| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:MDM2-NUP107 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: MDM2-NUP107 | FusionPDB ID: 52413 | FusionGDB2.0 ID: 52413 | Hgene | Tgene | Gene symbol | MDM2 | NUP107 | Gene ID | 4193 | 57122 |
| Gene name | MDM2 proto-oncogene | nucleoporin 107 | |
| Synonyms | ACTFS|HDMX|LSKB|hdm2 | NPHS11|NUP84|ODG6|ODG6; GAMOS7 | |
| Cytomap | 12q15 | 12q15 | |
| Type of gene | protein-coding | protein-coding | |
| Description | E3 ubiquitin-protein ligase Mdm2MDM2 oncogene, E3 ubiquitin protein ligaseMDM2 proto-oncogene, E3 ubiquitin protein ligaseMdm2, p53 E3 ubiquitin protein ligase homologMdm2, transformed 3T3 cell double minute 2, p53 binding proteindouble minute 2, hum | nuclear pore complex protein Nup107nucleoporin 107kDa | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | Q00987 | P57740 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000258149, ENST00000356290, ENST00000428863, ENST00000462284, ENST00000478070, ENST00000540827, ENST00000545204, ENST00000258148, ENST00000299252, ENST00000348801, ENST00000350057, ENST00000360430, ENST00000393410, ENST00000393412, ENST00000393413, ENST00000517852, ENST00000544125, ENST00000544561, | ENST00000229179, ENST00000378905, ENST00000539906, ENST00000401003, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 39 X 20 X 11=8580 | 27 X 22 X 9=5346 |
| # samples | 47 | 37 | |
| ** MAII score | log2(47/8580*10)=-4.19024498582191 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(37/5346*10)=-3.85286266172677 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: MDM2 [Title/Abstract] AND NUP107 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MDM2(69229764)-NUP107(69113121), # samples:2 MDM2(69229764)-NUP107(69135593), # samples:2 MDM2(69202271)-NUP107(69082742), # samples:2 NUP107(69084526)-MDM2(69242456), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MDM2-NUP107 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MDM2-NUP107 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MDM2-NUP107 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MDM2-NUP107 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MDM2-NUP107 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. MDM2-NUP107 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. MDM2-NUP107 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MDM2 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 9271120|17310983 |
| Hgene | MDM2 | GO:0006511 | ubiquitin-dependent protein catabolic process | 11278372|15314173|16173922|17310983 |
| Hgene | MDM2 | GO:0016567 | protein ubiquitination | 9450543|15878855|19656744|20153724 |
| Hgene | MDM2 | GO:0031648 | protein destabilization | 9529249|10360174|15314173 |
| Hgene | MDM2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process | 11278372 |
| Hgene | MDM2 | GO:0034504 | protein localization to nucleus | 10360174 |
| Hgene | MDM2 | GO:0042176 | regulation of protein catabolic process | 9153395 |
| Hgene | MDM2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | 9529249|10360174 |
| Hgene | MDM2 | GO:0045184 | establishment of protein localization | 10360174 |
| Hgene | MDM2 | GO:0045892 | negative regulation of transcription, DNA-templated | 9271120 |
| Hgene | MDM2 | GO:0065003 | protein-containing complex assembly | 10608892|12915590 |
| Hgene | MDM2 | GO:0071157 | negative regulation of cell cycle arrest | 9529249 |
| Hgene | MDM2 | GO:0071480 | cellular response to gamma radiation | 16213212 |
| Hgene | MDM2 | GO:0072717 | cellular response to actinomycin D | 15314173 |
| Hgene | MDM2 | GO:1901797 | negative regulation of signal transduction by p53 class mediator | 16173922 |
| Tgene | NUP107 | GO:0006406 | mRNA export from nucleus | 11684705 |
 Fusion gene breakpoints across MDM2 (5'-gene) Fusion gene breakpoints across MDM2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
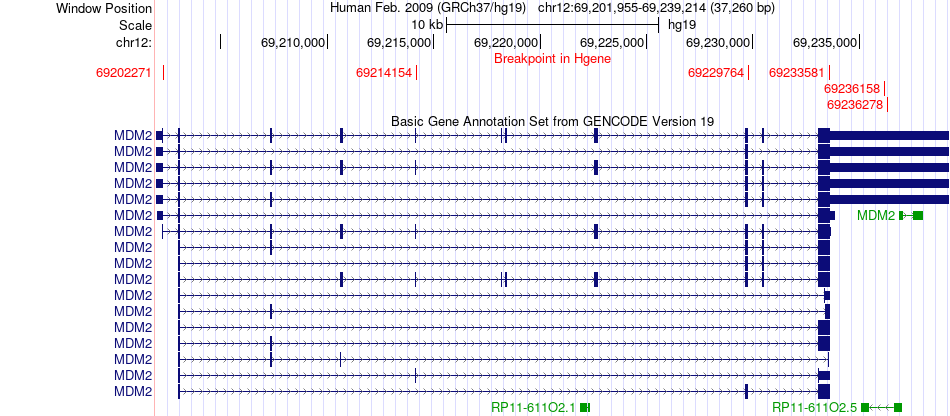 |
 Fusion gene breakpoints across NUP107 (3'-gene) Fusion gene breakpoints across NUP107 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
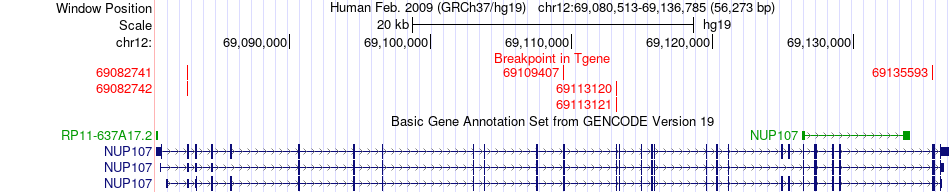 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-G2-A2EK-01A | MDM2 | chr12 | 69214154 | + | NUP107 | chr12 | 69109407 | + |
| ChimerDB4 | BRCA | TCGA-3C-AAAU-01A | MDM2 | chr12 | 69229764 | - | NUP107 | chr12 | 69113121 | + |
| ChimerDB4 | BRCA | TCGA-3C-AAAU-01A | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + |
| ChimerDB4 | BRCA | TCGA-3C-AAAU-01A | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + |
| ChimerDB4 | BRCA | TCGA-3C-AAAU-01A | MDM2 | chr12 | 69236158 | + | NUP107 | chr12 | 69113121 | + |
| ChimerDB4 | BRCA | TCGA-3C-AAAU-01A | MDM2 | chr12 | 69236278 | + | NUP107 | chr12 | 69113121 | + |
| ChimerDB4 | BRCA | TCGA-3C-AAAU | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + |
| ChimerDB4 | LUAD | TCGA-55-8085-01A | MDM2 | chr12 | 69229764 | - | NUP107 | chr12 | 69135593 | + |
| ChimerDB4 | LUAD | TCGA-55-8085-01A | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69135593 | + |
| ChimerDB4 | LUAD | TCGA-55-8085-01A | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69135593 | + |
| ChimerDB4 | LUSC | TCGA-NC-A5HE-01A | MDM2 | chr12 | 69202271 | - | NUP107 | chr12 | 69082742 | + |
| ChimerDB4 | LUSC | TCGA-NC-A5HE | MDM2 | chr12 | 69202271 | + | NUP107 | chr12 | 69082741 | + |
| ChimerDB4 | LUSC | TCGA-NC-A5HE | MDM2 | chr12 | 69202271 | + | NUP107 | chr12 | 69082742 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000462284 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113120 | + | 3380 | 1142 | 266 | 2836 | 856 |
| ENST00000462284 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113120 | + | 2794 | 1142 | 266 | 2572 | 768 |
| ENST00000462284 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113120 | + | 2906 | 1142 | 266 | 2836 | 856 |
| ENST00000356290 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113120 | + | 2855 | 617 | 251 | 2311 | 686 |
| ENST00000356290 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113120 | + | 2269 | 617 | 251 | 2047 | 598 |
| ENST00000356290 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113120 | + | 2381 | 617 | 251 | 2311 | 686 |
| ENST00000540827 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113120 | + | 2780 | 542 | 251 | 2236 | 661 |
| ENST00000540827 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113120 | + | 2194 | 542 | 251 | 1972 | 573 |
| ENST00000540827 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113120 | + | 2306 | 542 | 251 | 2236 | 661 |
| ENST00000258149 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113120 | + | 3200 | 962 | 251 | 2656 | 801 |
| ENST00000258149 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113120 | + | 2614 | 962 | 251 | 2392 | 713 |
| ENST00000258149 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113120 | + | 2726 | 962 | 251 | 2656 | 801 |
| ENST00000428863 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113120 | + | 2780 | 542 | 251 | 2236 | 661 |
| ENST00000428863 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113120 | + | 2194 | 542 | 251 | 1972 | 573 |
| ENST00000428863 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113120 | + | 2306 | 542 | 251 | 2236 | 661 |
| ENST00000258148 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113120 | + | 2927 | 689 | 14 | 2383 | 789 |
| ENST00000258148 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113120 | + | 2341 | 689 | 14 | 2119 | 701 |
| ENST00000258148 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113120 | + | 2453 | 689 | 14 | 2383 | 789 |
| ENST00000350057 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113120 | + | 2985 | 747 | 0 | 2441 | 813 |
| ENST00000350057 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113120 | + | 2399 | 747 | 0 | 2177 | 725 |
| ENST00000350057 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113120 | + | 2511 | 747 | 0 | 2441 | 813 |
| ENST00000360430 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113120 | + | 2475 | 237 | 0 | 1931 | 643 |
| ENST00000360430 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113120 | + | 1889 | 237 | 0 | 1667 | 555 |
| ENST00000360430 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113120 | + | 2001 | 237 | 0 | 1931 | 643 |
| ENST00000299252 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113120 | + | 2550 | 312 | 0 | 2006 | 668 |
| ENST00000299252 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113120 | + | 1964 | 312 | 0 | 1742 | 580 |
| ENST00000299252 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113120 | + | 2076 | 312 | 0 | 2006 | 668 |
| ENST00000348801 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113120 | + | 2460 | 222 | 0 | 1916 | 638 |
| ENST00000348801 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113120 | + | 1874 | 222 | 0 | 1652 | 550 |
| ENST00000348801 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113120 | + | 1986 | 222 | 0 | 1916 | 638 |
| ENST00000462284 | MDM2 | chr12 | 69202271 | + | ENST00000229179 | NUP107 | chr12 | 69082741 | + | 3629 | 316 | 266 | 3085 | 939 |
| ENST00000258148 | MDM2 | chr12 | 69202271 | + | ENST00000229179 | NUP107 | chr12 | 69082741 | + | 3341 | 28 | 14 | 2797 | 927 |
| ENST00000393412 | MDM2 | chr12 | 69233581 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2881 | 643 | 706 | 2337 | 543 |
| ENST00000393412 | MDM2 | chr12 | 69233581 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2295 | 643 | 706 | 2073 | 455 |
| ENST00000393412 | MDM2 | chr12 | 69233581 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2407 | 643 | 706 | 2337 | 543 |
| ENST00000258148 | MDM2 | chr12 | 69233581 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 3071 | 833 | 896 | 2527 | 543 |
| ENST00000258148 | MDM2 | chr12 | 69233581 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2485 | 833 | 896 | 2263 | 455 |
| ENST00000258148 | MDM2 | chr12 | 69233581 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2597 | 833 | 896 | 2527 | 543 |
| ENST00000393410 | MDM2 | chr12 | 69233581 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2442 | 204 | 267 | 1898 | 543 |
| ENST00000393410 | MDM2 | chr12 | 69233581 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 1856 | 204 | 267 | 1634 | 455 |
| ENST00000393410 | MDM2 | chr12 | 69233581 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 1968 | 204 | 267 | 1898 | 543 |
| ENST00000393413 | MDM2 | chr12 | 69233581 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2367 | 129 | 192 | 1823 | 543 |
| ENST00000393413 | MDM2 | chr12 | 69233581 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 1781 | 129 | 192 | 1559 | 455 |
| ENST00000393413 | MDM2 | chr12 | 69233581 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 1893 | 129 | 192 | 1823 | 543 |
| ENST00000517852 | MDM2 | chr12 | 69233581 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2442 | 204 | 267 | 1898 | 543 |
| ENST00000517852 | MDM2 | chr12 | 69233581 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 1856 | 204 | 267 | 1634 | 455 |
| ENST00000517852 | MDM2 | chr12 | 69233581 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 1968 | 204 | 267 | 1898 | 543 |
| ENST00000350057 | MDM2 | chr12 | 69233581 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 3111 | 873 | 936 | 2567 | 543 |
| ENST00000350057 | MDM2 | chr12 | 69233581 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2525 | 873 | 936 | 2303 | 455 |
| ENST00000350057 | MDM2 | chr12 | 69233581 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2637 | 873 | 936 | 2567 | 543 |
| ENST00000360430 | MDM2 | chr12 | 69233581 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2601 | 363 | 426 | 2057 | 543 |
| ENST00000360430 | MDM2 | chr12 | 69233581 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2015 | 363 | 426 | 1793 | 455 |
| ENST00000360430 | MDM2 | chr12 | 69233581 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2127 | 363 | 426 | 2057 | 543 |
| ENST00000299252 | MDM2 | chr12 | 69233581 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2676 | 438 | 501 | 2132 | 543 |
| ENST00000299252 | MDM2 | chr12 | 69233581 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2090 | 438 | 501 | 1868 | 455 |
| ENST00000299252 | MDM2 | chr12 | 69233581 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2202 | 438 | 501 | 2132 | 543 |
| ENST00000348801 | MDM2 | chr12 | 69233581 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2508 | 270 | 333 | 1964 | 543 |
| ENST00000348801 | MDM2 | chr12 | 69233581 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 1922 | 270 | 333 | 1700 | 455 |
| ENST00000348801 | MDM2 | chr12 | 69233581 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2034 | 270 | 333 | 1964 | 543 |
| ENST00000462284 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 3380 | 1142 | 266 | 2836 | 856 |
| ENST00000462284 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2794 | 1142 | 266 | 2572 | 768 |
| ENST00000462284 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2906 | 1142 | 266 | 2836 | 856 |
| ENST00000356290 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2855 | 617 | 251 | 2311 | 686 |
| ENST00000356290 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2269 | 617 | 251 | 2047 | 598 |
| ENST00000356290 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2381 | 617 | 251 | 2311 | 686 |
| ENST00000540827 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2780 | 542 | 251 | 2236 | 661 |
| ENST00000540827 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2194 | 542 | 251 | 1972 | 573 |
| ENST00000540827 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2306 | 542 | 251 | 2236 | 661 |
| ENST00000258149 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 3200 | 962 | 251 | 2656 | 801 |
| ENST00000258149 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2614 | 962 | 251 | 2392 | 713 |
| ENST00000258149 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2726 | 962 | 251 | 2656 | 801 |
| ENST00000428863 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2780 | 542 | 251 | 2236 | 661 |
| ENST00000428863 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2194 | 542 | 251 | 1972 | 573 |
| ENST00000428863 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2306 | 542 | 251 | 2236 | 661 |
| ENST00000258148 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2927 | 689 | 14 | 2383 | 789 |
| ENST00000258148 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2341 | 689 | 14 | 2119 | 701 |
| ENST00000258148 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2453 | 689 | 14 | 2383 | 789 |
| ENST00000350057 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2985 | 747 | 0 | 2441 | 813 |
| ENST00000350057 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 2399 | 747 | 0 | 2177 | 725 |
| ENST00000350057 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2511 | 747 | 0 | 2441 | 813 |
| ENST00000360430 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2475 | 237 | 0 | 1931 | 643 |
| ENST00000360430 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 1889 | 237 | 0 | 1667 | 555 |
| ENST00000360430 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2001 | 237 | 0 | 1931 | 643 |
| ENST00000299252 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2550 | 312 | 0 | 2006 | 668 |
| ENST00000299252 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 1964 | 312 | 0 | 1742 | 580 |
| ENST00000299252 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 2076 | 312 | 0 | 2006 | 668 |
| ENST00000348801 | MDM2 | chr12 | 69229764 | + | ENST00000229179 | NUP107 | chr12 | 69113121 | + | 2460 | 222 | 0 | 1916 | 638 |
| ENST00000348801 | MDM2 | chr12 | 69229764 | + | ENST00000378905 | NUP107 | chr12 | 69113121 | + | 1874 | 222 | 0 | 1652 | 550 |
| ENST00000348801 | MDM2 | chr12 | 69229764 | + | ENST00000539906 | NUP107 | chr12 | 69113121 | + | 1986 | 222 | 0 | 1916 | 638 |
| ENST00000462284 | MDM2 | chr12 | 69202271 | + | ENST00000229179 | NUP107 | chr12 | 69082742 | + | 3629 | 316 | 266 | 3085 | 939 |
| ENST00000258148 | MDM2 | chr12 | 69202271 | + | ENST00000229179 | NUP107 | chr12 | 69082742 | + | 3341 | 28 | 14 | 2797 | 927 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000462284 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.00055025 | 0.9994498 |
| ENST00000462284 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000999037 | 0.99900097 |
| ENST00000462284 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001122307 | 0.9988777 |
| ENST00000356290 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000663832 | 0.9993362 |
| ENST00000356290 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000862723 | 0.9991373 |
| ENST00000356290 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001576973 | 0.9984231 |
| ENST00000540827 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000673538 | 0.99932647 |
| ENST00000540827 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.00115321 | 0.99884677 |
| ENST00000540827 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001722858 | 0.9982772 |
| ENST00000258149 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000746172 | 0.9992538 |
| ENST00000258149 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001145424 | 0.9988545 |
| ENST00000258149 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001655022 | 0.998345 |
| ENST00000428863 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000673538 | 0.99932647 |
| ENST00000428863 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.00115321 | 0.99884677 |
| ENST00000428863 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001722858 | 0.9982772 |
| ENST00000258148 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000827246 | 0.99917275 |
| ENST00000258148 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001355768 | 0.9986443 |
| ENST00000258148 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.002003879 | 0.9979961 |
