotein receptorLDL receptorlow-density lipoprotein receptor class A domain-containing protein 3 | aminoacylase-1N-acyl-L-amino-acid amidohydrolaseacylaseepididymis secretory protein Li 5 | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | Q5SW96 | Q03154 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000455727, ENST00000535915, ENST00000545707, ENST00000557933, ENST00000558013, ENST00000558518, ENST00000560628, | ENST00000468068, ENST00000404366, ENST00000476351, ENST00000458031, ENST00000476854, ENST00000494103, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 23 X 17 X 15=5865 | 3 X 6 X 3=54 |
| # samples | 33 | 4 | |
| ** MAII score | log2(33/5865*10)=-4.15159317867005 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/54*10)=-0.432959407276106 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: LDLR [Title/Abstract] AND ACY1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | LDLR(11222315)-ACY1(52019222), # samples:1 LDLR(11222315)-ACY1(52019223), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | LDLR-ACY1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. LDLR-ACY1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. LDLR-ACY1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. LDLR-ACY1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across LDLR (5'-gene) Fusion gene breakpoints across LDLR (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
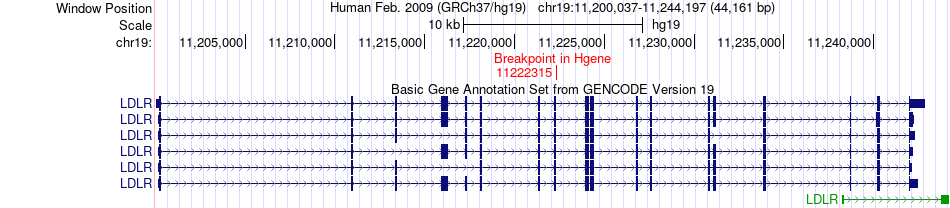 |
 Fusion gene breakpoints across ACY1 (3'-gene) Fusion gene breakpoints across ACY1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
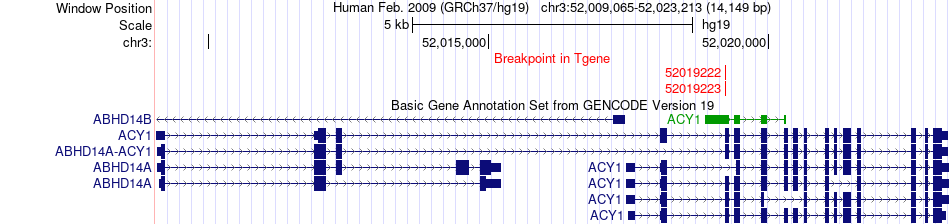 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | TCGA-CG-5730-11A | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + |
| ChimerDB4 | Non-Cancer | TCGA-CG-5730-11A | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000558518 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019222 | + | 2614 | 1373 | 169 | 2505 | 778 |
| ENST00000558518 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019222 | + | 2412 | 1373 | 169 | 2289 | 706 |
| ENST00000558518 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019222 | + | 2433 | 1373 | 169 | 2310 | 713 |
| ENST00000557933 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019222 | + | 2514 | 1273 | 69 | 2405 | 778 |
| ENST00000557933 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019222 | + | 2312 | 1273 | 69 | 2189 | 706 |
| ENST00000557933 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019222 | + | 2333 | 1273 | 69 | 2210 | 713 |
| ENST00000455727 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019222 | + | 2009 | 768 | 68 | 1900 | 610 |
| ENST00000455727 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019222 | + | 1807 | 768 | 68 | 1684 | 538 |
| ENST00000455727 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019222 | + | 1828 | 768 | 68 | 1705 | 545 |
| ENST00000535915 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019222 | + | 2390 | 1149 | 68 | 2281 | 737 |
| ENST00000535915 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019222 | + | 2188 | 1149 | 68 | 2065 | 665 |
| ENST00000535915 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019222 | + | 2209 | 1149 | 68 | 2086 | 672 |
| ENST00000545707 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019222 | + | 2132 | 891 | 68 | 2023 | 651 |
| ENST00000545707 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019222 | + | 1930 | 891 | 68 | 1807 | 579 |
| ENST00000545707 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019222 | + | 1951 | 891 | 68 | 1828 | 586 |
| ENST00000558013 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019222 | + | 2498 | 1257 | 53 | 2389 | 778 |
| ENST00000558013 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019222 | + | 2296 | 1257 | 53 | 2173 | 706 |
| ENST00000558013 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019222 | + | 2317 | 1257 | 53 | 2194 | 713 |
| ENST00000558518 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019223 | + | 2614 | 1373 | 169 | 2505 | 778 |
| ENST00000558518 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019223 | + | 2412 | 1373 | 169 | 2289 | 706 |
| ENST00000558518 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019223 | + | 2433 | 1373 | 169 | 2310 | 713 |
| ENST00000557933 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019223 | + | 2514 | 1273 | 69 | 2405 | 778 |
| ENST00000557933 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019223 | + | 2312 | 1273 | 69 | 2189 | 706 |
| ENST00000557933 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019223 | + | 2333 | 1273 | 69 | 2210 | 713 |
| ENST00000455727 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019223 | + | 2009 | 768 | 68 | 1900 | 610 |
| ENST00000455727 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019223 | + | 1807 | 768 | 68 | 1684 | 538 |
| ENST00000455727 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019223 | + | 1828 | 768 | 68 | 1705 | 545 |
| ENST00000535915 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019223 | + | 2390 | 1149 | 68 | 2281 | 737 |
| ENST00000535915 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019223 | + | 2188 | 1149 | 68 | 2065 | 665 |
| ENST00000535915 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019223 | + | 2209 | 1149 | 68 | 2086 | 672 |
| ENST00000545707 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019223 | + | 2132 | 891 | 68 | 2023 | 651 |
| ENST00000545707 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019223 | + | 1930 | 891 | 68 | 1807 | 579 |
| ENST00000545707 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019223 | + | 1951 | 891 | 68 | 1828 | 586 |
| ENST00000558013 | LDLR | chr19 | 11222315 | + | ENST00000458031 | ACY1 | chr3 | 52019223 | + | 2498 | 1257 | 53 | 2389 | 778 |
| ENST00000558013 | LDLR | chr19 | 11222315 | + | ENST00000494103 | ACY1 | chr3 | 52019223 | + | 2296 | 1257 | 53 | 2173 | 706 |
| ENST00000558013 | LDLR | chr19 | 11222315 | + | ENST00000476854 | ACY1 | chr3 | 52019223 | + | 2317 | 1257 | 53 | 2194 | 713 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000558518 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.011846315 | 0.98815376 |
| ENST00000558518 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.014507916 | 0.98549205 |
