| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:LPP-KMT2A |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: LPP-KMT2A | FusionPDB ID: 49568 | FusionGDB2.0 ID: 49568 | Hgene | Tgene | Gene symbol | LPP | KMT2A | Gene ID | 4026 | 4297 |
| Gene name | LIM domain containing preferred translocation partner in lipoma | lysine methyltransferase 2A | |
| Synonyms | - | ALL-1|CXXC7|HRX|HTRX1|MLL|MLL1|MLL1A|TRX1|WDSTS | |
| Cytomap | 3q27.3-q28 | 11q23.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | lipoma-preferred partnerLIM proteinlipoma preferred partner | histone-lysine N-methyltransferase 2ACXXC-type zinc finger protein 7lysine (K)-specific methyltransferase 2Alysine N-methyltransferase 2Amixed lineage leukemia 1myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)trithorax-like | |
| Modification date | 20200313 | 20200319 | |
| UniProtAcc | Q93052 | Q03164 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000312675, ENST00000543006, ENST00000448637, ENST00000471917, | ENST00000354520, ENST00000389506, ENST00000420751, ENST00000534358, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 41 X 30 X 15=18450 | 31 X 72 X 3=6696 |
| # samples | 52 | 79 | |
| ** MAII score | log2(52/18450*10)=-5.14896538280667 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(79/6696*10)=-3.08337496948588 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: LPP [Title/Abstract] AND KMT2A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | LPP-KMT2A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. LPP-KMT2A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. LPP-KMT2A seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor by not retaining the major functional domain in the partially deleted in-frame ORF. LPP-KMT2A seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor by not retaining the major functional domain in the partially deleted in-frame ORF. LPP-KMT2A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. LPP-KMT2A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. LPP-KMT2A seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target by not retaining the major functional domain in the partially deleted in-frame ORF. LPP-KMT2A seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target by not retaining the major functional domain in the partially deleted in-frame ORF. LPP-KMT2A seems lost the major protein functional domain in Hgene partner, which is a transcription factor by not retaining the major functional domain in the partially deleted in-frame ORF. LPP-KMT2A seems lost the major protein functional domain in Hgene partner, which is a transcription factor by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | KMT2A | GO:0044648 | histone H3-K4 dimethylation | 25561738 |
| Tgene | KMT2A | GO:0045944 | positive regulation of transcription by RNA polymerase II | 20861184 |
| Tgene | KMT2A | GO:0051568 | histone H3-K4 methylation | 19556245 |
| Tgene | KMT2A | GO:0065003 | protein-containing complex assembly | 15199122 |
| Tgene | KMT2A | GO:0080182 | histone H3-K4 trimethylation | 20861184 |
| Tgene | KMT2A | GO:0097692 | histone H3-K4 monomethylation | 25561738|26324722 |
 Fusion gene breakpoints across LPP (5'-gene) Fusion gene breakpoints across LPP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across KMT2A (3'-gene) Fusion gene breakpoints across KMT2A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | LPP | chr3 | 188478070 | + | KMT2A | chr11 | 118359328 | |
| ChimerKB3 | . | . | LPP | chr3 | 188584166 | + | KMT2A | chr11 | 118353210 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000312675 | LPP | chr3 | 188478070 | + | ENST00000534358 | KMT2A | chr11 | 118359328 | 13895 | 1656 | 123 | 9242 | 3039 | |
| ENST00000312675 | LPP | chr3 | 188478070 | + | ENST00000389506 | KMT2A | chr11 | 118359328 | 10979 | 1656 | 123 | 9233 | 3036 | |
| ENST00000312675 | LPP | chr3 | 188478070 | + | ENST00000354520 | KMT2A | chr11 | 118359328 | 11758 | 1656 | 123 | 9233 | 3036 | |
| ENST00000543006 | LPP | chr3 | 188478070 | + | ENST00000534358 | KMT2A | chr11 | 118359328 | 13838 | 1599 | 9 | 9185 | 3058 | |
| ENST00000543006 | LPP | chr3 | 188478070 | + | ENST00000389506 | KMT2A | chr11 | 118359328 | 10922 | 1599 | 9 | 9176 | 3055 | |
| ENST00000543006 | LPP | chr3 | 188478070 | + | ENST00000354520 | KMT2A | chr11 | 118359328 | 11701 | 1599 | 9 | 9176 | 3055 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >49568_49568_1_LPP-KMT2A_LPP_chr3_188478070_ENST00000312675_KMT2A_chr11_118359328_ENST00000354520_length(amino acids)=3036AA_BP=511 MRLRDRAEDRTWSSDSLCSAFFIVPLDAHQREDLFPQLIPTMSHPSWLPPKSTGEPLGHVPARMETTHSFGNPSISVSTQQPPKKFAPVV APKPKYNPYKQPGGEGDFLPPPPPPLDDSSALPSISGNFPPPPPLDEEAFKVQGNPGGKTLEERRSSLDAEIDSLTSILADLECSSPYKP RPPQSSTGSTASPPVSTPVTGHKRMVIPNQPPLTATKKSTLKPQPAPQAGPIPVAPIGTLKPQPQPVPASYTTASTSSRPTFNVQVKSAQ PSPHYMAAPSSGQIYGSGPQGYNTQPVPVSGQCPPPSTRGGMDYAYIPPPGLQPEPGYGYAPNQGRYYEGYYAAGPGYGGRNDSDPTYGQ QGHPNTWKREPGYTPPGAGNQNPPGMYPVTGPKKTYITDPVSAPCAPPLQPKGGHSGQLGPSSVAPSFRPEDELEHLTKKMLYDMENPPA DEYFGRCARCGENVVGEGTGCTAMDQVFHVDCFTCIICNNKLRGQPFYAVEKKAYCEPCYIFVYCQVCCEPFHKFCLEENERPLEDQLEN WCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLF AKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLN SRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGG QPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPG EPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHM AVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNG LEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFT ESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSR RHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGS SSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLR GQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPE FMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIEST SPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEE DIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTAT TRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDT YNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSS SISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADH ISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQT LPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSG LLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGS LNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVT LTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQ HVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRF QLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPK TVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGIL HDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMD LPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAAR -------------------------------------------------------------- >49568_49568_2_LPP-KMT2A_LPP_chr3_188478070_ENST00000312675_KMT2A_chr11_118359328_ENST00000389506_length(amino acids)=3036AA_BP=511 MRLRDRAEDRTWSSDSLCSAFFIVPLDAHQREDLFPQLIPTMSHPSWLPPKSTGEPLGHVPARMETTHSFGNPSISVSTQQPPKKFAPVV APKPKYNPYKQPGGEGDFLPPPPPPLDDSSALPSISGNFPPPPPLDEEAFKVQGNPGGKTLEERRSSLDAEIDSLTSILADLECSSPYKP RPPQSSTGSTASPPVSTPVTGHKRMVIPNQPPLTATKKSTLKPQPAPQAGPIPVAPIGTLKPQPQPVPASYTTASTSSRPTFNVQVKSAQ PSPHYMAAPSSGQIYGSGPQGYNTQPVPVSGQCPPPSTRGGMDYAYIPPPGLQPEPGYGYAPNQGRYYEGYYAAGPGYGGRNDSDPTYGQ QGHPNTWKREPGYTPPGAGNQNPPGMYPVTGPKKTYITDPVSAPCAPPLQPKGGHSGQLGPSSVAPSFRPEDELEHLTKKMLYDMENPPA DEYFGRCARCGENVVGEGTGCTAMDQVFHVDCFTCIICNNKLRGQPFYAVEKKAYCEPCYIFVYCQVCCEPFHKFCLEENERPLEDQLEN WCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLF AKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLN SRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINSDGG QPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPG EPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHM AVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNG LEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFT ESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSR RHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGS SSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLR GQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPE FMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIEST SPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSYGEE DIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTAT TRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDT YNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSS SISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADH ISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYVLQT LPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSG LLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGS LNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVT LTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQ HVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKTKRF QLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPK TVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGIL HDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMD LPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAAR -------------------------------------------------------------- >49568_49568_3_LPP-KMT2A_LPP_chr3_188478070_ENST00000312675_KMT2A_chr11_118359328_ENST00000534358_length(amino acids)=3039AA_BP=511 MRLRDRAEDRTWSSDSLCSAFFIVPLDAHQREDLFPQLIPTMSHPSWLPPKSTGEPLGHVPARMETTHSFGNPSISVSTQQPPKKFAPVV APKPKYNPYKQPGGEGDFLPPPPPPLDDSSALPSISGNFPPPPPLDEEAFKVQGNPGGKTLEERRSSLDAEIDSLTSILADLECSSPYKP RPPQSSTGSTASPPVSTPVTGHKRMVIPNQPPLTATKKSTLKPQPAPQAGPIPVAPIGTLKPQPQPVPASYTTASTSSRPTFNVQVKSAQ PSPHYMAAPSSGQIYGSGPQGYNTQPVPVSGQCPPPSTRGGMDYAYIPPPGLQPEPGYGYAPNQGRYYEGYYAAGPGYGGRNDSDPTYGQ QGHPNTWKREPGYTPPGAGNQNPPGMYPVTGPKKTYITDPVSAPCAPPLQPKGGHSGQLGPSSVAPSFRPEDELEHLTKKMLYDMENPPA DEYFGRCARCGENVVGEGTGCTAMDQVFHVDCFTCIICNNKLRGQPFYAVEKKAYCEPCYIFVYCQVCCEPFHKFCLEENERPLEDQLEN WCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLF AKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTA LLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKIIQAAINS DGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPK GPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKN VHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKF LNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPT SFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLLSSGLRSI GSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHL DGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHL HLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNL KPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQI ESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTPLYGVRSY GEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSV TATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNL LDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSS VSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMD ADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQNMQPLYV LQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNP PSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLD LGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPG HVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHY QLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGPTKPKPKT KRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESA EPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRML GILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRAT SMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGN -------------------------------------------------------------- >49568_49568_4_LPP-KMT2A_LPP_chr3_188478070_ENST00000543006_KMT2A_chr11_118359328_ENST00000354520_length(amino acids)=3055AA_BP=530 MRLRDRAEDRTWSSDSLCSAFFIVPLDAHQREDLFPQLHCSWLNLVSTIHSRSQLSEIPTMSHPSWLPPKSTGEPLGHVPARMETTHSFG NPSISVSTQQPPKKFAPVVAPKPKYNPYKQPGGEGDFLPPPPPPLDDSSALPSISGNFPPPPPLDEEAFKVQGNPGGKTLEERRSSLDAE IDSLTSILADLECSSPYKPRPPQSSTGSTASPPVSTPVTGHKRMVIPNQPPLTATKKSTLKPQPAPQAGPIPVAPIGTLKPQPQPVPASY TTASTSSRPTFNVQVKSAQPSPHYMAAPSSGQIYGSGPQGYNTQPVPVSGQCPPPSTRGGMDYAYIPPPGLQPEPGYGYAPNQGRYYEGY YAAGPGYGGRNDSDPTYGQQGHPNTWKREPGYTPPGAGNQNPPGMYPVTGPKKTYITDPVSAPCAPPLQPKGGHSGQLGPSSVAPSFRPE DELEHLTKKMLYDMENPPADEYFGRCARCGENVVGEGTGCTAMDQVFHVDCFTCIICNNKLRGQPFYAVEKKAYCEPCYIFVYCQVCCEP FHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKG WDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRL ALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVL EFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHT EQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCA LWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVF RRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDIN STVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHE IVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSS NLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEP SSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSK SFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQ SKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARAR SNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKI SQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSL ESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLF EVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPT PEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPA TEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHS FPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTL INFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVV SMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQT TGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQR SASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVL PEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNA RLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPN DEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYM -------------------------------------------------------------- >49568_49568_5_LPP-KMT2A_LPP_chr3_188478070_ENST00000543006_KMT2A_chr11_118359328_ENST00000389506_length(amino acids)=3055AA_BP=530 MRLRDRAEDRTWSSDSLCSAFFIVPLDAHQREDLFPQLHCSWLNLVSTIHSRSQLSEIPTMSHPSWLPPKSTGEPLGHVPARMETTHSFG NPSISVSTQQPPKKFAPVVAPKPKYNPYKQPGGEGDFLPPPPPPLDDSSALPSISGNFPPPPPLDEEAFKVQGNPGGKTLEERRSSLDAE IDSLTSILADLECSSPYKPRPPQSSTGSTASPPVSTPVTGHKRMVIPNQPPLTATKKSTLKPQPAPQAGPIPVAPIGTLKPQPQPVPASY TTASTSSRPTFNVQVKSAQPSPHYMAAPSSGQIYGSGPQGYNTQPVPVSGQCPPPSTRGGMDYAYIPPPGLQPEPGYGYAPNQGRYYEGY YAAGPGYGGRNDSDPTYGQQGHPNTWKREPGYTPPGAGNQNPPGMYPVTGPKKTYITDPVSAPCAPPLQPKGGHSGQLGPSSVAPSFRPE DELEHLTKKMLYDMENPPADEYFGRCARCGENVVGEGTGCTAMDQVFHVDCFTCIICNNKLRGQPFYAVEKKAYCEPCYIFVYCQVCCEP FHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKG WDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWRL ALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVL EFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHT EQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCA LWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVF RRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDIN STVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHE IVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSS NLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEP SSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKHSSK SFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENESQ SKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARAR SNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKI SQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSL ESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDMGLF EVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDPT PEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPA TEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHS FPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTL INFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVLNVV SMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQT TGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQR SASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVL PEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNA RLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEYNPN DEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCYM -------------------------------------------------------------- >49568_49568_6_LPP-KMT2A_LPP_chr3_188478070_ENST00000543006_KMT2A_chr11_118359328_ENST00000534358_length(amino acids)=3058AA_BP=530 MRLRDRAEDRTWSSDSLCSAFFIVPLDAHQREDLFPQLHCSWLNLVSTIHSRSQLSEIPTMSHPSWLPPKSTGEPLGHVPARMETTHSFG NPSISVSTQQPPKKFAPVVAPKPKYNPYKQPGGEGDFLPPPPPPLDDSSALPSISGNFPPPPPLDEEAFKVQGNPGGKTLEERRSSLDAE IDSLTSILADLECSSPYKPRPPQSSTGSTASPPVSTPVTGHKRMVIPNQPPLTATKKSTLKPQPAPQAGPIPVAPIGTLKPQPQPVPASY TTASTSSRPTFNVQVKSAQPSPHYMAAPSSGQIYGSGPQGYNTQPVPVSGQCPPPSTRGGMDYAYIPPPGLQPEPGYGYAPNQGRYYEGY YAAGPGYGGRNDSDPTYGQQGHPNTWKREPGYTPPGAGNQNPPGMYPVTGPKKTYITDPVSAPCAPPLQPKGGHSGQLGPSSVAPSFRPE DELEHLTKKMLYDMENPPADEYFGRCARCGENVVGEGTGCTAMDQVFHVDCFTCIICNNKLRGQPFYAVEKKAYCEPCYIFVYCQVCCEP FHKFCLEENERPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRCKSCGSTTPGKG WDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPAE WRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYT SVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREEN SHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHV NCALWSAEVFEDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKGEVVPENGF EVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEP DINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPT THEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTS TSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSF AEPSSVSFSSKEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSCKETFKEKH SSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMEN ESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSA RARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDL PKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQL SSLESSRRVHTSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGLDSNREKDM GLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGV DPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHL KPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQH LHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSN MTLINFTPSQLPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSGLASSSSVL NVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPS TQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSS GQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQV AVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEAR SNARLKQLSFAGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLASKHRQPPEY NPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIG -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:/chr11:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| LPP | KMT2A |
| FUNCTION: May play a structural role at sites of cell adhesion in maintaining cell shape and motility. In addition to these structural functions, it may also be implicated in signaling events and activation of gene transcription. May be involved in signal transduction from cell adhesion sites to the nucleus allowing successful integration of signals arising from soluble factors and cell-cell adhesion sites. Also suggested to serve as a scaffold protein upon which distinct protein complexes are assembled in the cytoplasm and in the nucleus. {ECO:0000269|PubMed:10637295}. | FUNCTION: Histone methyltransferase that plays an essential role in early development and hematopoiesis (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalytic subunit of the MLL1/MLL complex, a multiprotein complex that mediates both methylation of 'Lys-4' of histone H3 (H3K4me) complex and acetylation of 'Lys-16' of histone H4 (H4K16ac) (PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:24235145, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Catalyzes methyl group transfer from S-adenosyl-L-methionine to the epsilon-amino group of 'Lys-4' of histone H3 (H3K4) via a non-processive mechanism. Part of chromatin remodeling machinery predominantly forms H3K4me1 and H3K4me2 methylation marks at active chromatin sites where transcription and DNA repair take place (PubMed:25561738, PubMed:15960975, PubMed:12453419, PubMed:15960975, PubMed:19556245, PubMed:19187761, PubMed:20677832, PubMed:21220120, PubMed:26886794). Has weak methyltransferase activity by itself, and requires other component of the MLL1/MLL complex to obtain full methyltransferase activity (PubMed:19187761, PubMed:26886794). Has no activity toward histone H3 phosphorylated on 'Thr-3', less activity toward H3 dimethylated on 'Arg-8' or 'Lys-9', while it has higher activity toward H3 acetylated on 'Lys-9' (PubMed:19187761). Binds to unmethylated CpG elements in the promoter of target genes and helps maintain them in the nonmethylated state (PubMed:20010842). Required for transcriptional activation of HOXA9 (PubMed:12453419, PubMed:20677832, PubMed:20010842). Promotes PPP1R15A-induced apoptosis (PubMed:10490642). Plays a critical role in the control of circadian gene expression and is essential for the transcriptional activation mediated by the CLOCK-ARNTL/BMAL1 heterodimer (By similarity). Establishes a permissive chromatin state for circadian transcription by mediating a rhythmic methylation of 'Lys-4' of histone H3 (H3K4me) and this histone modification directs the circadian acetylation at H3K9 and H3K14 allowing the recruitment of CLOCK-ARNTL/BMAL1 to chromatin (By similarity). Also has auto-methylation activity on Cys-3882 in absence of histone H3 substrate (PubMed:24235145). {ECO:0000250|UniProtKB:P55200, ECO:0000269|PubMed:10490642, ECO:0000269|PubMed:12453419, ECO:0000269|PubMed:15960975, ECO:0000269|PubMed:19187761, ECO:0000269|PubMed:19556245, ECO:0000269|PubMed:20010842, ECO:0000269|PubMed:21220120, ECO:0000269|PubMed:24235145, ECO:0000269|PubMed:26886794, ECO:0000305|PubMed:20677832}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 138_156 | 470.0 | 613.0 | Compositional bias | Polar residues | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 185_202 | 470.0 | 613.0 | Compositional bias | Pro residues | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 204_219 | 470.0 | 613.0 | Compositional bias | Polar residues | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 26_42 | 470.0 | 613.0 | Compositional bias | Polar residues | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 64_78 | 470.0 | 613.0 | Compositional bias | Pro residues | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 138_156 | 470.0 | 613.0 | Compositional bias | Polar residues | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 185_202 | 470.0 | 613.0 | Compositional bias | Pro residues | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 204_219 | 470.0 | 613.0 | Compositional bias | Polar residues | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 26_42 | 470.0 | 613.0 | Compositional bias | Polar residues | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 64_78 | 470.0 | 613.0 | Compositional bias | Pro residues | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 414_473 | 470.0 | 613.0 | Domain | LIM zinc-binding 1 | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 414_473 | 470.0 | 613.0 | Domain | LIM zinc-binding 1 | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 132_219 | 470.0 | 613.0 | Region | Disordered | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 1_118 | 470.0 | 613.0 | Region | Disordered | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 307_387 | 470.0 | 613.0 | Region | Disordered | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 132_219 | 470.0 | 613.0 | Region | Disordered | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 1_118 | 470.0 | 613.0 | Region | Disordered | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 307_387 | 470.0 | 613.