| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:MAGI3-NOTCH2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: MAGI3-NOTCH2 | FusionPDB ID: 50820 | FusionGDB2.0 ID: 50820 | Hgene | Tgene | Gene symbol | MAGI3 | NOTCH2 | Gene ID | 260425 | 4853 |
| Gene name | membrane associated guanylate kinase, WW and PDZ domain containing 3 | notch receptor 2 | |
| Synonyms | MAGI-3|dJ730K3.2 | AGS2|HJCYS|hN2 | |
| Cytomap | 1p13.2 | 1p12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | membrane-associated guanylate kinase, WW and PDZ domain-containing protein 3membrane-associated guanylate kinase inverted 3membrane-associated guanylate kinase-related 3 | neurogenic locus notch homolog protein 2Notch homolog 2notch 2 | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | Q5TCQ9 | P0DPK3 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000486456, ENST00000307546, ENST00000369611, ENST00000369615, ENST00000369617, | ENST00000256646, ENST00000602566, ENST00000493703, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 12 X 8 X 5=480 | 13 X 10 X 6=780 |
| # samples | 12 | 14 | |
| ** MAII score | log2(12/480*10)=-2 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(14/780*10)=-2.47804729680464 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: MAGI3 [Title/Abstract] AND NOTCH2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MAGI3(113933971)-NOTCH2(120539955), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | MAGI3-NOTCH2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MAGI3-NOTCH2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MAGI3-NOTCH2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MAGI3-NOTCH2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | NOTCH2 | GO:0007050 | cell cycle arrest | 11306509 |
| Tgene | NOTCH2 | GO:0007219 | Notch signaling pathway | 11306509|25985737 |
| Tgene | NOTCH2 | GO:0010629 | negative regulation of gene expression | 11306509 |
| Tgene | NOTCH2 | GO:0010838 | positive regulation of keratinocyte proliferation | 18469519 |
| Tgene | NOTCH2 | GO:0045967 | negative regulation of growth rate | 11306509 |
| Tgene | NOTCH2 | GO:0046579 | positive regulation of Ras protein signal transduction | 11306509 |
| Tgene | NOTCH2 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 11306509 |
| Tgene | NOTCH2 | GO:2000249 | regulation of actin cytoskeleton reorganization | 18469519 |
 Fusion gene breakpoints across MAGI3 (5'-gene) Fusion gene breakpoints across MAGI3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
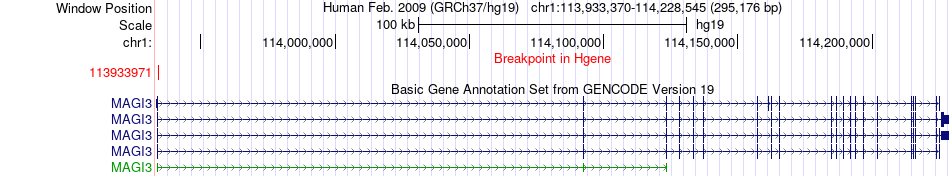 |
 Fusion gene breakpoints across NOTCH2 (3'-gene) Fusion gene breakpoints across NOTCH2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
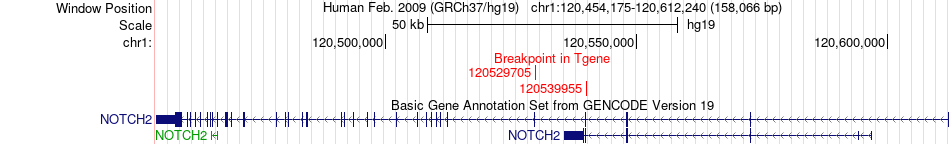 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-BH-A0W3-01A | MAGI3 | chr1 | 113933971 | - | NOTCH2 | chr1 | 120529705 | - |
| ChimerDB4 | BRCA | TCGA-BH-A0W3-01A | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120529705 | - |
| ChimerDB4 | BRCA | TCGA-BH-A0W3-01A | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120539955 | - |
| ChimerDB4 | BRCA | TCGA-BH-A0W3 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120539955 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000369617 | MAGI3 | chr1 | 113933971 | + | ENST00000256646 | NOTCH2 | chr1 | 120539955 | - | 11355 | 601 | 285 | 7601 | 2438 |
| ENST00000369617 | MAGI3 | chr1 | 113933971 | + | ENST00000602566 | NOTCH2 | chr1 | 120539955 | - | 4903 | 601 | 285 | 1010 | 241 |
| ENST00000307546 | MAGI3 | chr1 | 113933971 | + | ENST00000256646 | NOTCH2 | chr1 | 120539955 | - | 11145 | 391 | 75 | 7391 | 2438 |
| ENST00000307546 | MAGI3 | chr1 | 113933971 | + | ENST00000602566 | NOTCH2 | chr1 | 120539955 | - | 4693 | 391 | 75 | 800 | 241 |
| ENST00000369615 | MAGI3 | chr1 | 113933971 | + | ENST00000256646 | NOTCH2 | chr1 | 120539955 | - | 11132 | 378 | 62 | 7378 | 2438 |
| ENST00000369615 | MAGI3 | chr1 | 113933971 | + | ENST00000602566 | NOTCH2 | chr1 | 120539955 | - | 4680 | 378 | 62 | 787 | 241 |
| ENST00000369611 | MAGI3 | chr1 | 113933971 | + | ENST00000256646 | NOTCH2 | chr1 | 120539955 | - | 11115 | 361 | 45 | 7361 | 2438 |
| ENST00000369611 | MAGI3 | chr1 | 113933971 | + | ENST00000602566 | NOTCH2 | chr1 | 120539955 | - | 4663 | 361 | 45 | 770 | 241 |
| ENST00000369617 | MAGI3 | chr1 | 113933971 | + | ENST00000256646 | NOTCH2 | chr1 | 120529705 | - | 11019 | 601 | 285 | 7265 | 2326 |
| ENST00000307546 | MAGI3 | chr1 | 113933971 | + | ENST00000256646 | NOTCH2 | chr1 | 120529705 | - | 10809 | 391 | 75 | 7055 | 2326 |
| ENST00000369615 | MAGI3 | chr1 | 113933971 | + | ENST00000256646 | NOTCH2 | chr1 | 120529705 | - | 10796 | 378 | 62 | 7042 | 2326 |
| ENST00000369611 | MAGI3 | chr1 | 113933971 | + | ENST00000256646 | NOTCH2 | chr1 | 120529705 | - | 10779 | 361 | 45 | 7025 | 2326 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000369617 | ENST00000256646 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120539955 | - | 0.000944121 | 0.9990559 |
| ENST00000369617 | ENST00000602566 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120539955 | - | 0.008637389 | 0.99136263 |
| ENST00000307546 | ENST00000256646 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120539955 | - | 0.000761924 | 0.99923813 |
| ENST00000307546 | ENST00000602566 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120539955 | - | 0.007772417 | 0.9922276 |
| ENST00000369615 | ENST00000256646 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120539955 | - | 0.000747064 | 0.99925286 |
| ENST00000369615 | ENST00000602566 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120539955 | - | 0.007636217 | 0.9923638 |
| ENST00000369611 | ENST00000256646 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120539955 | - | 0.000734457 | 0.9992655 |
| ENST00000369611 | ENST00000602566 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120539955 | - | 0.007701264 | 0.9922988 |
| ENST00000369617 | ENST00000256646 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120529705 | - | 0.00095522 | 0.9990447 |
| ENST00000307546 | ENST00000256646 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120529705 | - | 0.000776845 | 0.9992231 |
| ENST00000369615 | ENST00000256646 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120529705 | - | 0.000762035 | 0.999238 |