| ENST00000350057 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000509246 | 0.99949074 |
| ENST00000350057 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000982023 | 0.99901795 |
| ENST00000350057 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001149335 | 0.9988507 |
| ENST00000360430 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000919971 | 0.99908006 |
| ENST00000360430 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001802795 | 0.99819726 |
| ENST00000360430 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.00262168 | 0.99737835 |
| ENST00000299252 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000649259 | 0.9993507 |
| ENST00000299252 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001038627 | 0.9989613 |
| ENST00000299252 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001657282 | 0.99834275 |
| ENST00000348801 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000578085 | 0.99942195 |
| ENST00000348801 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.000710322 | 0.99928963 |
| ENST00000348801 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113120 | + | 0.001684242 | 0.9983158 |
| ENST00000462284 | ENST00000229179 | MDM2 | chr12 | 69202271 | + | NUP107 | chr12 | 69082741 | + | 0.000188681 | 0.99981135 |
| ENST00000258148 | ENST00000229179 | MDM2 | chr12 | 69202271 | + | NUP107 | chr12 | 69082741 | + | 0.000198743 | 0.9998012 |
| ENST00000393412 | ENST00000229179 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000141389 | 0.9998586 |
| ENST00000393412 | ENST00000378905 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000168001 | 0.9998319 |
| ENST00000393412 | ENST00000539906 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.00025163 | 0.9997484 |
| ENST00000258148 | ENST00000229179 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000233391 | 0.99976665 |
| ENST00000258148 | ENST00000378905 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000310624 | 0.99968934 |
| ENST00000258148 | ENST00000539906 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000538181 | 0.9994618 |
| ENST00000393410 | ENST00000229179 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000148334 | 0.9998517 |
| ENST00000393410 | ENST00000378905 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000188009 | 0.99981207 |
| ENST00000393410 | ENST00000539906 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000360878 | 0.99963915 |
| ENST00000393413 | ENST00000229179 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000151171 | 0.99984884 |
| ENST00000393413 | ENST00000378905 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.0002053 | 0.99979466 |
| ENST00000393413 | ENST00000539906 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000398481 | 0.99960154 |
| ENST00000517852 | ENST00000229179 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000148334 | 0.9998517 |
| ENST00000517852 | ENST00000378905 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000188009 | 0.99981207 |
| ENST00000517852 | ENST00000539906 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000360878 | 0.99963915 |
| ENST00000350057 | ENST00000229179 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.00024659 | 0.9997534 |
| ENST00000350057 | ENST00000378905 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000338806 | 0.9996612 |
| ENST00000350057 | ENST00000539906 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000539268 | 0.99946076 |
| ENST00000360430 | ENST00000229179 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.00022958 | 0.99977046 |
| ENST00000360430 | ENST00000378905 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000338786 | 0.9996612 |
| ENST00000360430 | ENST00000539906 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000613114 | 0.9993869 |
| ENST00000299252 | ENST00000229179 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000215059 | 0.999785 |
| ENST00000299252 | ENST00000378905 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000293095 | 0.9997069 |
| ENST00000299252 | ENST00000539906 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000536207 | 0.99946386 |
| ENST00000348801 | ENST00000229179 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.00021225 | 0.99978775 |
| ENST00000348801 | ENST00000378905 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000317172 | 0.9996829 |
| ENST00000348801 | ENST00000539906 | MDM2 | chr12 | 69233581 | + | NUP107 | chr12 | 69113121 | + | 0.000601038 | 0.99939895 |
| ENST00000462284 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.00055025 | 0.9994498 |
| ENST00000462284 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000999037 | 0.99900097 |
| ENST00000462284 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001122307 | 0.9988777 |
| ENST00000356290 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000663832 | 0.9993362 |
| ENST00000356290 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000862723 | 0.9991373 |
| ENST00000356290 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001576973 | 0.9984231 |
| ENST00000540827 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000673538 | 0.99932647 |
| ENST00000540827 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.00115321 | 0.99884677 |
| ENST00000540827 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001722858 | 0.9982772 |
| ENST00000258149 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000746172 | 0.9992538 |
| ENST00000258149 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001145424 | 0.9988545 |
| ENST00000258149 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001655022 | 0.998345 |
| ENST00000428863 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000673538 | 0.99932647 |
| ENST00000428863 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.00115321 | 0.99884677 |
| ENST00000428863 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001722858 | 0.9982772 |
| ENST00000258148 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000827246 | 0.99917275 |
| ENST00000258148 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001355768 | 0.9986443 |
| ENST00000258148 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.002003879 | 0.9979961 |
| ENST00000350057 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000509246 | 0.99949074 |
| ENST00000350057 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000982023 | 0.99901795 |
| ENST00000350057 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001149335 | 0.9988507 |
| ENST00000360430 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000919971 | 0.99908006 |
| ENST00000360430 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001802795 | 0.99819726 |
| ENST00000360430 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.00262168 | 0.99737835 |
| ENST00000299252 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000649259 | 0.9993507 |
| ENST00000299252 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001038627 | 0.9989613 |
| ENST00000299252 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001657282 | 0.99834275 |
| ENST00000348801 | ENST00000229179 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000578085 | 0.99942195 |
| ENST00000348801 | ENST00000378905 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.000710322 | 0.99928963 |
| ENST00000348801 | ENST00000539906 | MDM2 | chr12 | 69229764 | + | NUP107 | chr12 | 69113121 | + | 0.001684242 | 0.9983158 |
| ENST00000462284 | ENST00000229179 | MDM2 | chr12 | 69202271 | + | NUP107 | chr12 | 69082742 | + | 0.000188681 | 0.99981135 |
| ENST00000258148 | ENST00000229179 | MDM2 | chr12 | 69202271 | + | NUP107 | chr12 | 69082742 | + | 0.000198743 | 0.9998012 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >52413_52413_1_MDM2-NUP107_MDM2_chr12_69202271_ENST00000258148_NUP107_chr12_69082741_ENST00000229179_length(amino acids)=927AA_BP=4 MVRSRSGFGEISSPVIREAEVTRTARKQSAQKRVLLQASQDENFGNTTPRNQVIPRTPSSFRQPFTPTSRSLLRQPDISCILGTGGKSPR LTQSSGFFGNLSMVTNLDDSNWAAAFSSQRSGLFTNTEPHSITEDVTISAVMLREDDPGEAASMSMFSDFLQSFLKHSSSTVFDLVEEYE NICGSQVNILSKIVSRATPGLQKFSKTASMLWLLQQEMVTWRLLASLYRDRIQSALEEESVFAVTAVNASEKTVVEALFQRDSLVRQSQL VVDWLESIAKDEIGEFSDNIEFYAKSVYWENTLHTLKQRQLTSYVGSVRPLVTELDPDAPIRQKMPLDDLDREDEVRLLKYLFTLIRAGM TEEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDT VWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKS RNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEA DLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQD SIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDA LTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELR -------------------------------------------------------------- >52413_52413_2_MDM2-NUP107_MDM2_chr12_69202271_ENST00000258148_NUP107_chr12_69082742_ENST00000229179_length(amino acids)=927AA_BP=4 MVRSRSGFGEISSPVIREAEVTRTARKQSAQKRVLLQASQDENFGNTTPRNQVIPRTPSSFRQPFTPTSRSLLRQPDISCILGTGGKSPR LTQSSGFFGNLSMVTNLDDSNWAAAFSSQRSGLFTNTEPHSITEDVTISAVMLREDDPGEAASMSMFSDFLQSFLKHSSSTVFDLVEEYE NICGSQVNILSKIVSRATPGLQKFSKTASMLWLLQQEMVTWRLLASLYRDRIQSALEEESVFAVTAVNASEKTVVEALFQRDSLVRQSQL VVDWLESIAKDEIGEFSDNIEFYAKSVYWENTLHTLKQRQLTSYVGSVRPLVTELDPDAPIRQKMPLDDLDREDEVRLLKYLFTLIRAGM TEEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDT VWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKS RNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEA DLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQD SIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDA LTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELR -------------------------------------------------------------- >52413_52413_3_MDM2-NUP107_MDM2_chr12_69202271_ENST00000462284_NUP107_chr12_69082741_ENST00000229179_length(amino acids)=939AA_BP=16 MRDPRLQARKPRMVRSRSGFGEISSPVIREAEVTRTARKQSAQKRVLLQASQDENFGNTTPRNQVIPRTPSSFRQPFTPTSRSLLRQPDI SCILGTGGKSPRLTQSSGFFGNLSMVTNLDDSNWAAAFSSQRSGLFTNTEPHSITEDVTISAVMLREDDPGEAASMSMFSDFLQSFLKHS SSTVFDLVEEYENICGSQVNILSKIVSRATPGLQKFSKTASMLWLLQQEMVTWRLLASLYRDRIQSALEEESVFAVTAVNASEKTVVEAL FQRDSLVRQSQLVVDWLESIAKDEIGEFSDNIEFYAKSVYWENTLHTLKQRQLTSYVGSVRPLVTELDPDAPIRQKMPLDDLDREDEVRL LKYLFTLIRAGMTEEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLK QLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDG LMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFE QRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHE AAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYE MDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERH -------------------------------------------------------------- >52413_52413_4_MDM2-NUP107_MDM2_chr12_69202271_ENST00000462284_NUP107_chr12_69082742_ENST00000229179_length(amino acids)=939AA_BP=16 MRDPRLQARKPRMVRSRSGFGEISSPVIREAEVTRTARKQSAQKRVLLQASQDENFGNTTPRNQVIPRTPSSFRQPFTPTSRSLLRQPDI SCILGTGGKSPRLTQSSGFFGNLSMVTNLDDSNWAAAFSSQRSGLFTNTEPHSITEDVTISAVMLREDDPGEAASMSMFSDFLQSFLKHS SSTVFDLVEEYENICGSQVNILSKIVSRATPGLQKFSKTASMLWLLQQEMVTWRLLASLYRDRIQSALEEESVFAVTAVNASEKTVVEAL FQRDSLVRQSQLVVDWLESIAKDEIGEFSDNIEFYAKSVYWENTLHTLKQRQLTSYVGSVRPLVTELDPDAPIRQKMPLDDLDREDEVRL LKYLFTLIRAGMTEEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLK QLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDG LMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFE QRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHE AAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYE MDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERH -------------------------------------------------------------- >52413_52413_5_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258148_NUP107_chr12_69113120_ENST00000229179_length(amino acids)=789AA_BP=225 MVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGD LFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVS EHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRI WKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEEL QATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHT NLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVID WLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKH MNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPM -------------------------------------------------------------- >52413_52413_6_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258148_NUP107_chr12_69113120_ENST00000378905_length(amino acids)=701AA_BP=225 MVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGD LFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVS EHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRI WKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEEL QATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDV IDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWF KHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCL -------------------------------------------------------------- >52413_52413_7_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258148_NUP107_chr12_69113120_ENST00000539906_length(amino acids)=789AA_BP=225 MVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGD LFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVS EHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRI WKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEEL QATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHT NLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVID WLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKH MNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPM -------------------------------------------------------------- >52413_52413_8_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258148_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=789AA_BP=225 MVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGD LFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVS EHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRI WKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEEL QATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHT NLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVID WLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKH MNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPM -------------------------------------------------------------- >52413_52413_9_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258148_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=701AA_BP=225 MVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGD LFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVS EHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRI WKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEEL QATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDV IDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWF KHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCL -------------------------------------------------------------- >52413_52413_10_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258148_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=789AA_BP=225 MVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGD LFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVS EHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRI WKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEEL QATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHT NLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVID WLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKH MNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPM -------------------------------------------------------------- >52413_52413_11_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258149_NUP107_chr12_69113120_ENST00000229179_length(amino acids)=801AA_BP=237 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTG TPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTE LEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLG ANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLK TYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGT TEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYL EAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTH -------------------------------------------------------------- >52413_52413_12_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258149_NUP107_chr12_69113120_ENST00000378905_length(amino acids)=713AA_BP=237 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTG TPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTE LEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLG ANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLK TYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRA YLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHER -------------------------------------------------------------- >52413_52413_13_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258149_NUP107_chr12_69113120_ENST00000539906_length(amino acids)=801AA_BP=237 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTG TPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTE LEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLG ANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLK TYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGT TEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYL EAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTH -------------------------------------------------------------- >52413_52413_14_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258149_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=801AA_BP=237 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTG TPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTE LEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLG ANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLK TYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGT TEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYL EAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTH -------------------------------------------------------------- >52413_52413_15_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258149_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=713AA_BP=237 