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1822_1847 | 1406.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2096_2118 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2146_2174 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2189_2232 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2275_2320 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2402_2418 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2419_2445 | 1406.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2526_2594 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2721_2746 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2747_2784 | 1406.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2785_2821 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3011_3064 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3166_3183 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3195_3222 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3230_3244 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3464_3530 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3563_3606 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1822_1847 | 1444.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2096_2118 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2146_2174 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2189_2232 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2275_2320 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2402_2418 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2419_2445 | 1444.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2526_2594 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2721_2746 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2747_2784 | 1444.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2785_2821 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3011_3064 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3166_3183 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3195_3222 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3230_3244 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3464_3530 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3563_3606 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1822_1847 | 1444.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2096_2118 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2146_2174 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2189_2232 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2275_2320 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2402_2418 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2419_2445 | 1444.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2526_2594 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2721_2746 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2747_2784 | 1444.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2785_2821 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3011_3064 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3166_3183 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3195_3222 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3230_3244 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3464_3530 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3563_3606 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1703_1748 | 1406.0 | 3932.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2018_2074 | 1406.0 | 3932.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3666_3747 | 1406.0 | 3932.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3829_3945 | 1406.0 | 3932.0 | Domain | SET | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3953_3969 | 1406.0 | 3932.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1703_1748 | 1444.0 | 3970.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2018_2074 | 1444.0 | 3970.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3666_3747 | 1444.0 | 3970.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3829_3945 | 1444.0 | 3970.0 | Domain | SET | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3953_3969 | 1444.0 | 3970.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1703_1748 | 1444.0 | 3973.0 | Domain | Bromo%3B divergent | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2018_2074 | 1444.0 | 3973.0 | Domain | FYR N-terminal | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3666_3747 | 1444.0 | 3973.0 | Domain | FYR C-terminal | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3829_3945 | 1444.0 | 3973.0 | Domain | SET | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3953_3969 | 1444.0 | 3973.0 | Domain | Post-SET | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1663_1713 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1806_1869 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2081_2133 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2145_2232 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2275_2333 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2373_2460 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2475_2618 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2647_2675 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2713_2821 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 2961_3064 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3166_3244 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3464_3608 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3620_3643 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3785_3808 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 3906_3907 | 1406.0 | 3932.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1663_1713 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1806_1869 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2081_2133 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2145_2232 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2275_2333 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2373_2460 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2475_2618 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2647_2675 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2713_2821 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 2961_3064 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3166_3244 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3464_3608 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3620_3643 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3785_3808 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 3906_3907 | 1444.0 | 3970.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1663_1713 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1806_1869 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2081_2133 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2145_2232 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2275_2333 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2373_2460 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2475_2618 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2647_2675 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2713_2821 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 2961_3064 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3166_3244 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3464_3608 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3620_3643 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3785_3808 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 3906_3907 | 1444.0 | 3973.