| ENST00000369611 | ENST00000256646 | MAGI3 | chr1 | 113933971 | + | NOTCH2 | chr1 | 120529705 | - | 0.000748808 | 0.9992512 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >50820_50820_1_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000307546_NOTCH2_chr1_120529705_ENST00000256646_length(amino acids)=2326AA_BP=618 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGFEGSTCERNIDDCPNHRCQNGGVCVDGVNTYNCRCPPQWTGQFCTEDVDECLLQPNACQNGGTCANRNGGYGCV CVNGWSGDDCSENIDDCAFASCTPGSTCIDRVASFSCMCPEGKAGLLCHLDDACISNPCHKGALCDTNPLNGQYICTCPQGYKGADCTED VDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEMDINECHSDPCQNDATCLDKIGGFTCLCMPGFKGVHCELEINECQSNPCVN NGQCVDKVNRFQCLCPPGFTGPVCQIDIDDCSSTPCLNGAKCIDHPNGYECQCATGFTGVLCEENIDNCDPDPCHHGQCQDGIDSYTCIC NPGYMGAICSDQIDECYSSPCLNDGRCIDLVNGYQCNCQPGTSGVNCEINFDDCASNPCIHGICMDGINRYSCVCSPGFTGQRCNIDIDE CASNPCRKGATCINGVNGFRCICPEGPHHPSCYSQVNECLSNPCIHGNCTGGLSGYKCLCDAGWVGINCEVDKNECLSNPCQNGGTCDNL VNGYRCTCKKGFKGYNCQVNIDECASNPCLNQGTCFDDISGYTCHCVLPYTGKNCQTVLAPCSPNPCENAAVCKESPNFESYTCLCAPGW QGQRCTIDIDECISKPCMNHGLCHNTQGSYMCECPPGFSGMDCEEDIDDCLANPCQNGGSCMDGVNTFSCLCLPGFTGDKCQTDMNECLS EPCKNGGTCSDYVNSYTCKCQAGFDGVHCENNINECTESSCFNGGTCVDGINSFSCLCPVGFTGSFCLHEINECSSHPCLNEGTCVDGLG TYRCSCPLGYTGKNCQTLVNLCSRSPCKNKGTCVQKKAESQCLCPSGWAGAYCDVPNVSCDIAASRRGVLVEHLCQHSGVCINAGNTHYC QCPLGYTGSYCEEQLDECASNPCQHGATCSDFIGGYRCECVPGYQGVNCEYEVDECQNQPCQNGGTCIDLVNHFKCSCPPGTRGLLCEEN IDDCARGPHCLNGGQCMDRIGGYSCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQLTNDYLCVCRSAFTGRHCETFVDVCPQMPCLN GGTCAVASNMPDGFICRCPPGFSGARCQSSCGQVKCRKGEQCVHTASGPRCFCPSPRDCESGCASSPCQHGGSCHPQRQPPYYSCQCAPP FSGSRCELYTAPPSTPPATCLSQYCADKARDGVCDEACNSHACQWDGGDCSLTMENPWANCSSPLPCWDYINNQCDELCNTVECLFDNFE CQGNSKTCKYDKYCADHFKDNHCDQGCNSEECGWDGLDCAADQPENLAEGTLVIVVLMPPEQLLQDARSFLRALGTLLHTNLRIKRDSQG ELMVYPYYGEKSAAMKKQRMTRRSLPGEQEQEVAGSKVFLEIDNRQCVQDSDHCFKNTDAAAALLASHAIQGTLSYPLVSVVSESLTPER TQLLYLLAVAVVIILFIILLGVIMAKRKRKHGSLWLPEGFTLRRDASNHKRREPVGQDAVGLKNLSVQVSEANLIGTGTSEHWVDDEGPQ PKKVKAEDEALLSEEDDPIDRRPWTQQHLEAADIRRTPSLALTPPQAEQEVDVLDVNVRGPDGCTPLMLASLRGGSSDLSDEDEDAEDSS ANIITDLVYQGASLQAQTDRTGEMALHLAARYSRADAAKRLLDAGADANAQDNMGRCPLHAAVAADAQGVFQILIRNRVTDLDARMNDGT TPLILAARLAVEGMVAELINCQADVNAVDDHGKSALHWAAAVNNVEATLLLLKNGANRDMQDNKEETPLFLAAREGSYEAAKILLDHFAN RDITDHMDRLPRDVARDRMHHDIVRLLDEYNVTPSPPGTVLTSALSPVICGPNRSFLSLKHTPMGKKSRRPSAKSTMPTSLPNLAKEAKD AKGSRRKKSLSEKVQLSESSVTLSPVDSLESPHTYVSDTTSSPMITSPGILQASPNPMLATAAPPAPVHAQHALSFSNLHEMQPLAHGAS TVLPSVSQLLSHHHIVSPGSGSAGSLSRLHPVPVPADWMNRMEVNETQYNEMFGMVLAPAEGTHPGIAPQSRPPEGKHITTPREPLPPIV TFQLIPKGSIAQPAGAPQPQSTCPPAVAGPLPTMYQIPEMARLPSVAFPTAMMPQQDGQVAQTILPAYHPFPASVGKYPTPPSQHSYASS -------------------------------------------------------------- >50820_50820_2_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000307546_NOTCH2_chr1_120539955_ENST00000256646_length(amino acids)=2438AA_BP=730 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGKECQWTDACLSHPCANGSTCTTVANQFSCKCLTGFTGQKCETDVNECDIPGHCQHGGTCLNLPGSYQCQCPQGF TGQYCDSLYVPCAPSPCVNGGTCRQTGDFTFECNCLPGFEGSTCERNIDDCPNHRCQNGGVCVDGVNTYNCRCPPQWTGQFCTEDVDECL LQPNACQNGGTCANRNGGYGCVCVNGWSGDDCSENIDDCAFASCTPGSTCIDRVASFSCMCPEGKAGLLCHLDDACISNPCHKGALCDTN PLNGQYICTCPQGYKGADCTEDVDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEMDINECHSDPCQNDATCLDKIGGFTCLCM PGFKGVHCELEINECQSNPCVNNGQCVDKVNRFQCLCPPGFTGPVCQIDIDDCSSTPCLNGAKCIDHPNGYECQCATGFTGVLCEENIDN CDPDPCHHGQCQDGIDSYTCICNPGYMGAICSDQIDECYSSPCLNDGRCIDLVNGYQCNCQPGTSGVNCEINFDDCASNPCIHGICMDGI NRYSCVCSPGFTGQRCNIDIDECASNPCRKGATCINGVNGFRCICPEGPHHPSCYSQVNECLSNPCIHGNCTGGLSGYKCLCDAGWVGIN CEVDKNECLSNPCQNGGTCDNLVNGYRCTCKKGFKGYNCQVNIDECASNPCLNQGTCFDDISGYTCHCVLPYTGKNCQTVLAPCSPNPCE NAAVCKESPNFESYTCLCAPGWQGQRCTIDIDECISKPCMNHGLCHNTQGSYMCECPPGFSGMDCEEDIDDCLANPCQNGGSCMDGVNTF SCLCLPGFTGDKCQTDMNECLSEPCKNGGTCSDYVNSYTCKCQAGFDGVHCENNINECTESSCFNGGTCVDGINSFSCLCPVGFTGSFCL HEINECSSHPCLNEGTCVDGLGTYRCSCPLGYTGKNCQTLVNLCSRSPCKNKGTCVQKKAESQCLCPSGWAGAYCDVPNVSCDIAASRRG VLVEHLCQHSGVCINAGNTHYCQCPLGYTGSYCEEQLDECASNPCQHGATCSDFIGGYRCECVPGYQGVNCEYEVDECQNQPCQNGGTCI DLVNHFKCSCPPGTRGLLCEENIDDCARGPHCLNGGQCMDRIGGYSCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQLTNDYLCVCR SAFTGRHCETFVDVCPQMPCLNGGTCAVASNMPDGFICRCPPGFSGARCQSSCGQVKCRKGEQCVHTASGPRCFCPSPRDCESGCASSPC QHGGSCHPQRQPPYYSCQCAPPFSGSRCELYTAPPSTPPATCLSQYCADKARDGVCDEACNSHACQWDGGDCSLTMENPWANCSSPLPCW DYINNQCDELCNTVECLFDNFECQGNSKTCKYDKYCADHFKDNHCDQGCNSEECGWDGLDCAADQPENLAEGTLVIVVLMPPEQLLQDAR SFLRALGTLLHTNLRIKRDSQGELMVYPYYGEKSAAMKKQRMTRRSLPGEQEQEVAGSKVFLEIDNRQCVQDSDHCFKNTDAAAALLASH AIQGTLSYPLVSVVSESLTPERTQLLYLLAVAVVIILFIILLGVIMAKRKRKHGSLWLPEGFTLRRDASNHKRREPVGQDAVGLKNLSVQ VSEANLIGTGTSEHWVDDEGPQPKKVKAEDEALLSEEDDPIDRRPWTQQHLEAADIRRTPSLALTPPQAEQEVDVLDVNVRGPDGCTPLM LASLRGGSSDLSDEDEDAEDSSANIITDLVYQGASLQAQTDRTGEMALHLAARYSRADAAKRLLDAGADANAQDNMGRCPLHAAVAADAQ GVFQILIRNRVTDLDARMNDGTTPLILAARLAVEGMVAELINCQADVNAVDDHGKSALHWAAAVNNVEATLLLLKNGANRDMQDNKEETP LFLAAREGSYEAAKILLDHFANRDITDHMDRLPRDVARDRMHHDIVRLLDEYNVTPSPPGTVLTSALSPVICGPNRSFLSLKHTPMGKKS RRPSAKSTMPTSLPNLAKEAKDAKGSRRKKSLSEKVQLSESSVTLSPVDSLESPHTYVSDTTSSPMITSPGILQASPNPMLATAAPPAPV HAQHALSFSNLHEMQPLAHGASTVLPSVSQLLSHHHIVSPGSGSAGSLSRLHPVPVPADWMNRMEVNETQYNEMFGMVLAPAEGTHPGIA PQSRPPEGKHITTPREPLPPIVTFQLIPKGSIAQPAGAPQPQSTCPPAVAGPLPTMYQIPEMARLPSVAFPTAMMPQQDGQVAQTILPAY HPFPASVGKYPTPPSQHSYASSNAAERTPSHSGHLQGEHPYLTPSPESPDQWSSSSPHSASDWSDVTTSPTPGGAGGGQRGPGTHMSEPP -------------------------------------------------------------- >50820_50820_3_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000307546_NOTCH2_chr1_120539955_ENST00000602566_length(amino acids)=241AA_BP=105 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGKECQWTDACLSHPCANGSTCTTVANQFSCKCLTGFTGQKCETDVNECDIPGHCQHGGTCLNLPGSYQCQCPQGF -------------------------------------------------------------- >50820_50820_4_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000369611_NOTCH2_chr1_120529705_ENST00000256646_length(amino acids)=2326AA_BP=618 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGFEGSTCERNIDDCPNHRCQNGGVCVDGVNTYNCRCPPQWTGQFCTEDVDECLLQPNACQNGGTCANRNGGYGCV CVNGWSGDDCSENIDDCAFASCTPGSTCIDRVASFSCMCPEGKAGLLCHLDDACISNPCHKGALCDTNPLNGQYICTCPQGYKGADCTED VDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEMDINECHSDPCQNDATCLDKIGGFTCLCMPGFKGVHCELEINECQSNPCVN NGQCVDKVNRFQCLCPPGFTGPVCQIDIDDCSSTPCLNGAKCIDHPNGYECQCATGFTGVLCEENIDNCDPDPCHHGQCQDGIDSYTCIC NPGYMGAICSDQIDECYSSPCLNDGRCIDLVNGYQCNCQPGTSGVNCEINFDDCASNPCIHGICMDGINRYSCVCSPGFTGQRCNIDIDE CASNPCRKGATCINGVNGFRCICPEGPHHPSCYSQVNECLSNPCIHGNCTGGLSGYKCLCDAGWVGINCEVDKNECLSNPCQNGGTCDNL VNGYRCTCKKGFKGYNCQVNIDECASNPCLNQGTCFDDISGYTCHCVLPYTGKNCQTVLAPCSPNPCENAAVCKESPNFESYTCLCAPGW QGQRCTIDIDECISKPCMNHGLCHNTQGSYMCECPPGFSGMDCEEDIDDCLANPCQNGGSCMDGVNTFSCLCLPGFTGDKCQTDMNECLS EPCKNGGTCSDYVNSYTCKCQAGFDGVHCENNINECTESSCFNGGTCVDGINSFSCLCPVGFTGSFCLHEINECSSHPCLNEGTCVDGLG