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTG TPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTE LEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLG ANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLK TYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRA YLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHER -------------------------------------------------------------- >52413_52413_16_MDM2-NUP107_MDM2_chr12_69229764_ENST00000258149_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=801AA_BP=237 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTG TPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTE LEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLG ANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLK TYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGT TEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYL EAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTH -------------------------------------------------------------- >52413_52413_17_MDM2-NUP107_MDM2_chr12_69229764_ENST00000299252_NUP107_chr12_69113120_ENST00000229179_length(amino acids)=668AA_BP=104 MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYS LSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQ LLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGL MDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQ RHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEA AKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEM DFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHK -------------------------------------------------------------- >52413_52413_18_MDM2-NUP107_MDM2_chr12_69229764_ENST00000299252_NUP107_chr12_69113120_ENST00000378905_length(amino acids)=580AA_BP=104 MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYS LSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQ LLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGL MDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKH EAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKY EMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSER -------------------------------------------------------------- >52413_52413_19_MDM2-NUP107_MDM2_chr12_69229764_ENST00000299252_NUP107_chr12_69113120_ENST00000539906_length(amino acids)=668AA_BP=104 MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYS LSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQ LLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGL MDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQ RHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEA AKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEM DFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHK -------------------------------------------------------------- >52413_52413_20_MDM2-NUP107_MDM2_chr12_69229764_ENST00000299252_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=668AA_BP=104 MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYS LSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQ LLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGL MDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQ RHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEA AKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEM DFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHK -------------------------------------------------------------- >52413_52413_21_MDM2-NUP107_MDM2_chr12_69229764_ENST00000299252_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=580AA_BP=104 MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYS LSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQ LLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGL MDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKH EAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKY EMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSER -------------------------------------------------------------- >52413_52413_22_MDM2-NUP107_MDM2_chr12_69229764_ENST00000299252_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=668AA_BP=104 MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYS LSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQ LLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGL MDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQ RHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEA AKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEM DFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHK -------------------------------------------------------------- >52413_52413_23_MDM2-NUP107_MDM2_chr12_69229764_ENST00000348801_NUP107_chr12_69113120_ENST00000229179_length(amino acids)=638AA_BP=74 MSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAAT LEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQT SVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFF RTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKD NGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLP AEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGG WMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLD -------------------------------------------------------------- >52413_52413_24_MDM2-NUP107_MDM2_chr12_69229764_ENST00000348801_NUP107_chr12_69113120_ENST00000378905_length(amino acids)=550AA_BP=74 MSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAAT LEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQT SVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFF RTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESP LPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVD GGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQG -------------------------------------------------------------- >52413_52413_25_MDM2-NUP107_MDM2_chr12_69229764_ENST00000348801_NUP107_chr12_69113120_ENST00000539906_length(amino acids)=638AA_BP=74 MSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAAT LEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQT SVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFF RTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKD NGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLP AEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGG WMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLD -------------------------------------------------------------- >52413_52413_26_MDM2-NUP107_MDM2_chr12_69229764_ENST00000348801_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=638AA_BP=74 MSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAAT LEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQT SVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFF RTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKD NGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLP AEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGG WMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLD -------------------------------------------------------------- >52413_52413_27_MDM2-NUP107_MDM2_chr12_69229764_ENST00000348801_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=550AA_BP=74 MSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAAT LEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQT SVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFF RTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESP LPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVD GGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQG -------------------------------------------------------------- >52413_52413_28_MDM2-NUP107_MDM2_chr12_69229764_ENST00000348801_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=638AA_BP=74 MSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAAT LEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQT SVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFF RTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKD NGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLP AEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGG WMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLD -------------------------------------------------------------- >52413_52413_29_MDM2-NUP107_MDM2_chr12_69229764_ENST00000350057_NUP107_chr12_69113120_ENST00000229179_length(amino acids)=813AA_BP=249 MCNTNMSVPTDGAVTTSQIPASEQETLVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQES SDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREI CCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWK LYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_30_MDM2-NUP107_MDM2_chr12_69229764_ENST00000350057_NUP107_chr12_69113120_ENST00000378905_length(amino acids)=725AA_BP=249 MCNTNMSVPTDGAVTTSQIPASEQETLVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQES SDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREI CCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWK LYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_31_MDM2-NUP107_MDM2_chr12_69229764_ENST00000350057_NUP107_chr12_69113120_ENST00000539906_length(amino acids)=813AA_BP=249 MCNTNMSVPTDGAVTTSQIPASEQETLVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQES SDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREI CCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWK LYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_32_MDM2-NUP107_MDM2_chr12_69229764_ENST00000350057_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=813AA_BP=249 MCNTNMSVPTDGAVTTSQIPASEQETLVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQES SDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREI CCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWK LYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_33_MDM2-NUP107_MDM2_chr12_69229764_ENST00000350057_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=725AA_BP=249 MCNTNMSVPTDGAVTTSQIPASEQETLVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQES SDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREI CCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWK LYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_34_MDM2-NUP107_MDM2_chr12_69229764_ENST00000350057_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=813AA_BP=249 MCNTNMSVPTDGAVTTSQIPASEQETLVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQES SDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREI CCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWK LYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_35_MDM2-NUP107_MDM2_chr12_69229764_ENST00000356290_NUP107_chr12_69113120_ENST00000229179_length(amino acids)=686AA_BP=122 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVS DQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDEL FNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQ EHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQD LAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEAL KQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQ PTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTG -------------------------------------------------------------- >52413_52413_36_MDM2-NUP107_MDM2_chr12_69229764_ENST00000356290_NUP107_chr12_69113120_ENST00000378905_length(amino acids)=598AA_BP=122 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVS DQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDEL FNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQ EHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAE ALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALI PQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHS -------------------------------------------------------------- >52413_52413_37_MDM2-NUP107_MDM2_chr12_69229764_ENST00000356290_NUP107_chr12_69113120_ENST00000539906_length(amino acids)=686AA_BP=122 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVS DQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDEL FNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQ EHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQD LAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEAL KQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQ PTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTG -------------------------------------------------------------- >52413_52413_38_MDM2-NUP107_MDM2_chr12_69229764_ENST00000356290_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=686AA_BP=122 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVS DQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDEL FNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQ EHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQD LAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEAL KQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQ PTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTG -------------------------------------------------------------- >52413_52413_39_MDM2-NUP107_MDM2_chr12_69229764_ENST00000356290_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=598AA_BP=122 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVS DQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDEL FNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQ EHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAE ALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALI PQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHS -------------------------------------------------------------- >52413_52413_40_MDM2-NUP107_MDM2_chr12_69229764_ENST00000356290_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=686AA_BP=122 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEDLDAGVSEHSGDWLDQDSVS DQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDEL FNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQ EHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQD LAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEAL KQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQ PTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTG -------------------------------------------------------------- >52413_52413_41_MDM2-NUP107_MDM2_chr12_69229764_ENST00000360430_NUP107_chr12_69113120_ENST00000229179_length(amino acids)=643AA_BP=79 MCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQA WRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVE QEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTH LILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVEN IRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGM ESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLL FVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLL -------------------------------------------------------------- >52413_52413_42_MDM2-NUP107_MDM2_chr12_69229764_ENST00000360430_NUP107_chr12_69113120_ENST00000378905_length(amino acids)=555AA_BP=79 MCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQA WRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVE QEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTH LILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQ GMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNV LLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLM -------------------------------------------------------------- >52413_52413_43_MDM2-NUP107_MDM2_chr12_69229764_ENST00000360430_NUP107_chr12_69113120_ENST00000539906_length(amino acids)=643AA_BP=79 MCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQA WRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVE QEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTH LILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVEN IRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGM ESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLL FVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLL -------------------------------------------------------------- >52413_52413_44_MDM2-NUP107_MDM2_chr12_69229764_ENST00000360430_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=643AA_BP=79 MCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQA WRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVE QEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTH LILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVEN IRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGM ESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLL FVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLL -------------------------------------------------------------- >52413_52413_45_MDM2-NUP107_MDM2_chr12_69229764_ENST00000360430_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=555AA_BP=79 MCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQA WRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVE QEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTH LILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQ GMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNV LLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLM -------------------------------------------------------------- >52413_52413_46_MDM2-NUP107_MDM2_chr12_69229764_ENST00000360430_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=643AA_BP=79 MCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQA WRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVE QEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTH LILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVEN IRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGM ESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLL FVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLL -------------------------------------------------------------- >52413_52413_47_MDM2-NUP107_MDM2_chr12_69229764_ENST00000428863_NUP107_chr12_69113120_ENST00000229179_length(amino acids)=661AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLEL AKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVK IPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKG HLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSK -------------------------------------------------------------- >52413_52413_48_MDM2-NUP107_MDM2_chr12_69229764_ENST00000428863_NUP107_chr12_69113120_ENST00000378905_length(amino