| ENST00000558518 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.016812818 | 0.9831872 |
| ENST00000557933 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.011880087 | 0.98811984 |
| ENST00000557933 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.014801628 | 0.9851983 |
| ENST00000557933 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.016738493 | 0.9832615 |
| ENST00000455727 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.016419258 | 0.98358077 |
| ENST00000455727 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.022035573 | 0.9779644 |
| ENST00000455727 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.024077492 | 0.97592247 |
| ENST00000535915 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.01108578 | 0.9889142 |
| ENST00000535915 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.01602114 | 0.98397887 |
| ENST00000535915 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.016221406 | 0.98377854 |
| ENST00000545707 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.01848895 | 0.98151106 |
| ENST00000545707 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.020890849 | 0.97910917 |
| ENST00000545707 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.022063768 | 0.97793627 |
| ENST00000558013 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.011970175 | 0.9880298 |
| ENST00000558013 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.014926088 | 0.9850739 |
| ENST00000558013 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019222 | + | 0.01697899 | 0.983021 |
| ENST00000558518 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.011846315 | 0.98815376 |
| ENST00000558518 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.014507916 | 0.98549205 |
| ENST00000558518 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.016812818 | 0.9831872 |
| ENST00000557933 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.011880087 | 0.98811984 |
| ENST00000557933 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.014801628 | 0.9851983 |
| ENST00000557933 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.016738493 | 0.9832615 |
| ENST00000455727 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.016419258 | 0.98358077 |
| ENST00000455727 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.022035573 | 0.9779644 |
| ENST00000455727 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.024077492 | 0.97592247 |
| ENST00000535915 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.01108578 | 0.9889142 |
| ENST00000535915 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.01602114 | 0.98397887 |
| ENST00000535915 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.016221406 | 0.98377854 |
| ENST00000545707 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.01848895 | 0.98151106 |
| ENST00000545707 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.020890849 | 0.97910917 |
| ENST00000545707 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.022063768 | 0.97793627 |
| ENST00000558013 | ENST00000458031 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.011970175 | 0.9880298 |
| ENST00000558013 | ENST00000494103 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.014926088 | 0.9850739 |
| ENST00000558013 | ENST00000476854 | LDLR | chr19 | 11222315 | + | ACY1 | chr3 | 52019223 | + | 0.01697899 | 0.983021 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >44351_44351_1_LDLR-ACY1_LDLR_chr19_11222315_ENST00000455727_ACY1_chr3_52019222_ENST00000458031_length(amino acids)=610AA_BP=233 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCLTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPD GFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGT NPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELF VQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSV TSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTL -------------------------------------------------------------- >44351_44351_2_LDLR-ACY1_LDLR_chr19_11222315_ENST00000455727_ACY1_chr3_52019222_ENST00000476854_length(amino acids)=545AA_BP=233 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCLTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPD GFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGT NPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELF VQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQ VTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMNRTPVLLHDHDERLHEAVFLRGVDIYTRLLPALASVPA -------------------------------------------------------------- >44351_44351_3_LDLR-ACY1_LDLR_chr19_11222315_ENST00000455727_ACY1_chr3_52019222_ENST00000494103_length(amino acids)=538AA_BP=233 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCLTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPD GFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGT NPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFR EKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDS -------------------------------------------------------------- >44351_44351_4_LDLR-ACY1_LDLR_chr19_11222315_ENST00000455727_ACY1_chr3_52019223_ENST00000458031_length(amino acids)=610AA_BP=233 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCLTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPD GFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGT NPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELF VQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSV TSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTL -------------------------------------------------------------- >44351_44351_5_LDLR-ACY1_LDLR_chr19_11222315_ENST00000455727_ACY1_chr3_52019223_ENST00000476854_length(amino acids)=545AA_BP=233 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCLTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPD GFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGT NPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELF VQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQ VTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMNRTPVLLHDHDERLHEAVFLRGVDIYTRLLPALASVPA -------------------------------------------------------------- >44351_44351_6_LDLR-ACY1_LDLR_chr19_11222315_ENST00000455727_ACY1_chr3_52019223_ENST00000494103_length(amino acids)=538AA_BP=233 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCLTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPD GFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGT NPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFR EKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDS -------------------------------------------------------------- >44351_44351_7_LDLR-ACY1_LDLR_chr19_11222315_ENST00000535915_ACY1_chr3_52019222_ENST00000458031_length(amino acids)=737AA_BP=360 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCSPKTCSQDEFRCHDGKCISRQ FVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCEDGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWR CDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDW SDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAV GAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQY LEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFM EDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGV TLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMNRTPVLLHDHDERLHEAVFLRGVDIY -------------------------------------------------------------- >44351_44351_8_LDLR-ACY1_LDLR_chr19_11222315_ENST00000535915_ACY1_chr3_52019222_ENST00000476854_length(amino acids)=672AA_BP=360 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCSPKTCSQDEFRCHDGKCISRQ FVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCEDGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWR CDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDW SDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAV GAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQY LEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFM EDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMN -------------------------------------------------------------- >44351_44351_9_LDLR-ACY1_LDLR_chr19_11222315_ENST00000535915_ACY1_chr3_52019222_ENST00000494103_length(amino acids)=665AA_BP=360 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCSPKTCSQDEFRCHDGKCISRQ FVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCEDGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWR CDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDW SDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAV GAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYSERSP WWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDF KAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMNRTPVLLH -------------------------------------------------------------- >44351_44351_10_LDLR-ACY1_LDLR_chr19_11222315_ENST00000535915_ACY1_chr3_52019223_ENST00000458031_length(amino acids)=737AA_BP=360 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCSPKTCSQDEFRCHDGKCISRQ FVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCEDGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWR CDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDW SDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAV GAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQY LEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFM EDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGV TLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMNRTPVLLHDHDERLHEAVFLRGVDIY -------------------------------------------------------------- >44351_44351_11_LDLR-ACY1_LDLR_chr19_11222315_ENST00000535915_ACY1_chr3_52019223_ENST00000476854_length(amino acids)=672AA_BP=360 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCSPKTCSQDEFRCHDGKCISRQ FVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCEDGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWR CDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDW SDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAV GAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQY LEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFM EDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMN -------------------------------------------------------------- >44351_44351_12_LDLR-ACY1_LDLR_chr19_11222315_ENST00000535915_ACY1_chr3_52019223_ENST00000494103_length(amino acids)=665AA_BP=360 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCSPKTCSQDEFRCHDGKCISRQ FVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCEDGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWR CDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDW SDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAV GAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYSERSP WWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDF KAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMNRTPVLLH -------------------------------------------------------------- >44351_44351_13_LDLR-ACY1_LDLR_chr19_11222315_ENST00000545707_ACY1_chr3_52019222_ENST00000458031_length(amino acids)=651AA_BP=274 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARD CRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKA CKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCV SIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHA SRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAA GEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMNRTPVLLHDHDERLHEAVFLRG -------------------------------------------------------------- >44351_44351_14_LDLR-ACY1_LDLR_chr19_11222315_ENST00000545707_ACY1_chr3_52019222_ENST00000476854_length(amino acids)=586AA_BP=274 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARD CRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKA CKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCV SIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHA SRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGF -------------------------------------------------------------- >44351_44351_15_LDLR-ACY1_LDLR_chr19_11222315_ENST00000545707_ACY1_chr3_52019222_ENST00000494103_length(amino acids)=579AA_BP=274 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARD CRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKA CKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYS ERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAP DVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMNRTP -------------------------------------------------------------- >44351_44351_16_LDLR-ACY1_LDLR_chr19_11222315_ENST00000545707_ACY1_chr3_52019223_ENST00000458031_length(amino acids)=651AA_BP=274 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARD CRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKA CKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCV SIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHA SRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAA GEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMNRTPVLLHDHDERLHEAVFLRG -------------------------------------------------------------- >44351_44351_17_LDLR-ACY1_LDLR_chr19_11222315_ENST00000545707_ACY1_chr3_52019223_ENST00000476854_length(amino acids)=586AA_BP=274 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARD CRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKA CKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCV SIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAGFALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHA SRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGF -------------------------------------------------------------- >44351_44351_18_LDLR-ACY1_LDLR_chr19_11222315_ENST00000545707_ACY1_chr3_52019223_ENST00000494103_length(amino acids)=579AA_BP=274 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPVATCRPDEFQCSDGNCIHGSRQCDREYDCKDMSDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARD CRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCEDIDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKA CKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSHTDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYS ERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAP DVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNRYIRAVGVPALGFSPMNRTP -------------------------------------------------------------- >44351_44351_19_LDLR-ACY1_LDLR_chr19_11222315_ENST00000557933_ACY1_chr3_52019222_ENST00000458031_length(amino acids)=778AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGV AYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNR -------------------------------------------------------------- >44351_44351_20_LDLR-ACY1_LDLR_chr19_11222315_ENST00000557933_ACY1_chr3_52019222_ENST00000476854_length(amino acids)=713AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWA -------------------------------------------------------------- >44351_44351_21_LDLR-ACY1_LDLR_chr19_11222315_ENST00000557933_ACY1_chr3_52019222_ENST00000494103_length(amino acids)=706AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPH LKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCK -------------------------------------------------------------- >44351_44351_22_LDLR-ACY1_LDLR_chr19_11222315_ENST00000557933_ACY1_chr3_52019223_ENST00000458031_length(amino acids)=778AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGV AYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNR -------------------------------------------------------------- >44351_44351_23_LDLR-ACY1_LDLR_chr19_11222315_ENST00000557933_ACY1_chr3_52019223_ENST00000476854_length(amino acids)=713AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWA -------------------------------------------------------------- >44351_44351_24_LDLR-ACY1_LDLR_chr19_11222315_ENST00000557933_ACY1_chr3_52019223_ENST00000494103_length(amino acids)=706AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPH LKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCK -------------------------------------------------------------- >44351_44351_25_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558013_ACY1_chr3_52019222_ENST00000458031_length(amino acids)=778AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGV AYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNR -------------------------------------------------------------- >44351_44351_26_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558013_ACY1_chr3_52019222_ENST00000476854_length(amino acids)=713AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWA -------------------------------------------------------------- >44351_44351_27_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558013_ACY1_chr3_52019222_ENST00000494103_length(amino acids)=706AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPH LKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCK -------------------------------------------------------------- >44351_44351_28_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558013_ACY1_chr3_52019223_ENST00000458031_length(amino acids)=778AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGV AYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNR -------------------------------------------------------------- >44351_44351_29_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558013_ACY1_chr3_52019223_ENST00000476854_length(amino acids)=713AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWA -------------------------------------------------------------- >44351_44351_30_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558013_ACY1_chr3_52019223_ENST00000494103_length(amino acids)=706AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPH LKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCK -------------------------------------------------------------- >44351_44351_31_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558518_ACY1_chr3_52019222_ENST00000458031_length(amino acids)=778AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGV AYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNR -------------------------------------------------------------- >44351_44351_32_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558518_ACY1_chr3_52019222_ENST00000476854_length(amino acids)=713AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWA -------------------------------------------------------------- >44351_44351_33_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558518_ACY1_chr3_52019222_ENST00000494103_length(amino acids)=706AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPH LKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCK -------------------------------------------------------------- >44351_44351_34_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558518_ACY1_chr3_52019223_ENST00000458031_length(amino acids)=778AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPHLKEGSVTSVNLTKLEGGV AYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCKDMNLTLEPEIMPAATDNR -------------------------------------------------------------- >44351_44351_35_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558518_ACY1_chr3_52019223_ENST00000476854_length(amino