TYRCSCPLGYTGKNCQTLVNLCSRSPCKNKGTCVQKKAESQCLCPSGWAGAYCDVPNVSCDIAASRRGVLVEHLCQHSGVCINAGNTHYC QCPLGYTGSYCEEQLDECASNPCQHGATCSDFIGGYRCECVPGYQGVNCEYEVDECQNQPCQNGGTCIDLVNHFKCSCPPGTRGLLCEEN IDDCARGPHCLNGGQCMDRIGGYSCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQLTNDYLCVCRSAFTGRHCETFVDVCPQMPCLN GGTCAVASNMPDGFICRCPPGFSGARCQSSCGQVKCRKGEQCVHTASGPRCFCPSPRDCESGCASSPCQHGGSCHPQRQPPYYSCQCAPP FSGSRCELYTAPPSTPPATCLSQYCADKARDGVCDEACNSHACQWDGGDCSLTMENPWANCSSPLPCWDYINNQCDELCNTVECLFDNFE CQGNSKTCKYDKYCADHFKDNHCDQGCNSEECGWDGLDCAADQPENLAEGTLVIVVLMPPEQLLQDARSFLRALGTLLHTNLRIKRDSQG ELMVYPYYGEKSAAMKKQRMTRRSLPGEQEQEVAGSKVFLEIDNRQCVQDSDHCFKNTDAAAALLASHAIQGTLSYPLVSVVSESLTPER TQLLYLLAVAVVIILFIILLGVIMAKRKRKHGSLWLPEGFTLRRDASNHKRREPVGQDAVGLKNLSVQVSEANLIGTGTSEHWVDDEGPQ PKKVKAEDEALLSEEDDPIDRRPWTQQHLEAADIRRTPSLALTPPQAEQEVDVLDVNVRGPDGCTPLMLASLRGGSSDLSDEDEDAEDSS ANIITDLVYQGASLQAQTDRTGEMALHLAARYSRADAAKRLLDAGADANAQDNMGRCPLHAAVAADAQGVFQILIRNRVTDLDARMNDGT TPLILAARLAVEGMVAELINCQADVNAVDDHGKSALHWAAAVNNVEATLLLLKNGANRDMQDNKEETPLFLAAREGSYEAAKILLDHFAN RDITDHMDRLPRDVARDRMHHDIVRLLDEYNVTPSPPGTVLTSALSPVICGPNRSFLSLKHTPMGKKSRRPSAKSTMPTSLPNLAKEAKD AKGSRRKKSLSEKVQLSESSVTLSPVDSLESPHTYVSDTTSSPMITSPGILQASPNPMLATAAPPAPVHAQHALSFSNLHEMQPLAHGAS TVLPSVSQLLSHHHIVSPGSGSAGSLSRLHPVPVPADWMNRMEVNETQYNEMFGMVLAPAEGTHPGIAPQSRPPEGKHITTPREPLPPIV TFQLIPKGSIAQPAGAPQPQSTCPPAVAGPLPTMYQIPEMARLPSVAFPTAMMPQQDGQVAQTILPAYHPFPASVGKYPTPPSQHSYASS -------------------------------------------------------------- >50820_50820_5_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000369611_NOTCH2_chr1_120539955_ENST00000256646_length(amino acids)=2438AA_BP=730 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGKECQWTDACLSHPCANGSTCTTVANQFSCKCLTGFTGQKCETDVNECDIPGHCQHGGTCLNLPGSYQCQCPQGF TGQYCDSLYVPCAPSPCVNGGTCRQTGDFTFECNCLPGFEGSTCERNIDDCPNHRCQNGGVCVDGVNTYNCRCPPQWTGQFCTEDVDECL LQPNACQNGGTCANRNGGYGCVCVNGWSGDDCSENIDDCAFASCTPGSTCIDRVASFSCMCPEGKAGLLCHLDDACISNPCHKGALCDTN PLNGQYICTCPQGYKGADCTEDVDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEMDINECHSDPCQNDATCLDKIGGFTCLCM PGFKGVHCELEINECQSNPCVNNGQCVDKVNRFQCLCPPGFTGPVCQIDIDDCSSTPCLNGAKCIDHPNGYECQCATGFTGVLCEENIDN CDPDPCHHGQCQDGIDSYTCICNPGYMGAICSDQIDECYSSPCLNDGRCIDLVNGYQCNCQPGTSGVNCEINFDDCASNPCIHGICMDGI NRYSCVCSPGFTGQRCNIDIDECASNPCRKGATCINGVNGFRCICPEGPHHPSCYSQVNECLSNPCIHGNCTGGLSGYKCLCDAGWVGIN CEVDKNECLSNPCQNGGTCDNLVNGYRCTCKKGFKGYNCQVNIDECASNPCLNQGTCFDDISGYTCHCVLPYTGKNCQTVLAPCSPNPCE NAAVCKESPNFESYTCLCAPGWQGQRCTIDIDECISKPCMNHGLCHNTQGSYMCECPPGFSGMDCEEDIDDCLANPCQNGGSCMDGVNTF SCLCLPGFTGDKCQTDMNECLSEPCKNGGTCSDYVNSYTCKCQAGFDGVHCENNINECTESSCFNGGTCVDGINSFSCLCPVGFTGSFCL HEINECSSHPCLNEGTCVDGLGTYRCSCPLGYTGKNCQTLVNLCSRSPCKNKGTCVQKKAESQCLCPSGWAGAYCDVPNVSCDIAASRRG VLVEHLCQHSGVCINAGNTHYCQCPLGYTGSYCEEQLDECASNPCQHGATCSDFIGGYRCECVPGYQGVNCEYEVDECQNQPCQNGGTCI DLVNHFKCSCPPGTRGLLCEENIDDCARGPHCLNGGQCMDRIGGYSCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQLTNDYLCVCR SAFTGRHCETFVDVCPQMPCLNGGTCAVASNMPDGFICRCPPGFSGARCQSSCGQVKCRKGEQCVHTASGPRCFCPSPRDCESGCASSPC QHGGSCHPQRQPPYYSCQCAPPFSGSRCELYTAPPSTPPATCLSQYCADKARDGVCDEACNSHACQWDGGDCSLTMENPWANCSSPLPCW DYINNQCDELCNTVECLFDNFECQGNSKTCKYDKYCADHFKDNHCDQGCNSEECGWDGLDCAADQPENLAEGTLVIVVLMPPEQLLQDAR SFLRALGTLLHTNLRIKRDSQGELMVYPYYGEKSAAMKKQRMTRRSLPGEQEQEVAGSKVFLEIDNRQCVQDSDHCFKNTDAAAALLASH AIQGTLSYPLVSVVSESLTPERTQLLYLLAVAVVIILFIILLGVIMAKRKRKHGSLWLPEGFTLRRDASNHKRREPVGQDAVGLKNLSVQ VSEANLIGTGTSEHWVDDEGPQPKKVKAEDEALLSEEDDPIDRRPWTQQHLEAADIRRTPSLALTPPQAEQEVDVLDVNVRGPDGCTPLM LASLRGGSSDLSDEDEDAEDSSANIITDLVYQGASLQAQTDRTGEMALHLAARYSRADAAKRLLDAGADANAQDNMGRCPLHAAVAADAQ GVFQILIRNRVTDLDARMNDGTTPLILAARLAVEGMVAELINCQADVNAVDDHGKSALHWAAAVNNVEATLLLLKNGANRDMQDNKEETP LFLAAREGSYEAAKILLDHFANRDITDHMDRLPRDVARDRMHHDIVRLLDEYNVTPSPPGTVLTSALSPVICGPNRSFLSLKHTPMGKKS RRPSAKSTMPTSLPNLAKEAKDAKGSRRKKSLSEKVQLSESSVTLSPVDSLESPHTYVSDTTSSPMITSPGILQASPNPMLATAAPPAPV HAQHALSFSNLHEMQPLAHGASTVLPSVSQLLSHHHIVSPGSGSAGSLSRLHPVPVPADWMNRMEVNETQYNEMFGMVLAPAEGTHPGIA PQSRPPEGKHITTPREPLPPIVTFQLIPKGSIAQPAGAPQPQSTCPPAVAGPLPTMYQIPEMARLPSVAFPTAMMPQQDGQVAQTILPAY HPFPASVGKYPTPPSQHSYASSNAAERTPSHSGHLQGEHPYLTPSPESPDQWSSSSPHSASDWSDVTTSPTPGGAGGGQRGPGTHMSEPP -------------------------------------------------------------- >50820_50820_6_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000369611_NOTCH2_chr1_120539955_ENST00000602566_length(amino acids)=241AA_BP=105 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGKECQWTDACLSHPCANGSTCTTVANQFSCKCLTGFTGQKCETDVNECDIPGHCQHGGTCLNLPGSYQCQCPQGF -------------------------------------------------------------- >50820_50820_7_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000369615_NOTCH2_chr1_120529705_ENST00000256646_length(amino acids)=2326AA_BP=618 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGFEGSTCERNIDDCPNHRCQNGGVCVDGVNTYNCRCPPQWTGQFCTEDVDECLLQPNACQNGGTCANRNGGYGCV CVNGWSGDDCSENIDDCAFASCTPGSTCIDRVASFSCMCPEGKAGLLCHLDDACISNPCHKGALCDTNPLNGQYICTCPQGYKGADCTED VDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEMDINECHSDPCQNDATCLDKIGGFTCLCMPGFKGVHCELEINECQSNPCVN NGQCVDKVNRFQCLCPPGFTGPVCQIDIDDCSSTPCLNGAKCIDHPNGYECQCATGFTGVLCEENIDNCDPDPCHHGQCQDGIDSYTCIC NPGYMGAICSDQIDECYSSPCLNDGRCIDLVNGYQCNCQPGTSGVNCEINFDDCASNPCIHGICMDGINRYSCVCSPGFTGQRCNIDIDE CASNPCRKGATCINGVNGFRCICPEGPHHPSCYSQVNECLSNPCIHGNCTGGLSGYKCLCDAGWVGINCEVDKNECLSNPCQNGGTCDNL VNGYRCTCKKGFKGYNCQVNIDECASNPCLNQGTCFDDISGYTCHCVLPYTGKNCQTVLAPCSPNPCENAAVCKESPNFESYTCLCAPGW QGQRCTIDIDECISKPCMNHGLCHNTQGSYMCECPPGFSGMDCEEDIDDCLANPCQNGGSCMDGVNTFSCLCLPGFTGDKCQTDMNECLS EPCKNGGTCSDYVNSYTCKCQAGFDGVHCENNINECTESSCFNGGTCVDGINSFSCLCPVGFTGSFCLHEINECSSHPCLNEGTCVDGLG TYRCSCPLGYTGKNCQTLVNLCSRSPCKNKGTCVQKKAESQCLCPSGWAGAYCDVPNVSCDIAASRRGVLVEHLCQHSGVCINAGNTHYC QCPLGYTGSYCEEQLDECASNPCQHGATCSDFIGGYRCECVPGYQGVNCEYEVDECQNQPCQNGGTCIDLVNHFKCSCPPGTRGLLCEEN IDDCARGPHCLNGGQCMDRIGGYSCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQLTNDYLCVCRSAFTGRHCETFVDVCPQMPCLN GGTCAVASNMPDGFICRCPPGFSGARCQSSCGQVKCRKGEQCVHTASGPRCFCPSPRDCESGCASSPCQHGGSCHPQRQPPYYSCQCAPP FSGSRCELYTAPPSTPPATCLSQYCADKARDGVCDEACNSHACQWDGGDCSLTMENPWANCSSPLPCWDYINNQCDELCNTVECLFDNFE CQGNSKTCKYDKYCADHFKDNHCDQGCNSEECGWDGLDCAADQPENLAEGTLVIVVLMPPEQLLQDARSFLRALGTLLHTNLRIKRDSQG ELMVYPYYGEKSAAMKKQRMTRRSLPGEQEQEVAGSKVFLEIDNRQCVQDSDHCFKNTDAAAALLASHAIQGTLSYPLVSVVSESLTPER TQLLYLLAVAVVIILFIILLGVIMAKRKRKHGSLWLPEGFTLRRDASNHKRREPVGQDAVGLKNLSVQVSEANLIGTGTSEHWVDDEGPQ PKKVKAEDEALLSEEDDPIDRRPWTQQHLEAADIRRTPSLALTPPQAEQEVDVLDVNVRGPDGCTPLMLASLRGGSSDLSDEDEDAEDSS ANIITDLVYQGASLQAQTDRTGEMALHLAARYSRADAAKRLLDAGADANAQDNMGRCPLHAAVAADAQGVFQILIRNRVTDLDARMNDGT TPLILAARLAVEGMVAELINCQADVNAVDDHGKSALHWAAAVNNVEATLLLLKNGANRDMQDNKEETPLFLAAREGSYEAAKILLDHFAN