acids)=573AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVF VKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIW KGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVF -------------------------------------------------------------- >52413_52413_49_MDM2-NUP107_MDM2_chr12_69229764_ENST00000428863_NUP107_chr12_69113120_ENST00000539906_length(amino acids)=661AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLEL AKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVK IPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKG HLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSK -------------------------------------------------------------- >52413_52413_50_MDM2-NUP107_MDM2_chr12_69229764_ENST00000428863_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=661AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLEL AKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVK IPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKG HLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSK -------------------------------------------------------------- >52413_52413_51_MDM2-NUP107_MDM2_chr12_69229764_ENST00000428863_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=573AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVF VKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIW KGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVF -------------------------------------------------------------- >52413_52413_52_MDM2-NUP107_MDM2_chr12_69229764_ENST00000428863_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=661AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLEL AKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVK IPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKG HLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSK -------------------------------------------------------------- >52413_52413_53_MDM2-NUP107_MDM2_chr12_69229764_ENST00000462284_NUP107_chr12_69113120_ENST00000229179_length(amino acids)=856AA_BP=292 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRR RAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVE SLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYA ALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFL ILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFL ESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKF LASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHE HKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLAD -------------------------------------------------------------- >52413_52413_54_MDM2-NUP107_MDM2_chr12_69229764_ENST00000462284_NUP107_chr12_69113120_ENST00000378905_length(amino acids)=768AA_BP=292 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRR RAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVE SLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYA ALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFL ILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMR KFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVA HEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQL -------------------------------------------------------------- >52413_52413_55_MDM2-NUP107_MDM2_chr12_69229764_ENST00000462284_NUP107_chr12_69113120_ENST00000539906_length(amino acids)=856AA_BP=292 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRR RAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVE SLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYA ALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFL ILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFL ESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKF LASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHE HKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLAD -------------------------------------------------------------- >52413_52413_56_MDM2-NUP107_MDM2_chr12_69229764_ENST00000462284_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=856AA_BP=292 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRR RAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVE SLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYA ALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFL ILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFL ESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKF LASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHE HKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLAD -------------------------------------------------------------- >52413_52413_57_MDM2-NUP107_MDM2_chr12_69229764_ENST00000462284_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=768AA_BP=292 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRR RAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVE SLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYA ALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFL ILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMR KFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVA HEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQL -------------------------------------------------------------- >52413_52413_58_MDM2-NUP107_MDM2_chr12_69229764_ENST00000462284_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=856AA_BP=292 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQ HIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRR RAISETEENSDELSGERQRKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDSVSDQFSVEFEVE SLDSEDYSLSEEGQELSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYA ALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFL ILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFL ESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKF LASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHE HKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLAD -------------------------------------------------------------- >52413_52413_59_MDM2-NUP107_MDM2_chr12_69229764_ENST00000540827_NUP107_chr12_69113120_ENST00000229179_length(amino acids)=661AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLEL AKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVK IPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKG HLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSK -------------------------------------------------------------- >52413_52413_60_MDM2-NUP107_MDM2_chr12_69229764_ENST00000540827_NUP107_chr12_69113120_ENST00000378905_length(amino acids)=573AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVF VKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIW KGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVF -------------------------------------------------------------- >52413_52413_61_MDM2-NUP107_MDM2_chr12_69229764_ENST00000540827_NUP107_chr12_69113120_ENST00000539906_length(amino acids)=661AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLEL AKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVK IPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKG HLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSK -------------------------------------------------------------- >52413_52413_62_MDM2-NUP107_MDM2_chr12_69229764_ENST00000540827_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=661AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLEL AKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVK IPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKG HLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSK -------------------------------------------------------------- >52413_52413_63_MDM2-NUP107_MDM2_chr12_69229764_ENST00000540827_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=573AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVF VKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIW KGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVF -------------------------------------------------------------- >52413_52413_64_MDM2-NUP107_MDM2_chr12_69229764_ENST00000540827_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=661AA_BP=97 MRDPRLQARKPRMVRSRQMCNTNMSVPTDGAVTTSQIPASEQETLDLDAGVSEHSGDWLDQDSVSDQFSVEFEVESLDSEDYSLSEEGQE LSDEDDEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDT WEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKW LSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLEL AKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVK IPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKG HLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSK -------------------------------------------------------------- >52413_52413_65_MDM2-NUP107_MDM2_chr12_69233581_ENST00000258148_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_66_MDM2-NUP107_MDM2_chr12_69233581_ENST00000258148_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=455AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_67_MDM2-NUP107_MDM2_chr12_69233581_ENST00000258148_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_68_MDM2-NUP107_MDM2_chr12_69233581_ENST00000299252_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_69_MDM2-NUP107_MDM2_chr12_69233581_ENST00000299252_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=455AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_70_MDM2-NUP107_MDM2_chr12_69233581_ENST00000299252_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_71_MDM2-NUP107_MDM2_chr12_69233581_ENST00000348801_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_72_MDM2-NUP107_MDM2_chr12_69233581_ENST00000348801_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=455AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_73_MDM2-NUP107_MDM2_chr12_69233581_ENST00000348801_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_74_MDM2-NUP107_MDM2_chr12_69233581_ENST00000350057_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_75_MDM2-NUP107_MDM2_chr12_69233581_ENST00000350057_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=455AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_76_MDM2-NUP107_MDM2_chr12_69233581_ENST00000350057_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_77_MDM2-NUP107_MDM2_chr12_69233581_ENST00000360430_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_78_MDM2-NUP107_MDM2_chr12_69233581_ENST00000360430_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=455AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_79_MDM2-NUP107_MDM2_chr12_69233581_ENST00000360430_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_80_MDM2-NUP107_MDM2_chr12_69233581_ENST00000393410_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_81_MDM2-NUP107_MDM2_chr12_69233581_ENST00000393410_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=455AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_82_MDM2-NUP107_MDM2_chr12_69233581_ENST00000393410_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_83_MDM2-NUP107_MDM2_chr12_69233581_ENST00000393412_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_84_MDM2-NUP107_MDM2_chr12_69233581_ENST00000393412_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=455AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_85_MDM2-NUP107_MDM2_chr12_69233581_ENST00000393412_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_86_MDM2-NUP107_MDM2_chr12_69233581_ENST00000393413_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_87_MDM2-NUP107_MDM2_chr12_69233581_ENST00000393413_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=455AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_88_MDM2-NUP107_MDM2_chr12_69233581_ENST00000393413_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_89_MDM2-NUP107_MDM2_chr12_69233581_ENST00000517852_NUP107_chr12_69113121_ENST00000229179_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- >52413_52413_90_MDM2-NUP107_MDM2_chr12_69233581_ENST00000517852_NUP107_chr12_69113121_ENST00000378905_length(amino acids)=455AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAED DNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMV DVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLG -------------------------------------------------------------- >52413_52413_91_MDM2-NUP107_MDM2_chr12_69233581_ENST00000517852_NUP107_chr12_69113121_ENST00000539906_length(amino acids)=543AA_BP= MYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLKQLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATL DETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDGLMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGL QTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFEQRHHCLELAKEADLDVATITKTVVENIRKKDNGEFS HHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHEAAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDN AIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYEMDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDV REDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERHKLYLVFSKEELRKLLQKLRESSLMLLDQGLDPLGYE -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:69229764/chr12:69113121) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MDM2 | NUP107 |
| FUNCTION: E3 ubiquitin-protein ligase that mediates ubiquitination of p53/TP53, leading to its degradation by the proteasome. Inhibits p53/TP53- and p73/TP73-mediated cell cycle arrest and apoptosis by binding its transcriptional activation domain. Also acts as a ubiquitin ligase E3 toward itself and ARRB1. Permits the nuclear export of p53/TP53. Promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma RB1 protein. Inhibits DAXX-mediated apoptosis by inducing its ubiquitination and degradation. Component of the TRIM28/KAP1-MDM2-p53/TP53 complex involved in stabilizing p53/TP53. Also component of the TRIM28/KAP1-ERBB4-MDM2 complex which links growth factor and DNA damage response pathways. Mediates ubiquitination and subsequent proteasome degradation of DYRK2 in nucleus. Ubiquitinates IGF1R and SNAI1 and promotes them to proteasomal degradation (PubMed:12821780, PubMed:15053880, PubMed:15195100, PubMed:15632057, PubMed:16337594, PubMed:17290220, PubMed:19098711, PubMed:19219073, PubMed:19837670, PubMed:19965871, PubMed:20173098, PubMed:20385133, PubMed:20858735, PubMed:22128911). Ubiquitinates DCX, leading to DCX degradation and reduction of the dendritic spine density of olfactory bulb granule cells (By similarity). Ubiquitinates DLG4, leading to proteasomal degradation of DLG4 which is required for AMPA receptor endocytosis (By similarity). Negatively regulates NDUFS1, leading to decreased mitochondrial respiration, marked oxidative stress, and commitment to the mitochondrial pathway of apoptosis (PubMed:30879903). Binds NDUFS1 leading to its cytosolic retention rather than mitochondrial localization resulting in decreased supercomplex assembly (interactions between complex I and complex III), decreased complex I activity, ROS production, and apoptosis (PubMed:30879903). {ECO:0000250|UniProtKB:P23804, ECO:0000269|PubMed:12821780, ECO:0000269|PubMed:15053880, ECO:0000269|PubMed:15195100, ECO:0000269|PubMed:15632057, ECO:0000269|PubMed:16337594, ECO:0000269|PubMed:17290220, ECO:0000269|PubMed:19098711, ECO:0000269|PubMed:19219073, ECO:0000269|PubMed:19837670, ECO:0000269|PubMed:19965871, ECO:0000269|PubMed:20173098, ECO:0000269|PubMed:20385133, ECO:0000269|PubMed:20858735, ECO:0000269|PubMed:22128911, ECO:0000269|PubMed:30879903}. | FUNCTION: Plays a role in the nuclear pore complex (NPC) assembly and/or maintenance (PubMed:12552102, PubMed:15229283, PubMed:30179222). Required for the assembly of peripheral proteins into the NPC (PubMed:15229283, PubMed:12552102). May anchor NUP62 to the NPC (PubMed:15229283). Involved in nephrogenesis (PubMed:30179222). {ECO:0000269|PubMed:12552102, ECO:0000269|PubMed:15229283, ECO:0000269|PubMed:30179222}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 210_215 | 219.0 | 437.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 210_215 | 280.0 | 498.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 210_215 | 219.0 | 437.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 210_215 | 280.0 | 498.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 210_215 | 219.0 | 437.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 210_215 | 280.0 | 498.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 26_109 | 219.0 | 437.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 26_109 | 280.0 | 498.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 26_109 | 219.0 | 437.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 26_109 | 280.0 | 498.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 26_109 | 219.0 | 437.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 26_109 | 280.0 | 498.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 179_185 | 219.0 | 437.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 190_202 | 219.0 | 437.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 179_185 | 280.0 | 498.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 190_202 | 280.0 | 498.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 179_185 | 219.0 | 437.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 190_202 | 219.0 | 437.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 179_185 | 280.0 | 498.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 190_202 | 280.0 | 498.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 179_185 | 219.0 | 437.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 190_202 | 219.0 | 437.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 179_185 | 280.0 | 498.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 190_202 | 280.0 | 498.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 1_101 | 219.0 | 437.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 1_101 | 104.0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 1_101 | 104.0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 1_101 | 280.0 | 498.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 1_101 | 219.0 | 437.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 1_101 | 104.0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 1_101 | 104.0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 1_101 | 280.0 | 498.