acids)=713AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGYIYARGAQDMKCVSIQYLEAVRRLKVEGHRFPRTIHMTFVPDEEVGGHQGMELFVQRPEFHALRAG FALDEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWA -------------------------------------------------------------- >44351_44351_36_LDLR-ACY1_LDLR_chr19_11222315_ENST00000558518_ACY1_chr3_52019223_ENST00000494103_length(amino acids)=706AA_BP=401 MAEAASMGPWGWKLRWTVALLLAAAGTAVGDRCERNEFQCQDGKCISYKWVCDGSAECQDGSDESQETCLSVTCKSGDFSCGGRVNRCIP QFWRCDGQVDCDNGSDEQGCPPKTCSQDEFRCHDGKCISRQFVCDSDRDCLDGSDEASCPVLTCGPASFQCNSSTCIPQLWACDNDPDCE DGSDEWPQRCRGLYVFQGDSSPCSAFEFHCLSGECIHSSWRCDGGPDCKDKSDEENCAVATCRPDEFQCSDGNCIHGSRQCDREYDCKDM SDEVGCVNVTLCEGPNKFKCHSGECITLDKVCNMARDCRDWSDEPIKECGTNECLDNNGGCSHVCNDLKIGYECLCPDGFQLVAQRRCED IDECQDPDTCSQLCVNLEGGYKCQCEEGFQLDPHTKACKAVGAAVAFFEETARQLGLGCQKVEVAPGYVVTVLTWPGTNPTLSSILLNSH TDVVPVFKEHWSHDPFEAFKDSEGIANPTDAFTVFYSERSPWWVRVTSTGRPGHASRFMEDTAAEKLHKVVNSILAFREKEWQRLQSNPH LKEGSVTSVNLTKLEGGVAYNVIPATMSASFDFRVAPDVDFKAFEEQLQSWCQAAGEGVTLEFAQKWMHPQVTPTDDSNPWWAAFSRVCK -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:11222315/chr3:52019222) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LDLR | ACY1 |
| FUNCTION: Adapter protein (clathrin-associated sorting protein (CLASP)) required for efficient endocytosis of the LDL receptor (LDLR) in polarized cells such as hepatocytes and lymphocytes, but not in non-polarized cells (fibroblasts). May be required for LDL binding and internalization but not for receptor clustering in coated pits. May facilitate the endocytocis of LDLR and LDLR-LDL complexes from coated pits by stabilizing the interaction between the receptor and the structural components of the pits. May also be involved in the internalization of other LDLR family members. Binds to phosphoinositides, which regulate clathrin bud assembly at the cell surface. Required for trafficking of LRP2 to the endocytic recycling compartment which is necessary for LRP2 proteolysis, releasing a tail fragment which translocates to the nucleus and mediates transcriptional repression (By similarity). {ECO:0000250|UniProtKB:D3ZAR1, ECO:0000269|PubMed:15728179}. | FUNCTION: Involved in the hydrolysis of N-acylated or N-acetylated amino acids (except L-aspartate). {ECO:0000269|PubMed:12933810}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 107_145 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 146_186 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 25_65 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 66_106 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 107_145 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 146_186 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 195_233 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 234_272 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 25_65 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 274_313 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 314_353 | 354.3333333333333 | 820.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 66_106 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 107_145 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 146_186 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 195_233 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 25_65 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 66_106 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 107_145 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 146_186 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 195_233 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 234_272 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 25_65 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 274_313 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 314_353 | 395.3333333333333 | 859.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 354_393 | 395.3333333333333 | 859.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 66_106 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 107_145 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 146_186 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 195_233 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 234_272 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 25_65 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 274_313 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 314_353 | 395.3333333333333 | 861.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 354_393 | 395.3333333333333 | 861.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 66_106 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 107_145 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 146_186 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 25_65 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 66_106 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 107_145 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 146_186 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 195_233 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 234_272 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 25_65 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 274_313 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 314_353 | 354.3333333333333 | 820.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 66_106 | 354.3333333333333 | 820.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 107_145 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 146_186 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 195_233 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 25_65 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 66_106 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 107_145 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 146_186 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 195_233 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 234_272 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 25_65 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 274_313 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 314_353 | 395.3333333333333 | 859.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 354_393 | 395.3333333333333 | 859.