RDITDHMDRLPRDVARDRMHHDIVRLLDEYNVTPSPPGTVLTSALSPVICGPNRSFLSLKHTPMGKKSRRPSAKSTMPTSLPNLAKEAKD AKGSRRKKSLSEKVQLSESSVTLSPVDSLESPHTYVSDTTSSPMITSPGILQASPNPMLATAAPPAPVHAQHALSFSNLHEMQPLAHGAS TVLPSVSQLLSHHHIVSPGSGSAGSLSRLHPVPVPADWMNRMEVNETQYNEMFGMVLAPAEGTHPGIAPQSRPPEGKHITTPREPLPPIV TFQLIPKGSIAQPAGAPQPQSTCPPAVAGPLPTMYQIPEMARLPSVAFPTAMMPQQDGQVAQTILPAYHPFPASVGKYPTPPSQHSYASS -------------------------------------------------------------- >50820_50820_8_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000369615_NOTCH2_chr1_120539955_ENST00000256646_length(amino acids)=2438AA_BP=730 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGKECQWTDACLSHPCANGSTCTTVANQFSCKCLTGFTGQKCETDVNECDIPGHCQHGGTCLNLPGSYQCQCPQGF TGQYCDSLYVPCAPSPCVNGGTCRQTGDFTFECNCLPGFEGSTCERNIDDCPNHRCQNGGVCVDGVNTYNCRCPPQWTGQFCTEDVDECL LQPNACQNGGTCANRNGGYGCVCVNGWSGDDCSENIDDCAFASCTPGSTCIDRVASFSCMCPEGKAGLLCHLDDACISNPCHKGALCDTN PLNGQYICTCPQGYKGADCTEDVDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEMDINECHSDPCQNDATCLDKIGGFTCLCM PGFKGVHCELEINECQSNPCVNNGQCVDKVNRFQCLCPPGFTGPVCQIDIDDCSSTPCLNGAKCIDHPNGYECQCATGFTGVLCEENIDN CDPDPCHHGQCQDGIDSYTCICNPGYMGAICSDQIDECYSSPCLNDGRCIDLVNGYQCNCQPGTSGVNCEINFDDCASNPCIHGICMDGI NRYSCVCSPGFTGQRCNIDIDECASNPCRKGATCINGVNGFRCICPEGPHHPSCYSQVNECLSNPCIHGNCTGGLSGYKCLCDAGWVGIN CEVDKNECLSNPCQNGGTCDNLVNGYRCTCKKGFKGYNCQVNIDECASNPCLNQGTCFDDISGYTCHCVLPYTGKNCQTVLAPCSPNPCE NAAVCKESPNFESYTCLCAPGWQGQRCTIDIDECISKPCMNHGLCHNTQGSYMCECPPGFSGMDCEEDIDDCLANPCQNGGSCMDGVNTF SCLCLPGFTGDKCQTDMNECLSEPCKNGGTCSDYVNSYTCKCQAGFDGVHCENNINECTESSCFNGGTCVDGINSFSCLCPVGFTGSFCL HEINECSSHPCLNEGTCVDGLGTYRCSCPLGYTGKNCQTLVNLCSRSPCKNKGTCVQKKAESQCLCPSGWAGAYCDVPNVSCDIAASRRG VLVEHLCQHSGVCINAGNTHYCQCPLGYTGSYCEEQLDECASNPCQHGATCSDFIGGYRCECVPGYQGVNCEYEVDECQNQPCQNGGTCI DLVNHFKCSCPPGTRGLLCEENIDDCARGPHCLNGGQCMDRIGGYSCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQLTNDYLCVCR SAFTGRHCETFVDVCPQMPCLNGGTCAVASNMPDGFICRCPPGFSGARCQSSCGQVKCRKGEQCVHTASGPRCFCPSPRDCESGCASSPC QHGGSCHPQRQPPYYSCQCAPPFSGSRCELYTAPPSTPPATCLSQYCADKARDGVCDEACNSHACQWDGGDCSLTMENPWANCSSPLPCW DYINNQCDELCNTVECLFDNFECQGNSKTCKYDKYCADHFKDNHCDQGCNSEECGWDGLDCAADQPENLAEGTLVIVVLMPPEQLLQDAR SFLRALGTLLHTNLRIKRDSQGELMVYPYYGEKSAAMKKQRMTRRSLPGEQEQEVAGSKVFLEIDNRQCVQDSDHCFKNTDAAAALLASH AIQGTLSYPLVSVVSESLTPERTQLLYLLAVAVVIILFIILLGVIMAKRKRKHGSLWLPEGFTLRRDASNHKRREPVGQDAVGLKNLSVQ VSEANLIGTGTSEHWVDDEGPQPKKVKAEDEALLSEEDDPIDRRPWTQQHLEAADIRRTPSLALTPPQAEQEVDVLDVNVRGPDGCTPLM LASLRGGSSDLSDEDEDAEDSSANIITDLVYQGASLQAQTDRTGEMALHLAARYSRADAAKRLLDAGADANAQDNMGRCPLHAAVAADAQ GVFQILIRNRVTDLDARMNDGTTPLILAARLAVEGMVAELINCQADVNAVDDHGKSALHWAAAVNNVEATLLLLKNGANRDMQDNKEETP LFLAAREGSYEAAKILLDHFANRDITDHMDRLPRDVARDRMHHDIVRLLDEYNVTPSPPGTVLTSALSPVICGPNRSFLSLKHTPMGKKS RRPSAKSTMPTSLPNLAKEAKDAKGSRRKKSLSEKVQLSESSVTLSPVDSLESPHTYVSDTTSSPMITSPGILQASPNPMLATAAPPAPV HAQHALSFSNLHEMQPLAHGASTVLPSVSQLLSHHHIVSPGSGSAGSLSRLHPVPVPADWMNRMEVNETQYNEMFGMVLAPAEGTHPGIA PQSRPPEGKHITTPREPLPPIVTFQLIPKGSIAQPAGAPQPQSTCPPAVAGPLPTMYQIPEMARLPSVAFPTAMMPQQDGQVAQTILPAY HPFPASVGKYPTPPSQHSYASSNAAERTPSHSGHLQGEHPYLTPSPESPDQWSSSSPHSASDWSDVTTSPTPGGAGGGQRGPGTHMSEPP -------------------------------------------------------------- >50820_50820_9_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000369615_NOTCH2_chr1_120539955_ENST00000602566_length(amino acids)=241AA_BP=105 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGKECQWTDACLSHPCANGSTCTTVANQFSCKCLTGFTGQKCETDVNECDIPGHCQHGGTCLNLPGSYQCQCPQGF -------------------------------------------------------------- >50820_50820_10_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000369617_NOTCH2_chr1_120529705_ENST00000256646_length(amino acids)=2326AA_BP=618 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGFEGSTCERNIDDCPNHRCQNGGVCVDGVNTYNCRCPPQWTGQFCTEDVDECLLQPNACQNGGTCANRNGGYGCV CVNGWSGDDCSENIDDCAFASCTPGSTCIDRVASFSCMCPEGKAGLLCHLDDACISNPCHKGALCDTNPLNGQYICTCPQGYKGADCTED VDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEMDINECHSDPCQNDATCLDKIGGFTCLCMPGFKGVHCELEINECQSNPCVN NGQCVDKVNRFQCLCPPGFTGPVCQIDIDDCSSTPCLNGAKCIDHPNGYECQCATGFTGVLCEENIDNCDPDPCHHGQCQDGIDSYTCIC NPGYMGAICSDQIDECYSSPCLNDGRCIDLVNGYQCNCQPGTSGVNCEINFDDCASNPCIHGICMDGINRYSCVCSPGFTGQRCNIDIDE CASNPCRKGATCINGVNGFRCICPEGPHHPSCYSQVNECLSNPCIHGNCTGGLSGYKCLCDAGWVGINCEVDKNECLSNPCQNGGTCDNL VNGYRCTCKKGFKGYNCQVNIDECASNPCLNQGTCFDDISGYTCHCVLPYTGKNCQTVLAPCSPNPCENAAVCKESPNFESYTCLCAPGW QGQRCTIDIDECISKPCMNHGLCHNTQGSYMCECPPGFSGMDCEEDIDDCLANPCQNGGSCMDGVNTFSCLCLPGFTGDKCQTDMNECLS EPCKNGGTCSDYVNSYTCKCQAGFDGVHCENNINECTESSCFNGGTCVDGINSFSCLCPVGFTGSFCLHEINECSSHPCLNEGTCVDGLG TYRCSCPLGYTGKNCQTLVNLCSRSPCKNKGTCVQKKAESQCLCPSGWAGAYCDVPNVSCDIAASRRGVLVEHLCQHSGVCINAGNTHYC QCPLGYTGSYCEEQLDECASNPCQHGATCSDFIGGYRCECVPGYQGVNCEYEVDECQNQPCQNGGTCIDLVNHFKCSCPPGTRGLLCEEN IDDCARGPHCLNGGQCMDRIGGYSCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQLTNDYLCVCRSAFTGRHCETFVDVCPQMPCLN GGTCAVASNMPDGFICRCPPGFSGARCQSSCGQVKCRKGEQCVHTASGPRCFCPSPRDCESGCASSPCQHGGSCHPQRQPPYYSCQCAPP FSGSRCELYTAPPSTPPATCLSQYCADKARDGVCDEACNSHACQWDGGDCSLTMENPWANCSSPLPCWDYINNQCDELCNTVECLFDNFE CQGNSKTCKYDKYCADHFKDNHCDQGCNSEECGWDGLDCAADQPENLAEGTLVIVVLMPPEQLLQDARSFLRALGTLLHTNLRIKRDSQG ELMVYPYYGEKSAAMKKQRMTRRSLPGEQEQEVAGSKVFLEIDNRQCVQDSDHCFKNTDAAAALLASHAIQGTLSYPLVSVVSESLTPER TQLLYLLAVAVVIILFIILLGVIMAKRKRKHGSLWLPEGFTLRRDASNHKRREPVGQDAVGLKNLSVQVSEANLIGTGTSEHWVDDEGPQ PKKVKAEDEALLSEEDDPIDRRPWTQQHLEAADIRRTPSLALTPPQAEQEVDVLDVNVRGPDGCTPLMLASLRGGSSDLSDEDEDAEDSS ANIITDLVYQGASLQAQTDRTGEMALHLAARYSRADAAKRLLDAGADANAQDNMGRCPLHAAVAADAQGVFQILIRNRVTDLDARMNDGT TPLILAARLAVEGMVAELINCQADVNAVDDHGKSALHWAAAVNNVEATLLLLKNGANRDMQDNKEETPLFLAAREGSYEAAKILLDHFAN RDITDHMDRLPRDVARDRMHHDIVRLLDEYNVTPSPPGTVLTSALSPVICGPNRSFLSLKHTPMGKKSRRPSAKSTMPTSLPNLAKEAKD AKGSRRKKSLSEKVQLSESSVTLSPVDSLESPHTYVSDTTSSPMITSPGILQASPNPMLATAAPPAPVHAQHALSFSNLHEMQPLAHGAS TVLPSVSQLLSHHHIVSPGSGSAGSLSRLHPVPVPADWMNRMEVNETQYNEMFGMVLAPAEGTHPGIAPQSRPPEGKHITTPREPLPPIV TFQLIPKGSIAQPAGAPQPQSTCPPAVAGPLPTMYQIPEMARLPSVAFPTAMMPQQDGQVAQTILPAYHPFPASVGKYPTPPSQHSYASS -------------------------------------------------------------- >50820_50820_11_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000369617_NOTCH2_chr1_120539955_ENST00000256646_length(amino acids)=2438AA_BP=730 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGKECQWTDACLSHPCANGSTCTTVANQFSCKCLTGFTGQKCETDVNECDIPGHCQHGGTCLNLPGSYQCQCPQGF TGQYCDSLYVPCAPSPCVNGGTCRQTGDFTFECNCLPGFEGSTCERNIDDCPNHRCQNGGVCVDGVNTYNCRCPPQWTGQFCTEDVDECL LQPNACQNGGTCANRNGGYGCVCVNGWSGDDCSENIDDCAFASCTPGSTCIDRVASFSCMCPEGKAGLLCHLDDACISNPCHKGALCDTN PLNGQYICTCPQGYKGADCTEDVDECAMANSNPCEHAGKCVNTDGAFHCECLKGYAGPRCEMDINECHSDPCQNDATCLDKIGGFTCLCM PGFKGVHCELEINECQSNPCVNNGQCVDKVNRFQCLCPPGFTGPVCQIDIDDCSSTPCLNGAKCIDHPNGYECQCATGFTGVLCEENIDN