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1431_1482 | 1406.0 | 3932.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1479_1533 | 1406.0 | 3932.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1566_1627 | 1406.0 | 3932.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1870_1910 | 1406.0 | 3932.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1931_1978 | 1406.0 | 3932.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1479_1533 | 1444.0 | 3970.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1566_1627 | 1444.0 | 3970.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1870_1910 | 1444.0 | 3970.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1931_1978 | 1444.0 | 3970.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1479_1533 | 1444.0 | 3973.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1566_1627 | 1444.0 | 3973.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1870_1910 | 1444.0 | 3973.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1931_1978 | 1444.0 | 3973.0 | Zinc finger | PHD-type 4 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 474_534 | 470.0 | 613.0 | Domain | LIM zinc-binding 2 | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000312675 | 8 | 11 | 535_603 | 470.0 | 613.0 | Domain | LIM zinc-binding 3 | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 474_534 | 470.0 | 613.0 | Domain | LIM zinc-binding 2 | |
| Hgene | LPP | chr3:188478070 | chr11:118359328 | ENST00000543006 | 8 | 11 | 535_603 | 470.0 | 613.0 | Domain | LIM zinc-binding 3 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1046_1061 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1249_1272 | 1406.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1278_1302 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1303_1317 | 1406.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 184_203 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 204_226 | 1406.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 236_253 | 1406.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 325_351 | 1406.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 37_58 | 1406.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 445_495 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 535_556 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 557_576 | 1406.0 | 3932.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 713_731 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 747_780 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 798_845 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 81_103 | 1406.0 | 3932.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 846_898 | 1406.0 | 3932.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1046_1061 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1249_1272 | 1444.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1278_1302 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1303_1317 | 1444.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 184_203 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 204_226 | 1444.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 236_253 | 1444.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 325_351 | 1444.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 37_58 | 1444.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 445_495 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 535_556 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 557_576 | 1444.0 | 3970.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 713_731 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 747_780 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 798_845 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 81_103 | 1444.0 | 3970.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 846_898 | 1444.0 | 3970.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1046_1061 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1249_1272 | 1444.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1278_1302 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1303_1317 | 1444.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 184_203 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 204_226 | 1444.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 236_253 | 1444.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 325_351 | 1444.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 37_58 | 1444.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 445_495 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 535_556 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 557_576 | 1444.0 | 3973.0 | Compositional bias | Pro residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 713_731 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 747_780 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 798_845 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 81_103 | 1444.0 | 3973.0 | Compositional bias | Polar residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 846_898 | 1444.0 | 3973.0 | Compositional bias | Basic and acidic residues | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 169_180 | 1406.0 | 3932.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 217_227 | 1406.0 | 3932.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 301_309 | 1406.0 | 3932.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 169_180 | 1444.0 | 3970.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 217_227 | 1444.0 | 3970.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 301_309 | 1444.0 | 3970.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 169_180 | 1444.0 | 3973.0 | DNA binding | Note=A.T hook 1 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 217_227 | 1444.0 | 3973.0 | DNA binding | Note=A.T hook 2 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 301_309 | 1444.0 | 3973.0 | DNA binding | Note=A.T hook 3 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 123_134 | 1406.0 | 3932.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 147_152 | 1406.0 | 3932.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 6_25 | 1406.0 | 3932.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 123_134 | 1444.0 | 3970.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 147_152 | 1444.0 | 3970.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 6_25 | 1444.0 | 3970.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 123_134 | 1444.0 | 3973.0 | Motif | Integrase domain-binding motif 1 (IBM1) | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 147_152 | 1444.0 | 3973.0 | Motif | Integrase domain-binding motif 2 (IBM2) | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 6_25 | 1444.0 | 3973.0 | Motif | Menin-binding motif (MBM) | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1038_1066 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1106_1166 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1200_1375 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 132_253 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1_108 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 301_352 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 445_585 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 713_780 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 798_949 | 1406.0 | 3932.