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 1_101 | 219.0 | 437.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 1_101 | 104.0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 1_101 | 104.0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 1_101 | 280.0 | 498.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 210_215 | 0 | 437.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 243_301 | 0 | 437.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 210_215 | 0.0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 243_301 | 0.0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 210_215 | 0.0 | 266.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 243_301 | 0.0 | 266.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 210_215 | 0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 243_301 | 0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 210_215 | 0.0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 243_301 | 0.0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 210_215 | 0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 243_301 | 0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 210_215 | 0.0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 243_301 | 0.0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 210_215 | 0 | 271.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 243_301 | 0 | 271.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 210_215 | 4.666666666666667 | 498.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 243_301 | 4.666666666666667 | 498.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 210_215 | 0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 243_301 | 0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 210_215 | 0 | 437.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 243_301 | 0 | 437.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 210_215 | 0.0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 243_301 | 0.0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 210_215 | 0.0 | 266.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 243_301 | 0.0 | 266.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 210_215 | 0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 243_301 | 0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 210_215 | 0.0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 243_301 | 0.0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 210_215 | 0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 243_301 | 0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 210_215 | 0.0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 243_301 | 0.0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 210_215 | 0 | 271.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 243_301 | 0 | 271.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 210_215 | 4.666666666666667 | 498.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 243_301 | 4.666666666666667 | 498.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 210_215 | 0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 243_301 | 0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 243_301 | 219.0 | 437.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 210_215 | 104.0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 243_301 | 104.0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 210_215 | 74.0 | 266.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 243_301 | 74.0 | 266.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 210_215 | 104.0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 243_301 | 104.0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 210_215 | 79.0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 243_301 | 79.0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 210_215 | 0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 243_301 | 0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 210_215 | 0.0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 243_301 | 0.0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 210_215 | 79.0 | 271.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 243_301 | 79.0 | 271.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 243_301 | 280.0 | 498.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 210_215 | 79.0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 243_301 | 79.0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 243_301 | 219.0 | 437.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 210_215 | 104.0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 243_301 | 104.0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 210_215 | 74.0 | 266.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 243_301 | 74.0 | 266.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 210_215 | 104.0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 243_301 | 104.0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 210_215 | 79.0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 243_301 | 79.0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 210_215 | 0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 243_301 | 0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 210_215 | 0.0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 243_301 | 0.0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 210_215 | 79.0 | 271.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 243_301 | 79.0 | 271.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 243_301 | 280.0 | 498.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 210_215 | 79.0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 243_301 | 79.0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 243_301 | 219.0 | 437.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 210_215 | 104.0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 243_301 | 104.0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 210_215 | 74.0 | 266.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 243_301 | 74.0 | 266.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 210_215 | 104.0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 243_301 | 104.0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 210_215 | 79.0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 243_301 | 79.0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 210_215 | 0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 243_301 | 0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 210_215 | 0.0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 243_301 | 0.0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 210_215 | 79.0 | 271.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 243_301 | 79.0 | 271.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 243_301 | 280.0 | 498.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 210_215 | 79.0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 243_301 | 79.0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 210_215 | 0 | 437.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 243_301 | 0 | 437.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 210_215 | 0.0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 243_301 | 0.0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 210_215 | 0.0 | 266.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 243_301 | 0.0 | 266.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 210_215 | 0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 243_301 | 0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 210_215 | 0.0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 243_301 | 0.0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 210_215 | 0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 243_301 | 0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 210_215 | 0.0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 243_301 | 0.0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 210_215 | 0 | 271.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 243_301 | 0 | 271.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 210_215 | 0 | 498.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 243_301 | 0 | 498.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 210_215 | 0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 243_301 | 0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 210_215 | 0 | 437.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 243_301 | 0 | 437.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 210_215 | 0.0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 243_301 | 0.0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 210_215 | 0.0 | 266.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 243_301 | 0.0 | 266.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 210_215 | 0 | 322.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 243_301 | 0 | 322.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 210_215 | 0.0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 243_301 | 0.0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 210_215 | 0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 243_301 | 0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 210_215 | 0.0 | 219.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 243_301 | 0.0 | 219.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 210_215 | 0 | 271.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 243_301 | 0 | 271.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 210_215 | 0 | 498.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 243_301 | 0 | 498.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 210_215 | 0 | 297.0 | Compositional bias | Note=Poly-Ser |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 243_301 | 0 | 297.0 | Compositional bias | Note=Asp/Glu-rich (acidic) |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 26_109 | 0 | 437.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 26_109 | 0.0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 26_109 | 0.0 | 266.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 26_109 | 0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 26_109 | 0.0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 26_109 | 0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 26_109 | 0.0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 26_109 | 0 | 271.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 26_109 | 4.666666666666667 | 498.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 26_109 | 0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 26_109 | 0 | 437.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 26_109 | 0.0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 26_109 | 0.0 | 266.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 26_109 | 0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 26_109 | 0.0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 26_109 | 0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 26_109 | 0.0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 26_109 | 0 | 271.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 26_109 | 4.666666666666667 | 498.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 26_109 | 0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 26_109 | 104.0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 26_109 | 74.0 | 266.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 26_109 | 104.0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 26_109 | 79.0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 26_109 | 0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 26_109 | 0.0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 26_109 | 79.0 | 271.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 26_109 | 79.0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 26_109 | 104.0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 26_109 | 74.0 | 266.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 26_109 | 104.0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 26_109 | 79.0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 26_109 | 0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 26_109 | 0.0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 26_109 | 79.0 | 271.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 26_109 | 79.0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 26_109 | 104.0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 26_109 | 74.0 | 266.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 26_109 | 104.0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 26_109 | 79.0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 26_109 | 0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 26_109 | 0.0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 26_109 | 79.0 | 271.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 26_109 | 79.0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 26_109 | 0 | 437.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 26_109 | 0.0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 26_109 | 0.0 | 266.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 26_109 | 0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 26_109 | 0.0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 26_109 | 0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 26_109 | 0.0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 26_109 | 0 | 271.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 26_109 | 0 | 498.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 26_109 | 0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 26_109 | 0 | 437.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 26_109 | 0.0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 26_109 | 0.0 | 266.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 26_109 | 0 | 322.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 26_109 | 0.0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 26_109 | 0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 26_109 | 0.0 | 219.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 26_109 | 0 | 271.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 26_109 | 0 | 498.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 26_109 | 0 | 297.0 | Domain | SWIB/MDM2 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 179_185 | 0 | 437.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 190_202 | 0 | 437.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 466_473 | 0 | 437.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 179_185 | 0.0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 190_202 | 0.0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 466_473 | 0.0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 179_185 | 0.0 | 266.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 190_202 | 0.0 | 266.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 466_473 | 0.0 | 266.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 179_185 | 0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 190_202 | 0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 466_473 | 0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 179_185 | 0.0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 190_202 | 0.0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 466_473 | 0.0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 179_185 | 0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 190_202 | 0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 466_473 | 0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 179_185 | 0.0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 190_202 | 0.0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 466_473 | 0.0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 179_185 | 0 | 271.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 190_202 | 0 | 271.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 466_473 | 0 | 271.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 179_185 | 4.666666666666667 | 498.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 190_202 | 4.666666666666667 | 498.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 466_473 | 4.666666666666667 | 498.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 179_185 | 0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 190_202 | 0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 466_473 | 0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 179_185 | 0 | 437.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 190_202 | 0 | 437.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 466_473 | 0 | 437.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 179_185 | 0.0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 190_202 | 0.0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 466_473 | 0.0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 179_185 | 0.0 | 266.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 190_202 | 0.0 | 266.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 466_473 | 0.0 | 266.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 179_185 | 0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 190_202 | 0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 466_473 | 0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 179_185 | 0.0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 190_202 | 0.0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 466_473 | 0.0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 179_185 | 0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 190_202 | 0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 466_473 | 0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 179_185 | 0.0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 190_202 | 0.0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 466_473 | 0.0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 179_185 | 0 | 271.