CDPDPCHHGQCQDGIDSYTCICNPGYMGAICSDQIDECYSSPCLNDGRCIDLVNGYQCNCQPGTSGVNCEINFDDCASNPCIHGICMDGI NRYSCVCSPGFTGQRCNIDIDECASNPCRKGATCINGVNGFRCICPEGPHHPSCYSQVNECLSNPCIHGNCTGGLSGYKCLCDAGWVGIN CEVDKNECLSNPCQNGGTCDNLVNGYRCTCKKGFKGYNCQVNIDECASNPCLNQGTCFDDISGYTCHCVLPYTGKNCQTVLAPCSPNPCE NAAVCKESPNFESYTCLCAPGWQGQRCTIDIDECISKPCMNHGLCHNTQGSYMCECPPGFSGMDCEEDIDDCLANPCQNGGSCMDGVNTF SCLCLPGFTGDKCQTDMNECLSEPCKNGGTCSDYVNSYTCKCQAGFDGVHCENNINECTESSCFNGGTCVDGINSFSCLCPVGFTGSFCL HEINECSSHPCLNEGTCVDGLGTYRCSCPLGYTGKNCQTLVNLCSRSPCKNKGTCVQKKAESQCLCPSGWAGAYCDVPNVSCDIAASRRG VLVEHLCQHSGVCINAGNTHYCQCPLGYTGSYCEEQLDECASNPCQHGATCSDFIGGYRCECVPGYQGVNCEYEVDECQNQPCQNGGTCI DLVNHFKCSCPPGTRGLLCEENIDDCARGPHCLNGGQCMDRIGGYSCRCLPGFAGERCEGDINECLSNPCSSEGSLDCIQLTNDYLCVCR SAFTGRHCETFVDVCPQMPCLNGGTCAVASNMPDGFICRCPPGFSGARCQSSCGQVKCRKGEQCVHTASGPRCFCPSPRDCESGCASSPC QHGGSCHPQRQPPYYSCQCAPPFSGSRCELYTAPPSTPPATCLSQYCADKARDGVCDEACNSHACQWDGGDCSLTMENPWANCSSPLPCW DYINNQCDELCNTVECLFDNFECQGNSKTCKYDKYCADHFKDNHCDQGCNSEECGWDGLDCAADQPENLAEGTLVIVVLMPPEQLLQDAR SFLRALGTLLHTNLRIKRDSQGELMVYPYYGEKSAAMKKQRMTRRSLPGEQEQEVAGSKVFLEIDNRQCVQDSDHCFKNTDAAAALLASH AIQGTLSYPLVSVVSESLTPERTQLLYLLAVAVVIILFIILLGVIMAKRKRKHGSLWLPEGFTLRRDASNHKRREPVGQDAVGLKNLSVQ VSEANLIGTGTSEHWVDDEGPQPKKVKAEDEALLSEEDDPIDRRPWTQQHLEAADIRRTPSLALTPPQAEQEVDVLDVNVRGPDGCTPLM LASLRGGSSDLSDEDEDAEDSSANIITDLVYQGASLQAQTDRTGEMALHLAARYSRADAAKRLLDAGADANAQDNMGRCPLHAAVAADAQ GVFQILIRNRVTDLDARMNDGTTPLILAARLAVEGMVAELINCQADVNAVDDHGKSALHWAAAVNNVEATLLLLKNGANRDMQDNKEETP LFLAAREGSYEAAKILLDHFANRDITDHMDRLPRDVARDRMHHDIVRLLDEYNVTPSPPGTVLTSALSPVICGPNRSFLSLKHTPMGKKS RRPSAKSTMPTSLPNLAKEAKDAKGSRRKKSLSEKVQLSESSVTLSPVDSLESPHTYVSDTTSSPMITSPGILQASPNPMLATAAPPAPV HAQHALSFSNLHEMQPLAHGASTVLPSVSQLLSHHHIVSPGSGSAGSLSRLHPVPVPADWMNRMEVNETQYNEMFGMVLAPAEGTHPGIA PQSRPPEGKHITTPREPLPPIVTFQLIPKGSIAQPAGAPQPQSTCPPAVAGPLPTMYQIPEMARLPSVAFPTAMMPQQDGQVAQTILPAY HPFPASVGKYPTPPSQHSYASSNAAERTPSHSGHLQGEHPYLTPSPESPDQWSSSSPHSASDWSDVTTSPTPGGAGGGQRGPGTHMSEPP -------------------------------------------------------------- >50820_50820_12_MAGI3-NOTCH2_MAGI3_chr1_113933971_ENST00000369617_NOTCH2_chr1_120539955_ENST00000602566_length(amino acids)=241AA_BP=105 MSKTLKKKKHWLSKVQECAVSWAGPPGDFGAEIRGGAERGEFPYLGRLREEPGGGTCCVVSGKAPSPGDVLLEVNGTPVSGLTNRDTLAV IRHFREPIRLKTVKPGKECQWTDACLSHPCANGSTCTTVANQFSCKCLTGFTGQKCETDVNECDIPGHCQHGGTCLNLPGSYQCQCPQGF -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:113933971/chr1:120539955) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MAGI3 | NOTCH2 |
| FUNCTION: Acts as a scaffolding protein at cell-cell junctions, thereby regulating various cellular and signaling processes. Cooperates with PTEN to modulate the kinase activity of AKT1. Its interaction with PTPRB and tyrosine phosphorylated proteins suggests that it may link receptor tyrosine phosphatase with its substrates at the plasma membrane. In polarized epithelial cells, involved in efficient trafficking of TGFA to the cell surface. Regulates the ability of LPAR2 to activate ERK and RhoA pathways. Regulates the JNK signaling cascade via its interaction with FZD4 and VANGL2. {ECO:0000269|PubMed:10748157}. | FUNCTION: Human-specific protein that promotes neural progenitor proliferation and evolutionary expansion of the brain neocortex by regulating the Notch signaling pathway (PubMed:29856954, PubMed:29856955, PubMed:29561261). Able to promote neural progenitor self-renewal, possibly by down-regulating neuronal differentiation genes, thereby delaying the differentiation of neuronal progenitors and leading to an overall final increase in neuronal production (PubMed:29856954, PubMed:29856955). Acts by enhancing the Notch signaling pathway via two different mechanisms that probably work in parallel to reach the same effect (PubMed:29856954, PubMed:29856955). Enhances Notch signaling pathway in a non-cell-autonomous manner via direct interaction with NOTCH2 (PubMed:29856954). Also promotes Notch signaling pathway in a cell-autonomous manner through inhibition of cis DLL1-NOTCH2 interactions, which promotes neuronal differentiation (PubMed:29856955). {ECO:0000269|PubMed:29561261, ECO:0000269|PubMed:29856954, ECO:0000269|PubMed:29856955}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 6_9 | 105.33333333333333 | 1482.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 6_9 | 105.33333333333333 | 1126.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 6_9 | 105.33333333333333 | 2009.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 6_9 | 105.33333333333333 | 1151.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 6_9 | 105.33333333333333 | 1482.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 6_9 | 105.33333333333333 | 1126.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 6_9 | 105.33333333333333 | 2009.0 | Compositional bias | Note=Poly-Lys |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 6_9 | 105.33333333333333 | 1151.0 | Compositional bias | Note=Poly-Lys |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1645_1648 | 250.33333333333334 | 2472.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1994_1997 | 250.33333333333334 | 2472.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 2426_2429 | 250.33333333333334 | 2472.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1645_1648 | 138.33333333333334 | 2472.0 | Compositional bias | Note=Poly-Ala | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1994_1997 | 138.33333333333334 | 2472.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 2426_2429 | 138.33333333333334 | 2472.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1025_1061 | 250.33333333333334 | 2472.0 | Domain | EGF-like 27%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1063_1099 | 250.33333333333334 | 2472.0 | Domain | EGF-like 28 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1101_1147 | 250.33333333333334 | 2472.0 | Domain | EGF-like 29 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1149_1185 | 250.33333333333334 | 2472.0 | Domain | EGF-like 30%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1187_1223 | 250.33333333333334 | 2472.0 | Domain | EGF-like 31%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1225_1262 | 250.33333333333334 | 2472.0 | Domain | EGF-like 32%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1264_1302 | 250.33333333333334 | 2472.0 | Domain | EGF-like 33 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1304_1343 | 250.33333333333334 | 2472.0 | Domain | EGF-like 34 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1374_1412 | 250.33333333333334 | 2472.0 | Domain | EGF-like 35 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 260_296 | 250.33333333333334 | 2472.0 | Domain | EGF-like 7%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 298_336 | 250.33333333333334 | 2472.0 | Domain | EGF-like 8%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 338_374 | 250.33333333333334 | 2472.0 | Domain | EGF-like 9%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 375_413 | 250.33333333333334 | 2472.0 | Domain | EGF-like 10 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 415_454 | 250.33333333333334 | 2472.0 | Domain | EGF-like 11%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 456_492 | 250.33333333333334 | 2472.0 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 494_530 | 250.33333333333334 | 2472.0 | Domain | EGF-like 13%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 532_568 | 250.33333333333334 | 2472.0 | Domain | EGF-like 14%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 570_605 | 250.33333333333334 | 2472.0 | Domain | EGF-like 15%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 607_643 | 250.33333333333334 | 2472.0 | Domain | EGF-like 16%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 645_680 | 250.33333333333334 | 2472.0 | Domain | EGF-like 17%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 682_718 | 250.33333333333334 | 2472.0 | Domain | EGF-like 18%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 720_755 | 250.33333333333334 | 2472.0 | Domain | EGF-like 19 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 757_793 | 250.33333333333334 | 2472.0 | Domain | EGF-like 20%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 795_831 | 250.33333333333334 | 2472.0 | Domain | EGF-like 21%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 833_871 | 250.33333333333334 | 2472.0 | Domain | EGF-like 22 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 873_909 | 250.33333333333334 | 2472.0 | Domain | EGF-like 23%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 911_947 | 250.33333333333334 | 2472.0 | Domain | EGF-like 24%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 949_985 | 250.33333333333334 | 2472.0 | Domain | EGF-like 25%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 987_1023 | 250.33333333333334 | 2472.0 | Domain | EGF-like 26%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1025_1061 | 138.33333333333334 | 2472.0 | Domain | EGF-like 27%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1063_1099 | 138.33333333333334 | 2472.0 | Domain | EGF-like 28 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1101_1147 | 138.33333333333334 | 2472.0 | Domain | EGF-like 29 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1149_1185 | 138.33333333333334 | 2472.0 | Domain | EGF-like 30%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1187_1223 | 138.33333333333334 | 2472.0 | Domain | EGF-like 31%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1225_1262 | 138.33333333333334 | 2472.0 | Domain | EGF-like 32%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1264_1302 | 138.33333333333334 | 2472.0 | Domain | EGF-like 33 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1304_1343 | 138.33333333333334 | 2472.0 | Domain | EGF-like 34 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1374_1412 | 138.33333333333334 | 2472.0 | Domain | EGF-like 35 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 144_180 | 138.33333333333334 | 2472.0 | Domain | EGF-like 4 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 182_219 | 138.33333333333334 | 2472.0 | Domain | EGF-like 5%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 221_258 | 138.33333333333334 | 2472.0 | Domain | EGF-like 6 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 260_296 | 138.33333333333334 | 2472.0 | Domain | EGF-like 7%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 298_336 | 138.33333333333334 | 2472.0 | Domain | EGF-like 8%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 338_374 | 138.33333333333334 | 2472.0 | Domain | EGF-like 9%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 375_413 | 138.33333333333334 | 2472.0 | Domain | EGF-like 10 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 415_454 | 138.33333333333334 | 2472.0 | Domain | EGF-like 11%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 456_492 | 138.33333333333334 | 2472.0 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 494_530 | 138.33333333333334 | 2472.0 | Domain | EGF-like 13%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 532_568 | 138.33333333333334 | 2472.0 | Domain | EGF-like 14%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 570_605 | 138.33333333333334 | 2472.0 | Domain | EGF-like 15%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 607_643 | 138.33333333333334 | 2472.0 | Domain | EGF-like 16%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 645_680 | 138.33333333333334 | 2472.0 | Domain | EGF-like 17%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 682_718 | 138.33333333333334 | 2472.0 | Domain | EGF-like 18%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 720_755 | 138.33333333333334 | 2472.0 | Domain | EGF-like 19 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 757_793 | 138.33333333333334 | 2472.0 | Domain | EGF-like 20%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 795_831 | 138.33333333333334 | 2472.0 | Domain | EGF-like 21%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 833_871 | 138.33333333333334 | 2472.0 | Domain | EGF-like 22 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 873_909 | 138.33333333333334 | 2472.0 | Domain | EGF-like 23%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 911_947 | 138.33333333333334 | 2472.0 | Domain | EGF-like 24%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 949_985 | 138.33333333333334 | 2472.0 | Domain | EGF-like 25%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 987_1023 | 138.33333333333334 | 2472.0 | Domain | EGF-like 26%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1425_1677 | 250.33333333333334 | 2472.0 | Region | Note=Negative regulatory region (NRR) | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1425_1677 | 138.33333333333334 | 2472.0 | Region | Note=Negative regulatory region (NRR) | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1425_1465 | 250.33333333333334 | 2472.0 | Repeat | Note=LNR 1 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1466_1502 | 250.33333333333334 | 2472.0 | Repeat | Note=LNR 2 