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1038_1066 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1106_1166 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1200_1375 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 132_253 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1_108 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 301_352 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 445_585 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 713_780 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 798_949 | 1444.0 | 3970.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1038_1066 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1106_1166 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1200_1375 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 132_253 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1_108 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 301_352 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 445_585 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 713_780 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 798_949 | 1444.0 | 3973.0 | Region | Disordered | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000354520 | 8 | 35 | 1147_1195 | 1406.0 | 3932.0 | Zinc finger | CXXC-type | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1147_1195 | 1444.0 | 3970.0 | Zinc finger | CXXC-type | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000389506 | 9 | 36 | 1431_1482 | 1444.0 | 3970.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1147_1195 | 1444.0 | 3973.0 | Zinc finger | CXXC-type | |
| Tgene | KMT2A | chr3:188478070 | chr11:118359328 | ENST00000534358 | 9 | 36 | 1431_1482 | 1444.0 | 3973.0 | Zinc finger | PHD-type 1 |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (1243) | LPP | chr3 | 188478070 | + | KMT2A | chr11 | 118359328 | MRLRDRAEDRTWSSDSLCSAFFIVPLDAHQREDLFPQLIPTMSHPSWLPP KSTGEPLGHVPARMETTHSFGNPSISVSTQQPPKKFAPVVAPKPKYNPYK QPGGEGDFLPPPPPPLDDSSALPSISGNFPPPPPLDEEAFKVQGNPGGKT LEERRSSLDAEIDSLTSILADLECSSPYKPRPPQSSTGSTASPPVSTPVT GHKRMVIPNQPPLTATKKSTLKPQPAPQAGPIPVAPIGTLKPQPQPVPAS YTTASTSSRPTFNVQVKSAQPSPHYMAAPSSGQIYGSGPQGYNTQPVPVS GQCPPPSTRGGMDYAYIPPPGLQPEPGYGYAPNQGRYYEGYYAAGPGYGG RNDSDPTYGQQGHPNTWKREPGYTPPGAGNQNPPGMYPVTGPKKTYITDP VSAPCAPPLQPKGGHSGQLGPSSVAPSFRPEDELEHLTKKMLYDMENPPA DEYFGRCARCGENVVGEGTGCTAMDQVFHVDCFTCIICNNKLRGQPFYAV EKKAYCEPCYIFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCH VCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRC KSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYES KMMQCGKCDRWVHSKCENLSDEMYEILSNLPESVAYTCVNCTERHPAEWR LALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESIPSR SSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDIVKI IQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPNKVS SNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPKGPG EPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYGDDS ANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQLRC EFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDLIKG EVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDCLGI LNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVEPDI NSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQTSGS CYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGDPLL SSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTTGTA TDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHLDGS SSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPKLAP QVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRDQHT DSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGKKSC KETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCNNVS SDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKMENES QSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSCQDS QSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFGLTP LYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLYYYN FTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSVTAT TRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKMDNC HSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLKSDS DNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEGLGL DSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSDLSV LTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPGVDP TPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPGLQV PVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIVVNQ NMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLTLTT GLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNPPSG LLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRKNKK LAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHRTVP NIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGSVSG LASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISNLLI KASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLPSTQ TTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQVSNF TQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSVPGP TKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSGTGT PGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESAEPK TVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQICA ESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFLIEQ LSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNFLAS KHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEAVGV YRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGIGCY MFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFAMRK | 3036 | |
| PDB file (1244) | LPP | chr3 | 188478070 | + | KMT2A | chr11 | 118359328 | MRLRDRAEDRTWSSDSLCSAFFIVPLDAHQREDLFPQLIPTMSHPSWLPP KSTGEPLGHVPARMETTHSFGNPSISVSTQQPPKKFAPVVAPKPKYNPYK QPGGEGDFLPPPPPPLDDSSALPSISGNFPPPPPLDEEAFKVQGNPGGKT LEERRSSLDAEIDSLTSILADLECSSPYKPRPPQSSTGSTASPPVSTPVT GHKRMVIPNQPPLTATKKSTLKPQPAPQAGPIPVAPIGTLKPQPQPVPAS YTTASTSSRPTFNVQVKSAQPSPHYMAAPSSGQIYGSGPQGYNTQPVPVS GQCPPPSTRGGMDYAYIPPPGLQPEPGYGYAPNQGRYYEGYYAAGPGYGG RNDSDPTYGQQGHPNTWKREPGYTPPGAGNQNPPGMYPVTGPKKTYITDP VSAPCAPPLQPKGGHSGQLGPSSVAPSFRPEDELEHLTKKMLYDMENPPA DEYFGRCARCGENVVGEGTGCTAMDQVFHVDCFTCIICNNKLRGQPFYAV EKKAYCEPCYIFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCKFCH VCGRQHQATKQLLECNKCRNSYHPECLGPNYPTKPTKKKKVWICTKCVRC KSCGSTTPGKGWDAQWSHDFSLCHDCAKLFAKGNFCPLCDKCYDDDDYES KMMQCGKCDRWVHSKCENLSGTEDEMYEILSNLPESVAYTCVNCTERHPA EWRLALEKELQISLKQVLTALLNSRTTSHLLRYRQAAKPPDLNPETEESI PSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRKMDQGNYTSVLEFSDDI VKIIQAAINSDGGQPEIKKANSMVKSFFIRQMERVFPWFSVKKSRFWEPN KVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHTEQPPLMKKIIPAPKPK GPGEPDSPTPLHPPTPPILSTDRSREDSPELNPPPGIEDNRQCALCLTYG DDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDDDGSLKNVHMAVIRGKQ LRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNCVFLDDKKVYCQRHRDL IKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGLEPENIHMMIGSMTIDC LGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKRCVYTCKIVECRPPVVE PDINSTVEHDENRTIAHSPTSFTESSSKESQNTAEIISPPSPDRPPHSQT SGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPSAGSPTPTTHEIVTVGD PLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMRTGNTYSRNNVSSVSTT GTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSSNLQRTVVTVGNKNSHL DGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSKSSEGSAHNVAYPGIPK LAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEALSFPHLHLRGQRNDRD QHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSIINEHMGSSSRDRRQKGK KSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEFMDEVLTPEYMGQRPCN NVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQAPRKRTVKVTLTPLKME NESQSKNALKESSPASPLQIESTSPTEPISASENPGDGPVAQPSPNNTSC QDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKRRYPRRSARARSNMFFG LTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQVDGADDLSTSDEDDLY YYNFTRTVISSGGEERLASHNLFREEEQCDLPKISQLDGVDDGTESDTSV TATTRKSSQIPKRNGKENGTENLKIDRPEDAGEKEHVTKSSVGHKNEPKM DNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTSTPSDKNLLDTYNTELLK SDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALGESPESSSSELLNLGEG LGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSSISAEEQFELPLELPSD LSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITEKSVASSESDPALLSPG VDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQGHGNNQDLTRNSSTPG LQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAVQTTPPHLKPATEKLIV VNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSVMETNTSVLGPMGGGLT LTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHSFPAATQSSFPPNISNP PSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATPSSGLKKRPISRLQTRK NKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPNHPSLLDLGSLNTSSHR TVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAAGTSTISQDTSHLTSGS VSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTLTNPRLLGTPDIGSISN LLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQTPSTAAITAASSICVLP STQTTGITAASPSGEADEHYQLQHVNQLLASKTGIHSSQRDLDSASGPQV SNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGGSPSSPSSGQRSASPSV PGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSSSEAHIPDQETTSLTSG TGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVLPEVQVTQNPANEQESA EPKTVEEEESNFSSPLMLWLQQEQKRKESITEKKPKKGLVFEISSDDGFQ ICAESIEDAWKSLTDKVQEARSNARLKQLSFAGVNGLRMLGILHDAVVFL IEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSARAEVHLRKSAFDMFNF LASKHRQPPEYNPNDEEEEEVQLKSARRATSMDLPMPMRFRHLKKTSKEA VGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIRSIQTDKREKYYDSKGI GCYMFRIDDSEVVDATMHGNAARFINHSCEPNCYSRVINIDGQKHIVIFA | 3039 | |
| PDB file (1246) | LPP | chr3 | 188478070 | + | KMT2A | chr11 | 118359328 | MRLRDRAEDRTWSSDSLCSAFFIVPLDAHQREDLFPQLHCSWLNLVSTIH SRSQLSEIPTMSHPSWLPPKSTGEPLGHVPARMETTHSFGNPSISVSTQQ PPKKFAPVVAPKPKYNPYKQPGGEGDFLPPPPPPLDDSSALPSISGNFPP PPPLDEEAFKVQGNPGGKTLEERRSSLDAEIDSLTSILADLECSSPYKPR PPQSSTGSTASPPVSTPVTGHKRMVIPNQPPLTATKKSTLKPQPAPQAGP IPVAPIGTLKPQPQPVPASYTTASTSSRPTFNVQVKSAQPSPHYMAAPSS GQIYGSGPQGYNTQPVPVSGQCPPPSTRGGMDYAYIPPPGLQPEPGYGYA PNQGRYYEGYYAAGPGYGGRNDSDPTYGQQGHPNTWKREPGYTPPGAGNQ NPPGMYPVTGPKKTYITDPVSAPCAPPLQPKGGHSGQLGPSSVAPSFRPE DELEHLTKKMLYDMENPPADEYFGRCARCGENVVGEGTGCTAMDQVFHVD CFTCIICNNKLRGQPFYAVEKKAYCEPCYIFVYCQVCCEPFHKFCLEENE RPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNY PTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFA KGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSDEMYEILSNLP ESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLLRYR QAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGVKRK MDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQMER VFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREENSHT EQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPELNPP PGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVFEDD DGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRAKNC VFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFLNGL EPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDARKR CVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQNTA EIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRPLPS AGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMSPMR TGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTSTSS NLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVLSSK SSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSSKEA LSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRSSII NEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLKPEF MDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKELQA PRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISASEN PGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGSFKR RYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSAEGQ VDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDLPKI SQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDAGEK EHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVHTST PSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQALG ESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSVSSS ISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVTITE KSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGSVEQ GHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISNAAV QTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSSTPSV METNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQHLHS FPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTVATP SSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQLPN HPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAATAA GTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGHVTL TNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGTSQT PSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLASKTG IHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTSPGG SPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLRTSS SEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQVAVL PEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESITEKK PKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSFAGV NGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPHGSA RAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATSMDL PMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGNVIR SIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEPNCY SRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCNCGAKKC | 3055 | |
| PDB file (1248) | LPP | chr3 | 188478070 | + | KMT2A | chr11 | 118359328 | MRLRDRAEDRTWSSDSLCSAFFIVPLDAHQREDLFPQLHCSWLNLVSTIH SRSQLSEIPTMSHPSWLPPKSTGEPLGHVPARMETTHSFGNPSISVSTQQ PPKKFAPVVAPKPKYNPYKQPGGEGDFLPPPPPPLDDSSALPSISGNFPP PPPLDEEAFKVQGNPGGKTLEERRSSLDAEIDSLTSILADLECSSPYKPR PPQSSTGSTASPPVSTPVTGHKRMVIPNQPPLTATKKSTLKPQPAPQAGP IPVAPIGTLKPQPQPVPASYTTASTSSRPTFNVQVKSAQPSPHYMAAPSS GQIYGSGPQGYNTQPVPVSGQCPPPSTRGGMDYAYIPPPGLQPEPGYGYA PNQGRYYEGYYAAGPGYGGRNDSDPTYGQQGHPNTWKREPGYTPPGAGNQ NPPGMYPVTGPKKTYITDPVSAPCAPPLQPKGGHSGQLGPSSVAPSFRPE DELEHLTKKMLYDMENPPADEYFGRCARCGENVVGEGTGCTAMDQVFHVD CFTCIICNNKLRGQPFYAVEKKAYCEPCYIFVYCQVCCEPFHKFCLEENE RPLEDQLENWCCRRCKFCHVCGRQHQATKQLLECNKCRNSYHPECLGPNY PTKPTKKKKVWICTKCVRCKSCGSTTPGKGWDAQWSHDFSLCHDCAKLFA KGNFCPLCDKCYDDDDYESKMMQCGKCDRWVHSKCENLSGTEDEMYEILS NLPESVAYTCVNCTERHPAEWRLALEKELQISLKQVLTALLNSRTTSHLL RYRQAAKPPDLNPETEESIPSRSSPEGPDPPVLTEVSKQDDQQPLDLEGV KRKMDQGNYTSVLEFSDDIVKIIQAAINSDGGQPEIKKANSMVKSFFIRQ MERVFPWFSVKKSRFWEPNKVSSNSGMLPNAVLPPSLDHNYAQWQEREEN SHTEQPPLMKKIIPAPKPKGPGEPDSPTPLHPPTPPILSTDRSREDSPEL NPPPGIEDNRQCALCLTYGDDSANDAGRLLYIGQNEWTHVNCALWSAEVF EDDDGSLKNVHMAVIRGKQLRCEFCQKPGATVGCCLTSCTSNYHFMCSRA KNCVFLDDKKVYCQRHRDLIKGEVVPENGFEVFRRVFVDFEGISLRRKFL NGLEPENIHMMIGSMTIDCLGILNDLSDCEDKLFPIGYQCSRVYWSTTDA RKRCVYTCKIVECRPPVVEPDINSTVEHDENRTIAHSPTSFTESSSKESQ NTAEIISPPSPDRPPHSQTSGSCYYHVISKVPRIRTPSYSPTQRSPGCRP LPSAGSPTPTTHEIVTVGDPLLSSGLRSIGSRRHSTSSLSPQRSKLRIMS PMRTGNTYSRNNVSSVSTTGTATDLESSAKVVDHVLGPLNSSTSLGQNTS TSSNLQRTVVTVGNKNSHLDGSSSSEMKQSSASDLVSKSSSLKGEKTKVL SSKSSEGSAHNVAYPGIPKLAPQVHNTTSRELNVSKIGSFAEPSSVSFSS KEALSFPHLHLRGQRNDRDQHTDSTQSANSSPDEDTEVKTLKLSGMSNRS SIINEHMGSSSRDRRQKGKKSCKETFKEKHSSKSFLEPGQVTTGEEGNLK PEFMDEVLTPEYMGQRPCNNVSSDKIGDKGLSMPGVPKAPPMQVEGSAKE LQAPRKRTVKVTLTPLKMENESQSKNALKESSPASPLQIESTSPTEPISA SENPGDGPVAQPSPNNTSCQDSQSNNYQNLPVQDRNLMLPDGPKPQEDGS FKRRYPRRSARARSNMFFGLTPLYGVRSYGEEDIPFYSSSTGKKRGKRSA EGQVDGADDLSTSDEDDLYYYNFTRTVISSGGEERLASHNLFREEEQCDL PKISQLDGVDDGTESDTSVTATTRKSSQIPKRNGKENGTENLKIDRPEDA GEKEHVTKSSVGHKNEPKMDNCHSVSRVKTQGQDSLEAQLSSLESSRRVH TSTPSDKNLLDTYNTELLKSDSDNNNSDDCGNILPSDIMDFVLKNTPSMQ ALGESPESSSSELLNLGEGLGLDSNREKDMGLFEVFSQQLPTTEPVDSSV SSSISAEEQFELPLELPSDLSVLTTRSPTVPSQNPSRLAVISDSGEKRVT ITEKSVASSESDPALLSPGVDPTPEGHMTPDHFIQGHMDADHISSPPCGS VEQGHGNNQDLTRNSSTPGLQVPVSPTVPIQNQKYVPNSTDSPGPSQISN AAVQTTPPHLKPATEKLIVVNQNMQPLYVLQTLPNGVTQKIQLTSSVSST PSVMETNTSVLGPMGGGLTLTTGLNPSLPTSQSLFPSASKGLLPMSHHQH LHSFPAATQSSFPPNISNPPSGLLIGVQPPPDPQLLVSESSQRTDLSTTV ATPSSGLKKRPISRLQTRKNKKLAPSSTPSNIAPSDVVSNMTLINFTPSQ LPNHPSLLDLGSLNTSSHRTVPNIIKRSKSSIMYFEPAPLLPQSVGGTAA TAAGTSTISQDTSHLTSGSVSGLASSSSVLNVVSMQTTTTPTSSASVPGH VTLTNPRLLGTPDIGSISNLLIKASQQSLGIQDQPVALPPSSGMFPQLGT SQTPSTAAITAASSICVLPSTQTTGITAASPSGEADEHYQLQHVNQLLAS KTGIHSSQRDLDSASGPQVSNFTQTVDAPNSMGLEQNKALSSAVQASPTS PGGSPSSPSSGQRSASPSVPGPTKPKPKTKRFQLPLDKGNGKKHKVSHLR TSSSEAHIPDQETTSLTSGTGTPGAEAEQQDTASVEQSSQKECGQPAGQV AVLPEVQVTQNPANEQESAEPKTVEEEESNFSSPLMLWLQQEQKRKESIT EKKPKKGLVFEISSDDGFQICAESIEDAWKSLTDKVQEARSNARLKQLSF AGVNGLRMLGILHDAVVFLIEQLSGAKHCRNYKFRFHKPEEANEPPLNPH GSARAEVHLRKSAFDMFNFLASKHRQPPEYNPNDEEEEEVQLKSARRATS MDLPMPMRFRHLKKTSKEAVGVYRSPIHGRGLFCKRNIDAGEMVIEYAGN VIRSIQTDKREKYYDSKGIGCYMFRIDDSEVVDATMHGNAARFINHSCEP NCYSRVINIDGQKHIVIFAMRKIYRGEELTYDYKFPIEDASNKLPCNCGA | 3058 |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