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 66_106 | 395.3333333333333 | 859.0 | Domain | LDL-receptor class A 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 107_145 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 146_186 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 195_233 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 234_272 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 25_65 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 274_313 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 314_353 | 395.3333333333333 | 861.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 354_393 | 395.3333333333333 | 861.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 66_106 | 395.3333333333333 | 861.0 | Domain | LDL-receptor class A 2 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 195_233 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 234_272 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 274_313 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 314_353 | 227.33333333333334 | 693.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 354_393 | 227.33333333333334 | 693.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 663_712 | 227.33333333333334 | 693.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 354_393 | 354.3333333333333 | 820.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 663_712 | 354.3333333333333 | 820.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 234_272 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 274_313 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 314_353 | 268.3333333333333 | 683.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 354_393 | 268.3333333333333 | 683.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 663_712 | 268.3333333333333 | 683.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 663_712 | 395.3333333333333 | 859.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 663_712 | 395.3333333333333 | 861.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 195_233 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 234_272 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 274_313 | 227.33333333333334 | 693.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 314_353 | 227.33333333333334 | 693.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 354_393 | 227.33333333333334 | 693.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 663_712 | 227.33333333333334 | 693.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 354_393 | 354.3333333333333 | 820.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 663_712 | 354.3333333333333 | 820.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 234_272 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 274_313 | 268.3333333333333 | 683.0 | Domain | LDL-receptor class A 7 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 314_353 | 268.3333333333333 | 683.0 | Domain | EGF-like 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 354_393 | 268.3333333333333 | 683.0 | Domain | EGF-like 2%3B calcium-binding |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 663_712 | 268.3333333333333 | 683.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 663_712 | 395.3333333333333 | 859.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 663_712 | 395.3333333333333 | 861.0 | Domain | EGF-like 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 823_828 | 227.33333333333334 | 693.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 823_828 | 354.3333333333333 | 820.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 823_828 | 268.3333333333333 | 683.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 823_828 | 395.3333333333333 | 859.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 823_828 | 395.3333333333333 | 861.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 823_828 | 227.33333333333334 | 693.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 823_828 | 354.3333333333333 | 820.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 823_828 | 268.3333333333333 | 683.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 823_828 | 395.3333333333333 | 859.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 823_828 | 395.3333333333333 | 861.0 | Motif | NPXY motif |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 721_768 | 227.33333333333334 | 693.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 811_860 | 227.33333333333334 | 693.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 721_768 | 354.3333333333333 | 820.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 811_860 | 354.3333333333333 | 820.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 721_768 | 268.3333333333333 | 683.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 811_860 | 268.3333333333333 | 683.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 721_768 | 395.3333333333333 | 859.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 811_860 | 395.3333333333333 | 859.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 721_768 | 395.3333333333333 | 861.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 811_860 | 395.3333333333333 | 861.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 721_768 | 227.33333333333334 | 693.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 811_860 | 227.33333333333334 | 693.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 721_768 | 354.3333333333333 | 820.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 811_860 | 354.3333333333333 | 820.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 721_768 | 268.3333333333333 | 683.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 811_860 | 268.3333333333333 | 683.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 721_768 | 395.3333333333333 | 859.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 811_860 | 395.3333333333333 | 859.