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 190_202 | 0 | 271.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 466_473 | 0 | 271.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 179_185 | 4.666666666666667 | 498.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 190_202 | 4.666666666666667 | 498.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 466_473 | 4.666666666666667 | 498.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 179_185 | 0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 190_202 | 0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 466_473 | 0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 466_473 | 219.0 | 437.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 179_185 | 104.0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 190_202 | 104.0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 466_473 | 104.0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 179_185 | 74.0 | 266.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 190_202 | 74.0 | 266.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 466_473 | 74.0 | 266.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 179_185 | 104.0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 190_202 | 104.0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 466_473 | 104.0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 179_185 | 79.0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 190_202 | 79.0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 466_473 | 79.0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 179_185 | 0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 190_202 | 0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 466_473 | 0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 179_185 | 0.0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 190_202 | 0.0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 466_473 | 0.0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 179_185 | 79.0 | 271.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 190_202 | 79.0 | 271.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 466_473 | 79.0 | 271.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 466_473 | 280.0 | 498.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 179_185 | 79.0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 190_202 | 79.0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 466_473 | 79.0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 466_473 | 219.0 | 437.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 179_185 | 104.0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 190_202 | 104.0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 466_473 | 104.0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 179_185 | 74.0 | 266.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 190_202 | 74.0 | 266.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 466_473 | 74.0 | 266.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 179_185 | 104.0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 190_202 | 104.0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 466_473 | 104.0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 179_185 | 79.0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 190_202 | 79.0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 466_473 | 79.0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 179_185 | 0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 190_202 | 0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 466_473 | 0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 179_185 | 0.0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 190_202 | 0.0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 466_473 | 0.0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 179_185 | 79.0 | 271.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 190_202 | 79.0 | 271.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 466_473 | 79.0 | 271.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 466_473 | 280.0 | 498.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 179_185 | 79.0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 190_202 | 79.0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 466_473 | 79.0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 466_473 | 219.0 | 437.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 179_185 | 104.0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 190_202 | 104.0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 466_473 | 104.0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 179_185 | 74.0 | 266.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 190_202 | 74.0 | 266.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 466_473 | 74.0 | 266.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 179_185 | 104.0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 190_202 | 104.0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 466_473 | 104.0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 179_185 | 79.0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 190_202 | 79.0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 466_473 | 79.0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 179_185 | 0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 190_202 | 0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 466_473 | 0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 179_185 | 0.0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 190_202 | 0.0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 466_473 | 0.0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 179_185 | 79.0 | 271.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 190_202 | 79.0 | 271.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 466_473 | 79.0 | 271.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 466_473 | 280.0 | 498.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 179_185 | 79.0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 190_202 | 79.0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 466_473 | 79.0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 179_185 | 0 | 437.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 190_202 | 0 | 437.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 466_473 | 0 | 437.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 179_185 | 0.0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 190_202 | 0.0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 466_473 | 0.0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 179_185 | 0.0 | 266.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 190_202 | 0.0 | 266.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 466_473 | 0.0 | 266.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 179_185 | 0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 190_202 | 0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 466_473 | 0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 179_185 | 0.0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 190_202 | 0.0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 466_473 | 0.0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 179_185 | 0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 190_202 | 0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 466_473 | 0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 179_185 | 0.0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 190_202 | 0.0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 466_473 | 0.0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 179_185 | 0 | 271.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 190_202 | 0 | 271.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 466_473 | 0 | 271.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 179_185 | 0 | 498.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 190_202 | 0 | 498.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 466_473 | 0 | 498.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 179_185 | 0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 190_202 | 0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 466_473 | 0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 179_185 | 0 | 437.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 190_202 | 0 | 437.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 466_473 | 0 | 437.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 179_185 | 0.0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 190_202 | 0.0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 466_473 | 0.0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 179_185 | 0.0 | 266.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 190_202 | 0.0 | 266.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 466_473 | 0.0 | 266.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 179_185 | 0 | 322.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 190_202 | 0 | 322.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 466_473 | 0 | 322.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 179_185 | 0.0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 190_202 | 0.0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 466_473 | 0.0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 179_185 | 0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 190_202 | 0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 466_473 | 0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 179_185 | 0.0 | 219.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 190_202 | 0.0 | 219.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 466_473 | 0.0 | 219.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 179_185 | 0 | 271.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 190_202 | 0 | 271.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 466_473 | 0 | 271.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 179_185 | 0 | 498.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 190_202 | 0 | 498.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 466_473 | 0 | 498.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 179_185 | 0 | 297.0 | Motif | Nuclear localization signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 190_202 | 0 | 297.0 | Motif | Note=Nuclear export signal |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 466_473 | 0 | 297.0 | Motif | Nucleolar localization signal |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 1_101 | 0 | 437.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 210_304 | 0 | 437.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 242_331 | 0 | 437.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 1_101 | 0.0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 210_304 | 0.0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 242_331 | 0.0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 1_101 | 0.0 | 266.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 210_304 | 0.0 | 266.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 242_331 | 0.0 | 266.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 1_101 | 0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 210_304 | 0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 242_331 | 0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 1_101 | 0.0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 210_304 | 0.0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 242_331 | 0.0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 1_101 | 0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 210_304 | 0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 242_331 | 0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 1_101 | 0.0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 210_304 | 0.0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 242_331 | 0.0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 1_101 | 0 | 271.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 210_304 | 0 | 271.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 242_331 | 0 | 271.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 1_101 | 4.666666666666667 | 498.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 210_304 | 4.666666666666667 | 498.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 242_331 | 4.666666666666667 | 498.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 1_101 | 0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 210_304 | 0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 242_331 | 0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 1_101 | 0 | 437.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 210_304 | 0 | 437.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 242_331 | 0 | 437.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 1_101 | 0.0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 210_304 | 0.0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 242_331 | 0.0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 1_101 | 0.0 | 266.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 210_304 | 0.0 | 266.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 242_331 | 0.0 | 266.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 1_101 | 0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 210_304 | 0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 242_331 | 0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 1_101 | 0.0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 210_304 | 0.0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 242_331 | 0.0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 1_101 | 0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 210_304 | 0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 242_331 | 0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 1_101 | 0.0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 210_304 | 0.0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 242_331 | 0.0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 1_101 | 0 | 271.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 210_304 | 0 | 271.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 242_331 | 0 | 271.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 1_101 | 4.666666666666667 | 498.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 210_304 | 4.666666666666667 | 498.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 242_331 | 4.666666666666667 | 498.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 1_101 | 0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 210_304 | 0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 242_331 | 0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 210_304 | 219.0 | 437.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 242_331 | 219.0 | 437.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 210_304 | 104.0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 242_331 | 104.0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 1_101 | 74.0 | 266.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 210_304 | 74.0 | 266.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 242_331 | 74.0 | 266.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 210_304 | 104.0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 242_331 | 104.0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 1_101 | 79.0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 210_304 | 79.0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 242_331 | 79.0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 1_101 | 0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 210_304 | 0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 242_331 | 0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 1_101 | 0.0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 210_304 | 0.0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 242_331 | 0.0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 1_101 | 79.0 | 271.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 210_304 | 79.0 | 271.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 242_331 | 79.0 | 271.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 210_304 | 280.0 | 498.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 242_331 | 280.0 | 498.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 1_101 | 79.0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 210_304 | 79.0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 242_331 | 79.0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 210_304 | 219.0 | 437.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 242_331 | 219.0 | 437.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 210_304 | 104.0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 242_331 | 104.0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 1_101 | 74.0 | 266.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 210_304 | 74.0 | 266.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 242_331 | 74.0 | 266.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 210_304 | 104.0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 242_331 | 104.0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 1_101 | 79.0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 210_304 | 79.0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 242_331 | 79.0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 1_101 | 0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 210_304 | 0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 242_331 | 0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 1_101 | 0.0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 210_304 | 0.0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 242_331 | 0.0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 1_101 | 79.0 | 271.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 210_304 | 79.0 | 271.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 242_331 | 79.0 | 271.