LPP_pLDDT.png |
KMT2A_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
LPP_KMT2A_1246_pLDDT_and_active_sites.png (AA BP:)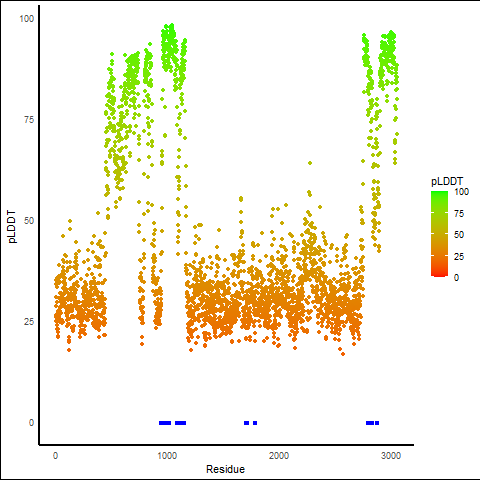 |
LPP_KMT2A_1248_pLDDT_and_active_sites.png (AA BP:)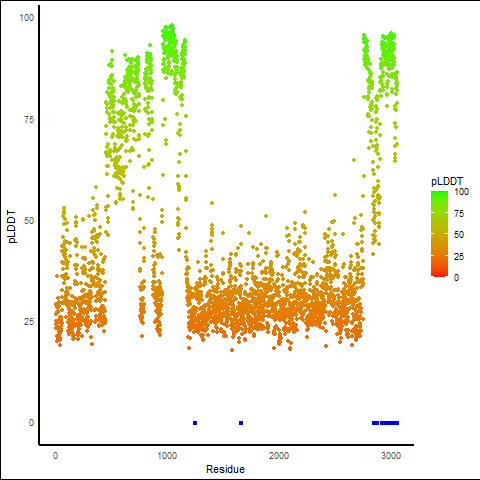 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 1246 | 1.021 | 303 | 1.075 | 1138.417 | 0.694 | 0.662 | 0.783 | 0.587 | 0.804 | 0.729 | 0.709 | Chain A: 946,947,948,949,954,955,956,957,958,960,9 65,977,978,980,981,982,983,984,1015,1084,1085,1087 ,1088,1089,1090,1091,1092,1093,1096,1100,1104,1105 ,1106,1107,1108,1109,1110,1111,1112,1113,1138,1139 ,1140,1141,1142,1150,1152,1700,1701,1702,1703,1704 ,1708,1783,1784,1785,1786,1787,2795,2796,2797,2798 ,2799,2800,2811,2812,2815,2816,2833,2871 |
| 1248 | 1.001 | 349 | 1.013 | 1085.595 | 0.566 | 0.7 | 0.933 | 0.402 | 1.063 | 0.378 | 0.848 | Chain A: 1662,1663,2850,2856,2857,2858,2859,2860,2 861,2862,2864,2865,2866,2867,2868,2871,2873,2875,2 876,2877,2924,2925,2926,2927,2928,2929,2930,2947,2 954,2957,2968,2969,2970,2971,2972,2973,2974,2975,2 981,2985,2986,2991,2992,2993,2994,2995,2996,3000,3 003,3004,3005,3006,3015,3031,3032,3033,3034,3045,3 046,3047,3048,3057 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| KMT2A | PPIE, PPP1R15A, KMT2A, ASH2L, HCFC1, HCFC2, MEN1, RBBP5, WDR5, AVP, INS, OXT, MAP3K5, HDAC1, CTBP1, CBX4, BMI1, CREBBP, SMARCB1, CXXC1, MYB, CTNNB1, SNW1, E2F2, E2F4, E2F6, PSIP1, MLLT4, POLR2A, KAT8, RNF2, TP53, tat, SBF1, MTM1, SET, HIST1H3A, HIST1H4A, KAT6A, ELL, AFF1, AFF4, CDK9, CCNT1, CTR9, LEO1, PAF1, CDC73, WDR61, MLLT3, DOT1L, SKP2, HIST3H3, SVIL, HIST2H3C, SIN3A, MLLT1, RUNX1, CBFB, H3F3A, SIRT7, ASB2, TCEB1, TCEB2, CBX8, TOP1, TAF6, NCL, HECW2, LGR4, CSNK2A2, SENP3, SYMPK, PKN1, PIH1D1, KRAS, TAF1, CHD3, SMARCA2, SMARCC2, SMARCC1, HDAC2, RBBP4, RBBP7, TBP, MBD3, SAP30, RAN, TAF9, TASP1, HIST1H2BG, EWSR1, DYNLT1, KIF11, ING4, Cbx1, Set, ZNF131, ASB7, AR, CSNK2A1, OGT, BRD4, KDM5B, KDM5D, KDM6B, CKS1B, CKS2, CHD4, ESR2, AGR2, EZH2, FBXW7, SOX2, MED17, WDR82, ACTR8, GEMIN5, RUVBL1, MCM2, RTF1, MED15, MED8, MRGBP, MED14, HIST1H2BB, HIST1H2AB, KIAA1429, SP1, MYC, NR2C2, IRF2, PLEKHA4, DPY30, PTEN, NPM1, RPL13, CCT6A, TMCO1, ITGB1, ESR1, MYCN, SUPT5H, CIT, Pik3r2, FASN, LDLR, CDC27, KMT2B, SETD1A, KDM6A, KANSL2, BUB3, SETD1B, KANSL1, RERE, MCRS1, KMT2C, ZZZ3, MBIP, YEATS2, PAXIP1, KANSL3, KMT2D, BOD1L1, CSRP2BP, ZNF608, ATN1, PHF20L1, PHF20, NCOA6, TADA3, APEX1, ASF1A, CBX3, CD3EAP, CENPA, NUP50, POLR1E, TERF2IP, ZNF330, NAA40, PRKRA, SRSF4, KRR1, RPL13A, ABT1, CSNK2B, OASL, SRSF5, SRSF6, RPS9, FGFBP1, RPLP0, RPL36, KAL1, RPL3, PML, ELF1, ELF2, FEV, NHLH1, EN1, KLF12, KLF3, NFIX, NFYC, YY1, BRD3, |
| LPP | ATP5J2, PPP2R1A, ELAVL1, EGLN2, VHL, CLNS1A, PNPO, HNRNPL, CPT1A, HSPA5, AGFG1, ALAD, ARPC3, ARPC4, CALR, DBNL, HSPBP1, IPO11, P3H1, DPP9, EFTUD1, FEN1, FKBP10, RPRD1B, SAE1, SURF2, UFM1, NIF3L1, NPLOC4, OGT, PPME1, PPP2R5D, RANGAP1, RDX, SH3GLB1, TPRKB, UBE3A, ZPR1, SUZ12, RNF2, COPS5, GMPPA, PICALM, NINL, XPO1, Sestd1, Ppp2r3a, CDH1, PTPN21, PPP2R3A, WDYHV1, SERPINB2, PDLIM7, LPXN, CBLC, MRPL38, WASF3, MTNR1A, G3BP1, TES, EFTUD2, ESR2, KIAA1429, HDAC2, DYRK1A, SH2D3C, DCP1A, RIN3, ACTB, EZR, GFAP, IMPDH2, KRT19, MAPRE1, PEX14, PFN1, PRPH, SQSTM1, SYNE3, TJP2, VASP, VCL, VIM, ZYX, NAA40, KIAA0408, SLC9A3R2, AJUBA, PLEKHG2, COQ3, SLU7, FHL2, FBXW7, nsp14, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| LPP |  |
| KMT2A |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to LPP-KMT2A |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to LPP-KMT2A |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | LPP | C0087031 | Juvenile-Onset Still Disease | 1 | CTD_human |
| Hgene | LPP | C3495559 | Juvenile arthritis | 1 | CTD_human |
| Hgene | LPP | C3714758 | Juvenile psoriatic arthritis | 1 | CTD_human |
| Hgene | LPP | C4552091 | Polyarthritis, Juvenile, Rheumatoid Factor Negative | 1 | CTD_human |
| Hgene | LPP | C4704862 | Polyarthritis, Juvenile, Rheumatoid Factor Positive | 1 | CTD_human |
| Tgene | KMT2A | C2826025 | Mixed phenotype acute leukemia | 3 | ORPHANET |
| Tgene | KMT2A | C0023418 | leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023452 | Childhood Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023453 | L2 Acute Lymphoblastic Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0023466 | Leukemia, Monocytic, Chronic | 2 | CTD_human |
| Tgene | KMT2A | C0023467 | Leukemia, Myelocytic, Acute | 2 | CTD_human |
| Tgene | KMT2A | C0023470 | Myeloid Leukemia | 2 | CTD_human |
| Tgene | KMT2A | C0026998 | Acute Myeloid Leukemia, M1 | 2 | CTD_human |
| Tgene | KMT2A | C1854630 | Growth Deficiency and Mental Retardation with Facial Dysmorphism | 2 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | KMT2A | C1879321 | Acute Myeloid Leukemia (AML-M2) | 2 | CTD_human |
| Tgene | KMT2A | C1961102 | Precursor Cell Lymphoblastic Leukemia Lymphoma | 2 | CTD_human |
| Tgene | KMT2A | C0001418 | Adenocarcinoma | 1 | CTD_human |
| Tgene | KMT2A | C0004403 | Autosome Abnormalities | 1 | CTD_human |
| Tgene | KMT2A | C0005684 | Malignant neoplasm of urinary bladder | 1 | CTD_human |
| Tgene | KMT2A | C0005695 | Bladder Neoplasm | 1 | CTD_human |
| Tgene | KMT2A | C0007138 | Carcinoma, Transitional Cell | 1 | CTD_human |
| Tgene | KMT2A | C0008625 | Chromosome Aberrations | 1 | CTD_human |
| Tgene | KMT2A | C0023448 | Lymphoid leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0023465 | Acute monocytic leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0023479 | Acute myelomonocytic leukemia | 1 | CTD_human |
| Tgene | KMT2A | C0024623 | Malignant neoplasm of stomach | 1 | CTD_human |
| Tgene | KMT2A | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Tgene | KMT2A | C0036341 | Schizophrenia | 1 | PSYGENET |
| Tgene | KMT2A | C0038356 | Stomach Neoplasms | 1 | CTD_human |
| Tgene | KMT2A | C0149925 | Small cell carcinoma of lung | 1 | CTD_human |
| Tgene | KMT2A | C0205641 | Adenocarcinoma, Basal Cell | 1 | CTD_human |
| Tgene | KMT2A | C0205642 | Adenocarcinoma, Oxyphilic | 1 | CTD_human |
| Tgene | KMT2A | C0205643 | Carcinoma, Cribriform | 1 | CTD_human |
| Tgene | KMT2A | C0205644 | Carcinoma, Granular Cell | 1 | CTD_human |
| Tgene | KMT2A | C0205645 | Adenocarcinoma, Tubular | 1 | CTD_human |
| Tgene | KMT2A | C0270972 | Cornelia De Lange Syndrome | 1 | ORPHANET |
| Tgene | KMT2A | C0280141 | Acute Undifferentiated Leukemia | 1 | ORPHANET |
| Tgene | KMT2A | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| Tgene | KMT2A | C0856823 | Undifferentiated type acute leukemia | 1 | ORPHANET |
| Tgene | KMT2A | C1535926 | Neurodevelopmental Disorders | 1 | CTD_human |
| Tgene | KMT2A | C1708349 | Hereditary Diffuse Gastric Cancer | 1 | CTD_human |
| Tgene | KMT2A | C2239176 | Liver carcinoma | 1 | CTD_human |
| Tgene | KMT2A | C2930974 | Acute erythroleukemia | 1 | CTD_human |
| Tgene | KMT2A | C2930975 | Acute erythroleukemia - M6a subtype | 1 | CTD_human |
| Tgene | KMT2A | C2930976 | Acute myeloid leukemia FAB-M6 | 1 | CTD_human |
| Tgene | KMT2A | C2930977 | Acute erythroleukemia - M6b subtype | 1 | CTD_human |