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1503_1544 | 250.33333333333334 | 2472.0 | Repeat | Note=LNR 3 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1827_1871 | 250.33333333333334 | 2472.0 | Repeat | Note=ANK 1 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1876_1905 | 250.33333333333334 | 2472.0 | Repeat | Note=ANK 2 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1909_1939 | 250.33333333333334 | 2472.0 | Repeat | Note=ANK 3 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1943_1972 | 250.33333333333334 | 2472.0 | Repeat | Note=ANK 4 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1976_2005 | 250.33333333333334 | 2472.0 | Repeat | Note=ANK 5 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 2009_2038 | 250.33333333333334 | 2472.0 | Repeat | Note=ANK 6 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1425_1465 | 138.33333333333334 | 2472.0 | Repeat | Note=LNR 1 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1466_1502 | 138.33333333333334 | 2472.0 | Repeat | Note=LNR 2 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1503_1544 | 138.33333333333334 | 2472.0 | Repeat | Note=LNR 3 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1827_1871 | 138.33333333333334 | 2472.0 | Repeat | Note=ANK 1 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1876_1905 | 138.33333333333334 | 2472.0 | Repeat | Note=ANK 2 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1909_1939 | 138.33333333333334 | 2472.0 | Repeat | Note=ANK 3 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1943_1972 | 138.33333333333334 | 2472.0 | Repeat | Note=ANK 4 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1976_2005 | 138.33333333333334 | 2472.0 | Repeat | Note=ANK 5 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 2009_2038 | 138.33333333333334 | 2472.0 | Repeat | Note=ANK 6 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1699_2471 | 250.33333333333334 | 2472.0 | Topological domain | Cytoplasmic | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1699_2471 | 138.33333333333334 | 2472.0 | Topological domain | Cytoplasmic | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 1678_1698 | 250.33333333333334 | 2472.0 | Transmembrane | Helical | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 1678_1698 | 138.33333333333334 | 2472.0 | Transmembrane | Helical |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 238_243 | 105.33333333333333 | 1482.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 238_243 | 105.33333333333333 | 1126.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 238_243 | 105.33333333333333 | 2009.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 238_243 | 105.33333333333333 | 1151.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 238_243 | 105.33333333333333 | 1482.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 238_243 | 105.33333333333333 | 1126.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 238_243 | 105.33333333333333 | 2009.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 238_243 | 105.33333333333333 | 1151.0 | Compositional bias | Note=Poly-Glu |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 1021_1103 | 105.33333333333333 | 1482.0 | Domain | PDZ 6 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 114_288 | 105.33333333333333 | 1482.0 | Domain | Guanylate kinase-like |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 293_326 | 105.33333333333333 | 1482.0 | Domain | WW 1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 339_372 | 105.33333333333333 | 1482.0 | Domain | WW 2 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 578_654 | 105.33333333333333 | 1482.0 | Domain | PDZ 3 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 1021_1103 | 105.33333333333333 | 1126.0 | Domain | PDZ 6 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 114_288 | 105.33333333333333 | 1126.0 | Domain | Guanylate kinase-like |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 293_326 | 105.33333333333333 | 1126.0 | Domain | WW 1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 339_372 | 105.33333333333333 | 1126.0 | Domain | WW 2 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 578_654 | 105.33333333333333 | 1126.0 | Domain | PDZ 3 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 1021_1103 | 105.33333333333333 | 2009.0 | Domain | PDZ 6 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 114_288 | 105.33333333333333 | 2009.0 | Domain | Guanylate kinase-like |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 293_326 | 105.33333333333333 | 2009.0 | Domain | WW 1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 339_372 | 105.33333333333333 | 2009.0 | Domain | WW 2 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 578_654 | 105.33333333333333 | 2009.0 | Domain | PDZ 3 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 1021_1103 | 105.33333333333333 | 1151.0 | Domain | PDZ 6 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 114_288 | 105.33333333333333 | 1151.0 | Domain | Guanylate kinase-like |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 293_326 | 105.33333333333333 | 1151.0 | Domain | WW 1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 339_372 | 105.33333333333333 | 1151.0 | Domain | WW 2 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 578_654 | 105.33333333333333 | 1151.0 | Domain | PDZ 3 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 1021_1103 | 105.33333333333333 | 1482.0 | Domain | PDZ 6 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 114_288 | 105.33333333333333 | 1482.0 | Domain | Guanylate kinase-like |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 293_326 | 105.33333333333333 | 1482.0 | Domain | WW 1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 339_372 | 105.33333333333333 | 1482.0 | Domain | WW 2 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 578_654 | 105.33333333333333 | 1482.0 | Domain | PDZ 3 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 1021_1103 | 105.33333333333333 | 1126.0 | Domain | PDZ 6 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 114_288 | 105.33333333333333 | 1126.0 | Domain | Guanylate kinase-like |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 293_326 | 105.33333333333333 | 1126.0 | Domain | WW 1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 339_372 | 105.33333333333333 | 1126.0 | Domain | WW 2 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 578_654 | 105.33333333333333 | 1126.0 | Domain | PDZ 3 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 