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 721_768 | 395.3333333333333 | 861.0 | Region | Note=Clustered O-linked oligosaccharides |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 811_860 | 395.3333333333333 | 861.0 | Region | Required for MYLIP-triggered down-regulation of LDLR |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 397_438 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 439_485 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 486_528 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 529_572 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 573_615 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 616_658 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 397_438 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 439_485 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 486_528 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 529_572 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 573_615 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 616_658 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 397_438 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 439_485 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 486_528 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 529_572 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 573_615 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 616_658 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 397_438 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 439_485 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 486_528 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 529_572 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 573_615 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 616_658 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 397_438 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 439_485 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 486_528 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 529_572 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 573_615 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 616_658 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 397_438 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 439_485 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 486_528 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 529_572 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 573_615 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 616_658 | 227.33333333333334 | 693.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 397_438 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 439_485 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 486_528 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 529_572 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 573_615 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 616_658 | 354.3333333333333 | 820.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 397_438 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 439_485 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 486_528 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 529_572 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 573_615 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 616_658 | 268.3333333333333 | 683.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 397_438 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 439_485 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 486_528 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 529_572 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 573_615 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 616_658 | 395.3333333333333 | 859.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 397_438 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 1 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 439_485 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 2 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 486_528 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 3 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 529_572 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 4 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 573_615 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 5 |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 616_658 | 395.3333333333333 | 861.0 | Repeat | Note=LDL-receptor class B 6 |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 22_788 | 227.33333333333334 | 693.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 22_788 | 354.3333333333333 | 820.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 22_788 | 268.3333333333333 | 683.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 22_788 | 395.3333333333333 | 859.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 22_788 | 395.3333333333333 | 861.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 22_788 | 227.33333333333334 | 693.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 22_788 | 354.3333333333333 | 820.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 22_788 | 268.3333333333333 | 683.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 22_788 | 395.3333333333333 | 859.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 22_788 | 395.3333333333333 | 861.0 | Topological domain | Extracellular |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000455727 | + | 6 | 16 | 789_810 | 227.33333333333334 | 693.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000535915 | + | 7 | 17 | 789_810 | 354.3333333333333 | 820.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000545707 | + | 7 | 16 | 789_810 | 268.3333333333333 | 683.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558013 | + | 8 | 18 | 789_810 | 395.3333333333333 | 859.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11222315 | chr3:52019222 | ENST00000558518 | + | 8 | 18 | 789_810 | 395.3333333333333 | 861.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000455727 | + | 6 | 16 | 789_810 | 227.33333333333334 | 693.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000535915 | + | 7 | 17 | 789_810 | 354.3333333333333 | 820.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000545707 | + | 7 | 16 | 789_810 | 268.3333333333333 | 683.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558013 | + | 8 | 18 | 789_810 | 395.3333333333333 | 859.0 | Transmembrane | Helical |
| Hgene | LDLR | chr19:11222315 | chr3:52019223 | ENST00000558518 | + | 8 | 18 | 789_810 | 395.3333333333333 | 861.0 | Transmembrane | Helical |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| LDLR | |
| ACY1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to LDLR-ACY1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to LDLR-ACY1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