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 210_304 | 280.0 | 498.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 242_331 | 280.0 | 498.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 1_101 | 79.0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 210_304 | 79.0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 242_331 | 79.0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 210_304 | 219.0 | 437.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 242_331 | 219.0 | 437.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 210_304 | 104.0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 242_331 | 104.0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 1_101 | 74.0 | 266.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 210_304 | 74.0 | 266.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 242_331 | 74.0 | 266.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 210_304 | 104.0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 242_331 | 104.0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 1_101 | 79.0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 210_304 | 79.0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 242_331 | 79.0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 1_101 | 0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 210_304 | 0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 242_331 | 0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 1_101 | 0.0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 210_304 | 0.0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 242_331 | 0.0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 1_101 | 79.0 | 271.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 210_304 | 79.0 | 271.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 242_331 | 79.0 | 271.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 210_304 | 280.0 | 498.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 242_331 | 280.0 | 498.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 1_101 | 79.0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 210_304 | 79.0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 242_331 | 79.0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 1_101 | 0 | 437.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 210_304 | 0 | 437.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 242_331 | 0 | 437.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 1_101 | 0.0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 210_304 | 0.0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 242_331 | 0.0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 1_101 | 0.0 | 266.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 210_304 | 0.0 | 266.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 242_331 | 0.0 | 266.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 1_101 | 0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 210_304 | 0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 242_331 | 0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 1_101 | 0.0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 210_304 | 0.0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 242_331 | 0.0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 1_101 | 0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 210_304 | 0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 242_331 | 0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 1_101 | 0.0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 210_304 | 0.0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 242_331 | 0.0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 1_101 | 0 | 271.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 210_304 | 0 | 271.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 242_331 | 0 | 271.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 1_101 | 0 | 498.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 210_304 | 0 | 498.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 242_331 | 0 | 498.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 1_101 | 0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 210_304 | 0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 242_331 | 0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 1_101 | 0 | 437.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 210_304 | 0 | 437.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 242_331 | 0 | 437.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 1_101 | 0.0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 210_304 | 0.0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 242_331 | 0.0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 1_101 | 0.0 | 266.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 210_304 | 0.0 | 266.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 242_331 | 0.0 | 266.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 1_101 | 0 | 322.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 210_304 | 0 | 322.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 242_331 | 0 | 322.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 1_101 | 0.0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 210_304 | 0.0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 242_331 | 0.0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 1_101 | 0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 210_304 | 0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 242_331 | 0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 1_101 | 0.0 | 219.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 210_304 | 0.0 | 219.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 242_331 | 0.0 | 219.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 1_101 | 0 | 271.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 210_304 | 0 | 271.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 242_331 | 0 | 271.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 1_101 | 0 | 498.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 210_304 | 0 | 498.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 242_331 | 0 | 498.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 1_101 | 0 | 297.0 | Region | Sufficient to promote the mitochondrial pathway of apoptosis |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 210_304 | 0 | 297.0 | Region | Note=ARF-binding |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 242_331 | 0 | 297.0 | Region | Note=Region II |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 299_328 | 0 | 437.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 438_479 | 0 | 437.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 299_328 | 0.0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 438_479 | 0.0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 299_328 | 0.0 | 266.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 438_479 | 0.0 | 266.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 299_328 | 0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 438_479 | 0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 299_328 | 0.0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 438_479 | 0.0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 299_328 | 0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 438_479 | 0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 299_328 | 0.0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 438_479 | 0.0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 299_328 | 0 | 271.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 438_479 | 0 | 271.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 299_328 | 4.666666666666667 | 498.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 438_479 | 4.666666666666667 | 498.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 299_328 | 0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 438_479 | 0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 299_328 | 0 | 437.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 438_479 | 0 | 437.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 299_328 | 0.0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 438_479 | 0.0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 299_328 | 0.0 | 266.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 438_479 | 0.0 | 266.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 299_328 | 0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 438_479 | 0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 299_328 | 0.0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 438_479 | 0.0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 299_328 | 0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 438_479 | 0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 299_328 | 0.0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 438_479 | 0.0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 299_328 | 0 | 271.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 438_479 | 0 | 271.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 299_328 | 4.666666666666667 | 498.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 438_479 | 4.666666666666667 | 498.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 299_328 | 0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 438_479 | 0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 299_328 | 219.0 | 437.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 438_479 | 219.0 | 437.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 299_328 | 104.0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 438_479 | 104.0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 299_328 | 74.0 | 266.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 438_479 | 74.0 | 266.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 299_328 | 104.0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 438_479 | 104.0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 299_328 | 79.0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 438_479 | 79.0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 299_328 | 0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 438_479 | 0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 299_328 | 0.0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 438_479 | 0.0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 299_328 | 79.0 | 271.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 438_479 | 79.0 | 271.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 299_328 | 280.0 | 498.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 438_479 | 280.0 | 498.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 299_328 | 79.0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 438_479 | 79.0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 299_328 | 219.0 | 437.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 438_479 | 219.0 | 437.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 299_328 | 104.0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 438_479 | 104.0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 299_328 | 74.0 | 266.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 438_479 | 74.0 | 266.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 299_328 | 104.0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 438_479 | 104.0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 299_328 | 79.0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 438_479 | 79.0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 299_328 | 0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 438_479 | 0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 299_328 | 0.0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 438_479 | 0.0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 299_328 | 79.0 | 271.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 438_479 | 79.0 | 271.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 299_328 | 280.0 | 498.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 438_479 | 280.0 | 498.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 299_328 | 79.0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 438_479 | 79.0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 299_328 | 219.0 | 437.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 438_479 | 219.0 | 437.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 299_328 | 104.0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 438_479 | 104.0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 299_328 | 74.0 | 266.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 438_479 | 74.0 | 266.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 299_328 | 104.0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 438_479 | 104.0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 299_328 | 79.0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 438_479 | 79.0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 299_328 | 0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 438_479 | 0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 299_328 | 0.0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 438_479 | 0.0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 299_328 | 79.0 | 271.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 438_479 | 79.0 | 271.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 299_328 | 280.0 | 498.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 438_479 | 280.0 | 498.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 299_328 | 79.0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 438_479 | 79.0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 299_328 | 0 | 437.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 438_479 | 0 | 437.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 299_328 | 0.0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 438_479 | 0.0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 299_328 | 0.0 | 266.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 438_479 | 0.0 | 266.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 299_328 | 0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 438_479 | 0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 299_328 | 0.0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 438_479 | 0.0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 299_328 | 0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 438_479 | 0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 299_328 | 0.0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 438_479 | 0.0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 299_328 | 0 | 271.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 438_479 | 0 | 271.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 299_328 | 0 | 498.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 438_479 | 0 | 498.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 299_328 | 0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 438_479 | 0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 299_328 | 0 | 437.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 438_479 | 0 | 437.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 299_328 | 0.0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 438_479 | 0.0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 299_328 | 0.0 | 266.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 438_479 | 0.0 | 266.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 299_328 | 0 | 322.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 438_479 | 0 | 322.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 299_328 | 0.0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 438_479 | 0.0 | 297.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 299_328 | 0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 438_479 | 0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 299_328 | 0.0 | 219.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 438_479 | 0.0 | 219.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 299_328 | 0 | 271.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 438_479 | 0 | 271.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 299_328 | 0 | 498.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 438_479 | 0 | 498.0 | Zinc finger | RING-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 299_328 | 0 | 297.0 | Zinc finger | RanBP2-type |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 438_479 | 0 | 297.0 | Zinc finger | RING-type |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file >>>1731_MDM2_69202271_NUP107_69082742_ranked_0.pdb | MDM2 | 69202271 | 69202271 | ENST00000229179 | NUP107 | chr12 | 69082742 | + | MRDPRLQARKPRMVRSRSGFGEISSPVIREAEVTRTARKQSAQKRVLLQASQDENFGNTTPRNQVIPRTPSSFRQPFTPTSRSLLRQPDI SCILGTGGKSPRLTQSSGFFGNLSMVTNLDDSNWAAAFSSQRSGLFTNTEPHSITEDVTISAVMLREDDPGEAASMSMFSDFLQSFLKHS SSTVFDLVEEYENICGSQVNILSKIVSRATPGLQKFSKTASMLWLLQQEMVTWRLLASLYRDRIQSALEEESVFAVTAVNASEKTVVEAL FQRDSLVRQSQLVVDWLESIAKDEIGEFSDNIEFYAKSVYWENTLHTLKQRQLTSYVGSVRPLVTELDPDAPIRQKMPLDDLDREDEVRL LKYLFTLIRAGMTEEAQRLCKRCGQAWRAATLEGWKLYHDPNVNGGTELEPVEGNPYRRIWKISCWRMAEDELFNRYERAIYAALSGNLK QLLPVCDTWEDTVWAYFRVMVDSLVEQEIQTSVATLDETEELPREYLGANWTLEKVFEELQATDKKRVLEENQEHYHIVQKFLILGDIDG LMDEFSKWLSKSRNNLPGHLLRFMTHLILFFRTLGLQTKEEVSIEVLKTYIQLLIREKHTNLIAFYTCHLPQDLAVAQYALFLESVTEFE QRHHCLELAKEADLDVATITKTVVENIRKKDNGEFSHHDLAPALDTGTTEEDRLKIDVIDWLVFDPAQRAEALKQGNAIMRKFLASKKHE AAKEVFVKIPQDSIAEIYNQCEEQGMESPLPAEDDNAIREHLCIRAYLEAHETFNEWFKHMNSVPQKPALIPQPTFTEKVAHEHKEKKYE MDFGIWKGHLDALTADVKEKMYNVLLFVDGGWMVDVREDAKEDHERTHQMVLLRKLCLPMLCFLLHTILHSTGQYQECLQLADMVSSERH | 939 |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
MDM2_pLDDT.png |