1021_1103 | 105.33333333333333 | 2009.0 | Domain | PDZ 6 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 114_288 | 105.33333333333333 | 2009.0 | Domain | Guanylate kinase-like |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 293_326 | 105.33333333333333 | 2009.0 | Domain | WW 1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 339_372 | 105.33333333333333 | 2009.0 | Domain | WW 2 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 578_654 | 105.33333333333333 | 2009.0 | Domain | PDZ 3 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 1021_1103 | 105.33333333333333 | 1151.0 | Domain | PDZ 6 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 114_288 | 105.33333333333333 | 1151.0 | Domain | Guanylate kinase-like |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 293_326 | 105.33333333333333 | 1151.0 | Domain | WW 1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 339_372 | 105.33333333333333 | 1151.0 | Domain | WW 2 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 578_654 | 105.33333333333333 | 1151.0 | Domain | PDZ 3 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 121_128 | 105.33333333333333 | 1482.0 | Nucleotide binding | ATP |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 121_128 | 105.33333333333333 | 1126.0 | Nucleotide binding | ATP |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 121_128 | 105.33333333333333 | 2009.0 | Nucleotide binding | ATP |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 121_128 | 105.33333333333333 | 1151.0 | Nucleotide binding | ATP |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 121_128 | 105.33333333333333 | 1482.0 | Nucleotide binding | ATP |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 121_128 | 105.33333333333333 | 1126.0 | Nucleotide binding | ATP |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 121_128 | 105.33333333333333 | 2009.0 | Nucleotide binding | ATP |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 121_128 | 105.33333333333333 | 1151.0 | Nucleotide binding | ATP |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 105_143 | 250.33333333333334 | 2472.0 | Domain | EGF-like 3 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 144_180 | 250.33333333333334 | 2472.0 | Domain | EGF-like 4 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 182_219 | 250.33333333333334 | 2472.0 | Domain | EGF-like 5%3B calcium-binding | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 221_258 | 250.33333333333334 | 2472.0 | Domain | EGF-like 6 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 26_63 | 250.33333333333334 | 2472.0 | Domain | EGF-like 1 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 64_102 | 250.33333333333334 | 2472.0 | Domain | EGF-like 2 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 105_143 | 138.33333333333334 | 2472.0 | Domain | EGF-like 3 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 26_63 | 138.33333333333334 | 2472.0 | Domain | EGF-like 1 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 64_102 | 138.33333333333334 | 2472.0 | Domain | EGF-like 2 | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120529705 | ENST00000256646 | 3 | 34 | 26_1677 | 250.33333333333334 | 2472.0 | Topological domain | Extracellular | |
| Tgene | NOTCH2 | chr1:113933971 | chr1:120539955 | ENST00000256646 | 2 | 34 | 26_1677 | 138.33333333333334 | 2472.0 | Topological domain | Extracellular |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
MAGI3_pLDDT.png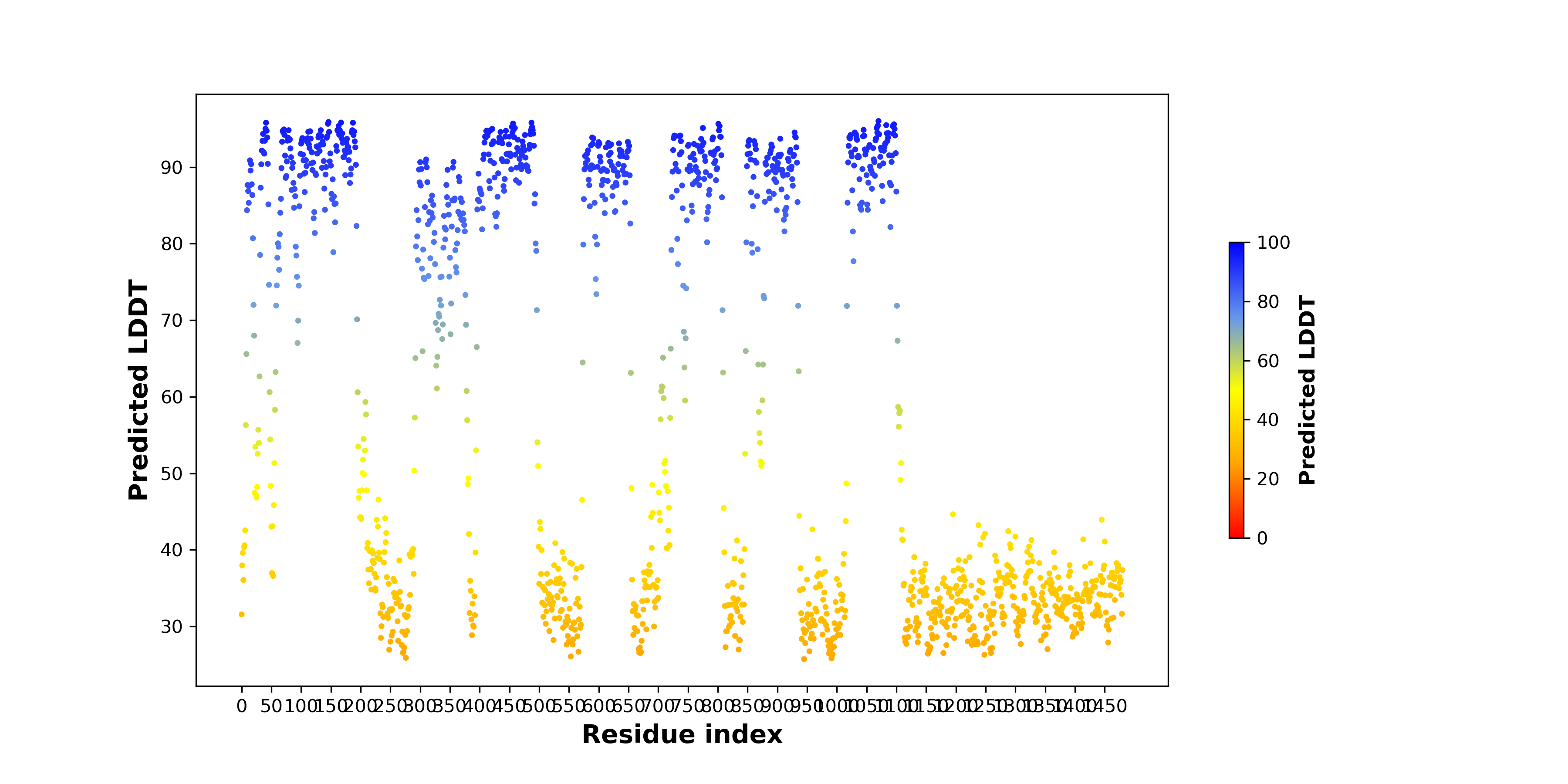 |
NOTCH2_pLDDT.png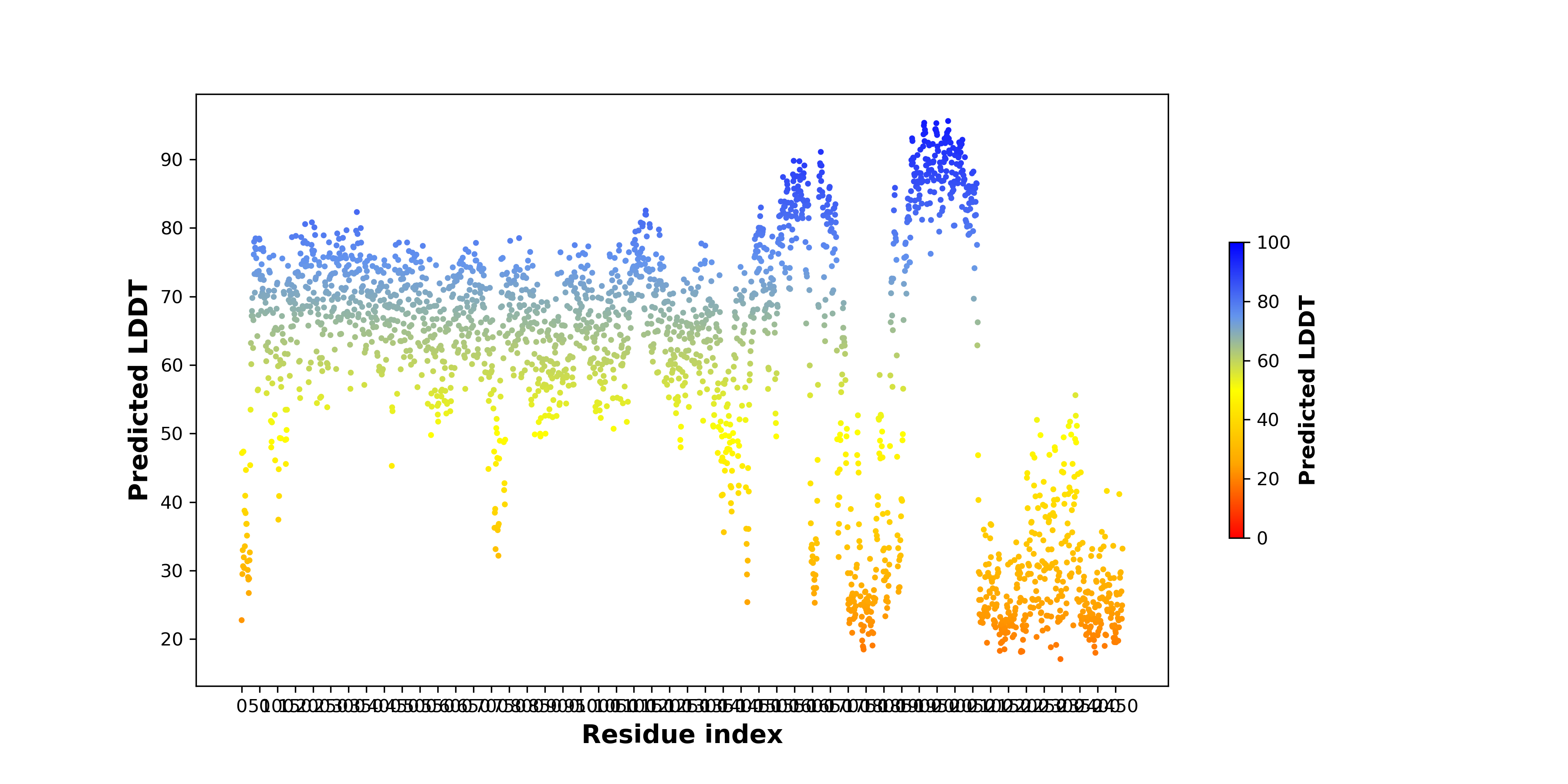 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| MAGI3 | |
| NOTCH2 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 726_808 | 105.33333333333333 | 1482.0 | ADGRB1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 726_808 | 105.33333333333333 | 1126.0 | ADGRB1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 726_808 | 105.33333333333333 | 2009.0 | ADGRB1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 726_808 | 105.33333333333333 | 1151.0 | ADGRB1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 726_808 | 105.33333333333333 | 1482.0 | ADGRB1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 726_808 | 105.33333333333333 | 1126.0 | ADGRB1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 726_808 | 105.33333333333333 | 2009.0 | ADGRB1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 726_808 | 105.33333333333333 | 1151.0 | ADGRB1 |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 851_938 | 105.33333333333333 | 1482.0 | LPAR2 and GRIN2B |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 851_938 | 105.33333333333333 | 1126.0 | LPAR2 and GRIN2B |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 851_938 | 105.33333333333333 | 2009.0 | LPAR2 and GRIN2B |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 851_938 | 105.33333333333333 | 1151.0 | LPAR2 and GRIN2B |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 851_938 | 105.33333333333333 | 1482.0 | LPAR2 and GRIN2B |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 851_938 | 105.33333333333333 | 1126.0 | LPAR2 and GRIN2B |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 851_938 | 105.33333333333333 | 2009.0 | LPAR2 and GRIN2B |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 851_938 | 105.33333333333333 | 1151.0 | LPAR2 and GRIN2B |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000307546 | + | 1 | 21 | 410_492 | 105.33333333333333 | 1482.0 | PTEN |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369611 | + | 1 | 21 | 410_492 | 105.33333333333333 | 1126.0 | PTEN |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369615 | + | 1 | 22 | 410_492 | 105.33333333333333 | 2009.0 | PTEN |
| Hgene | MAGI3 | chr1:113933971 | chr1:120529705 | ENST00000369617 | + | 1 | 22 | 410_492 | 105.33333333333333 | 1151.0 | PTEN |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000307546 | + | 1 | 21 | 410_492 | 105.33333333333333 | 1482.0 | PTEN |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369611 | + | 1 | 21 | 410_492 | 105.33333333333333 | 1126.0 | PTEN |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369615 | + | 1 | 22 | 410_492 | 105.33333333333333 | 2009.0 | PTEN |
| Hgene | MAGI3 | chr1:113933971 | chr1:120539955 | ENST00000369617 | + | 1 | 22 | 410_492 | 105.33333333333333 | 1151.0 | PTEN |
Top |
Related Drugs to MAGI3-NOTCH2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to MAGI3-NOTCH2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