NUP107_pLDDT.png |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| NUP107 | SEH1L, NUP133, NUP85, NUP160, NUP43, NUP37, SEC13, TPR, NUP214, KPNB1, Nup107, Nup98, SIRT7, NUP98, WRNIP1, CUL3, CENPF, NUP153, TP53BP1, PNPT1, EIF4B, CORO1B, MYL1, C1QBP, CPSF6, VCP, PYCARD, CUL7, OBSL1, SUZ12, EED, PNKD, NTRK1, IFI16, SYNE1, APC, PRKDC, RRP1B, NALCN, CD1E, ACTBL2, HSP90AB1, XPO1, SENP1, SENP2, AHCTF1, RANGAP1, H2AFV, NUP50, RANBP2, B9D2, ACLY, RCC1, GLE1, ITPR2, KPNA4, NUP88, RAN, UBE2I, SUMO1, AAAS, RAE1, NUP155, NUP93, NUPL1, WDR1, NXF1, IPO7, NUPL2, NUP205, NUP210, NUP188, NUP62, NXT1, SEC61A1, NUP54, TMEM214, SMPD4, NDC1, KRAS, IP6K3, TMEM209, NUP35, RPUSD3, RGPD8, POM121C, Ranbp2, Ube2i, Rcc1, Kifc1, Nup155, Nup214, FOXA3, FOXI2, FOXK2, FOXL1, FOXP3, FOXQ1, CDC5L, ZNF746, DUSP13, SPAST, UXS1, SIGLECL1, CD70, RAF1, PDHA1, TRIM25, BRCA1, EGLN3, TGFB1, NKX2-1, RNF4, CDC34, BPLF1, EZH2, DCPS, MYC, CDK9, CANX, EZR, LMNA, TOMM20, MGST3, PTRH2, KIAA1429, HIST1H4A, Dppa3, BIRC3, LMBR1L, PLEKHA4, GAPDH, M, nsp16, nsp4, nsp6, ORF6, MAU2, LRRC31, CIT, KIF14, PRC1, TRIM66, FKBP8, PTPN1, RHOT2, SLC25A46, SUMO2, NUPR1, CIC, LGALS9, DYRK1A, BKRF1, DNAJC1, ANAPC2, COX8A, DERL1, EMD, KRT8, LMNB1, LRRC59, RPN1, SEC61B, STIM1, SYNE3, TMPO, NAA40, VPS33A, RGPD1, EFNA4, COMTD1, SLFN11, FBXW7, FBXO32, RCHY1, SIRT6, GLI3, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| MDM2 | |
| NUP107 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 170_306 | 0 | 437.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 170_306 | 0.0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 170_306 | 0.0 | 266.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 170_306 | 0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 170_306 | 0.0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 170_306 | 0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 170_306 | 0.0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 170_306 | 0 | 271.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 170_306 | 4.666666666666667 | 498.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 170_306 | 0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 170_306 | 0 | 437.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 170_306 | 0.0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 170_306 | 0.0 | 266.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 170_306 | 0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 170_306 | 0.0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 170_306 | 0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 170_306 | 0.0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 170_306 | 0 | 271.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 170_306 | 4.666666666666667 | 498.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 170_306 | 0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 170_306 | 219.0 | 437.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 170_306 | 104.0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 170_306 | 74.0 | 266.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 170_306 | 104.0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 170_306 | 79.0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 170_306 | 0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 170_306 | 0.0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 170_306 | 79.0 | 271.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000462284 | + | 9 | 11 | 170_306 | 280.0 | 498.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 170_306 | 79.0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 170_306 | 219.0 | 437.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 170_306 | 104.0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 170_306 | 74.0 | 266.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 170_306 | 104.0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 170_306 | 79.0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 170_306 | 0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 170_306 | 0.0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 170_306 | 79.0 | 271.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000462284 | + | 9 | 11 | 170_306 | 280.0 | 498.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 170_306 | 79.0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 170_306 | 219.0 | 437.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 170_306 | 104.0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 170_306 | 74.0 | 266.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 170_306 | 104.0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 170_306 | 79.0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 170_306 | 0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 170_306 | 0.0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 170_306 | 79.0 | 271.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000462284 | + | 9 | 11 | 170_306 | 280.0 | 498.0 | MTBP |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 170_306 | 79.0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 170_306 | 0 | 437.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 170_306 | 0.0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 170_306 | 0.0 | 266.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 170_306 | 0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 170_306 | 0.0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 170_306 | 0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 170_306 | 0.0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 170_306 | 0 | 271.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 170_306 | 0 | 498.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 170_306 | 0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 170_306 | 0 | 437.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 170_306 | 0.0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 170_306 | 0.0 | 266.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 170_306 | 0 | 322.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 170_306 | 0.0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 170_306 | 0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 170_306 | 0.0 | 219.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 170_306 | 0 | 271.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 170_306 | 0 | 498.0 | MTBP |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 170_306 | 0 | 297.0 | MTBP |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 150_230 | 0 | 437.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 150_230 | 0.0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 150_230 | 0.0 | 266.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 150_230 | 0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 150_230 | 0.0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 150_230 | 0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 150_230 | 0.0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 150_230 | 0 | 271.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 150_230 | 4.666666666666667 | 498.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 150_230 | 0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 150_230 | 0 | 437.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 150_230 | 0.0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 150_230 | 0.0 | 266.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 150_230 | 0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 150_230 | 0.0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 150_230 | 0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 150_230 | 0.0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 150_230 | 0 | 271.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 150_230 | 4.666666666666667 | 498.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 150_230 | 0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 150_230 | 219.0 | 437.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 150_230 | 104.0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 150_230 | 74.0 | 266.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 150_230 | 104.0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 150_230 | 79.0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 150_230 | 0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 150_230 | 0.0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 150_230 | 79.0 | 271.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 150_230 | 79.0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 150_230 | 219.0 | 437.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 150_230 | 104.0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 150_230 | 74.0 | 266.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 150_230 | 104.0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 150_230 | 79.0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 150_230 | 0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 150_230 | 0.0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 150_230 | 79.0 | 271.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 150_230 | 79.0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 150_230 | 219.0 | 437.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 150_230 | 104.0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 150_230 | 74.0 | 266.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 150_230 | 104.0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 150_230 | 79.0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 150_230 | 0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 150_230 | 0.0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 150_230 | 79.0 | 271.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 150_230 | 79.0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 150_230 | 0 | 437.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 150_230 | 0.0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 150_230 | 0.0 | 266.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 150_230 | 0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 150_230 | 0.0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 150_230 | 0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 150_230 | 0.0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 150_230 | 0 | 271.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 150_230 | 0 | 498.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 150_230 | 0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 150_230 | 0 | 437.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 150_230 | 0.0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 150_230 | 0.0 | 266.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 150_230 | 0 | 322.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 150_230 | 0.0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 150_230 | 0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 150_230 | 0.0 | 219.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 150_230 | 0 | 271.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 150_230 | 0 | 498.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 150_230 | 0 | 297.0 | PYHIN1 and necessary for interaction with RFFL and RNF34 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000258149 | + | 1 | 9 | 223_232 | 0 | 437.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000299252 | + | 1 | 5 | 223_232 | 0.0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000348801 | + | 1 | 3 | 223_232 | 0.0 | 266.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000356290 | + | 1 | 6 | 223_232 | 0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000360430 | + | 1 | 4 | 223_232 | 0.0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393412 | + | 1 | 3 | 223_232 | 0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000393413 | + | 1 | 2 | 223_232 | 0.0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000428863 | + | 1 | 4 | 223_232 | 0 | 271.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000462284 | + | 1 | 11 | 223_232 | 4.666666666666667 | 498.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082741 | ENST00000540827 | + | 1 | 5 | 223_232 | 0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000258149 | + | 1 | 9 | 223_232 | 0 | 437.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000299252 | + | 1 | 5 | 223_232 | 0.0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000348801 | + | 1 | 3 | 223_232 | 0.0 | 266.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000356290 | + | 1 | 6 | 223_232 | 0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000360430 | + | 1 | 4 | 223_232 | 0.0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393412 | + | 1 | 3 | 223_232 | 0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000393413 | + | 1 | 2 | 223_232 | 0.0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000428863 | + | 1 | 4 | 223_232 | 0 | 271.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000462284 | + | 1 | 11 | 223_232 | 4.666666666666667 | 498.0 | USP7 |
| Hgene | MDM2 | chr12:69202271 | chr12:69082742 | ENST00000540827 | + | 1 | 5 | 223_232 | 0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000258149 | + | 7 | 9 | 223_232 | 219.0 | 437.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000299252 | + | 3 | 5 | 223_232 | 104.0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000348801 | + | 2 | 3 | 223_232 | 74.0 | 266.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000356290 | + | 4 | 6 | 223_232 | 104.0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000360430 | + | 2 | 4 | 223_232 | 79.0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393412 | + | 1 | 3 | 223_232 | 0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000393413 | + | 1 | 2 | 223_232 | 0.0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000428863 | + | 3 | 4 | 223_232 | 79.0 | 271.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113120 | ENST00000540827 | + | 3 | 5 | 223_232 | 79.0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000258149 | + | 7 | 9 | 223_232 | 219.0 | 437.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000299252 | + | 3 | 5 | 223_232 | 104.0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000348801 | + | 2 | 3 | 223_232 | 74.0 | 266.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000356290 | + | 4 | 6 | 223_232 | 104.0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000360430 | + | 2 | 4 | 223_232 | 79.0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 223_232 | 0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 223_232 | 0.0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000428863 | + | 3 | 4 | 223_232 | 79.0 | 271.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69113121 | ENST00000540827 | + | 3 | 5 | 223_232 | 79.0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000258149 | + | 7 | 9 | 223_232 | 219.0 | 437.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000299252 | + | 3 | 5 | 223_232 | 104.0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000348801 | + | 2 | 3 | 223_232 | 74.0 | 266.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000356290 | + | 4 | 6 | 223_232 | 104.0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000360430 | + | 2 | 4 | 223_232 | 79.0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 223_232 | 0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 223_232 | 0.0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000428863 | + | 3 | 4 | 223_232 | 79.0 | 271.0 | USP7 |
| Hgene | MDM2 | chr12:69229764 | chr12:69135593 | ENST00000540827 | + | 3 | 5 | 223_232 | 79.0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000258149 | + | 1 | 9 | 223_232 | 0 | 437.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000299252 | + | 1 | 5 | 223_232 | 0.0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000348801 | + | 1 | 3 | 223_232 | 0.0 | 266.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000356290 | + | 1 | 6 | 223_232 | 0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000360430 | + | 1 | 4 | 223_232 | 0.0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393412 | + | 1 | 3 | 223_232 | 0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000393413 | + | 1 | 2 | 223_232 | 0.0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000428863 | + | 1 | 4 | 223_232 | 0 | 271.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000462284 | + | 1 | 11 | 223_232 | 0 | 498.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69113121 | ENST00000540827 | + | 1 | 5 | 223_232 | 0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000258149 | + | 1 | 9 | 223_232 | 0 | 437.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000299252 | + | 1 | 5 | 223_232 | 0.0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000348801 | + | 1 | 3 | 223_232 | 0.0 | 266.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000356290 | + | 1 | 6 | 223_232 | 0 | 322.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000360430 | + | 1 | 4 | 223_232 | 0.0 | 297.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393412 | + | 1 | 3 | 223_232 | 0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000393413 | + | 1 | 2 | 223_232 | 0.0 | 219.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000428863 | + | 1 | 4 | 223_232 | 0 | 271.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000462284 | + | 1 | 11 | 223_232 | 0 | 498.0 | USP7 |
| Hgene | MDM2 | chr12:69233581 | chr12:69135593 | ENST00000540827 | + | 1 | 5 | 223_232 | 0 | 297.0 | USP7 |
Top |
Related Drugs to MDM2-NUP107 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to MDM2-NUP107 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | NUP107 | C4225228 | NEPHROTIC SYNDROME, TYPE 11 | 2 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | NUP107 | C0018051 | Gonadal Dysgenesis | 1 | GENOMICS_ENGLAND |
| Tgene | NUP107 | C0685837 | Pure Gonadal Dysgenesis, 46, XX | 1 | ORPHANET |
| Tgene | NUP107 | C0795949 | Galloway Mowat syndrome | 1 | ORPHANET |
| Tgene | NUP107 | C0949595 | Gonadal Dysgenesis, 46,XX | 1 | ORPHANET |
| Tgene | NUP107 | C1868672 | NEPHROTIC SYNDROME, STEROID-RESISTANT, AUTOSOMAL RECESSIVE | 1 | ORPHANET |
| Tgene | NUP107 | C4748084 | OVARIAN DYSGENESIS 6 | 1 | UNIPROT |

