| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:MFGE8-CRTC1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: MFGE8-CRTC1 | FusionPDB ID: 53177 | FusionGDB2.0 ID: 53177 | Hgene | Tgene | Gene symbol | MFGE8 | CRTC1 | Gene ID | 4240 | 23373 |
| Gene name | milk fat globule EGF and factor V/VIII domain containing | CREB regulated transcription coactivator 1 | |
| Synonyms | BA46|EDIL1|HMFG|HsT19888|MFG-E8|MFGM|OAcGD3S|SED1|SPAG10|hP47 | MAML2|MECT1|Mam-2|TORC-1|TORC1|WAMTP1 | |
| Cytomap | 15q26.1 | 19p13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | lactadherinO-acetyl disialoganglioside synthasebreast epithelial antigen BA46medinmilk fat globule-EGF factor 8 proteinsperm associated antigen 10sperm surface protein hP47 | CREB-regulated transcription coactivator 1Mastermind-like protein 2mucoepidermoid carcinoma translocated protein 1transducer of regulated cAMP response element-binding protein (CREB) 1 | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | Q08431 | Q6UUV9 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000268150, ENST00000268151, ENST00000539437, ENST00000542878, ENST00000566497, ENST00000559997, | ENST00000601916, ENST00000321949, ENST00000338797, ENST00000594658, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 6 X 5 X 4=120 | 9 X 8 X 8=576 |
| # samples | 6 | 10 | |
| ** MAII score | log2(6/120*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/576*10)=-2.52606881166759 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: MFGE8 [Title/Abstract] AND CRTC1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MFGE8(89444781)-CRTC1(18882251), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MFGE8-CRTC1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MFGE8-CRTC1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MFGE8-CRTC1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MFGE8-CRTC1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MFGE8-CRTC1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. MFGE8-CRTC1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CRTC1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 14536081 |
 Fusion gene breakpoints across MFGE8 (5'-gene) Fusion gene breakpoints across MFGE8 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
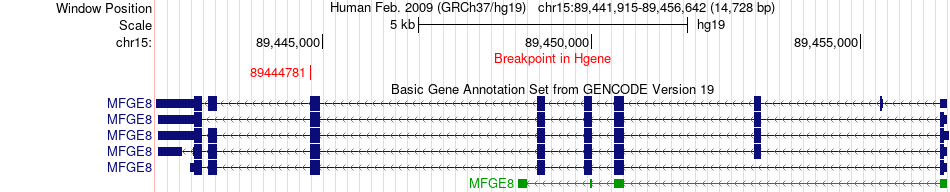 |
 Fusion gene breakpoints across CRTC1 (3'-gene) Fusion gene breakpoints across CRTC1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
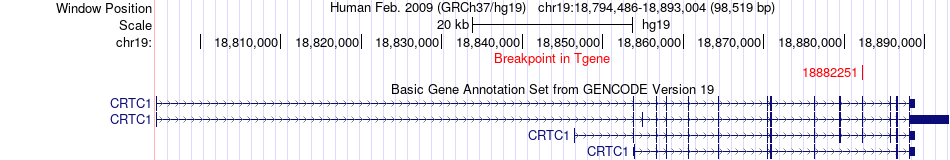 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-13-0762 | MFGE8 | chr15 | 89444781 | - | CRTC1 | chr19 | 18882251 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000268151 | MFGE8 | chr15 | 89444781 | - | ENST00000321949 | CRTC1 | chr19 | 18882251 | + | 2087 | 932 | 17 | 1516 | 499 |
| ENST00000268151 | MFGE8 | chr15 | 89444781 | - | ENST00000338797 | CRTC1 | chr19 | 18882251 | + | 6329 | 932 | 17 | 1516 | 499 |
| ENST00000268151 | MFGE8 | chr15 | 89444781 | - | ENST00000594658 | CRTC1 | chr19 | 18882251 | + | 2087 | 932 | 17 | 1516 | 499 |
| ENST00000268150 | MFGE8 | chr15 | 89444781 | - | ENST00000321949 | CRTC1 | chr19 | 18882251 | + | 2117 | 962 | 47 | 1546 | 499 |
| ENST00000268150 | MFGE8 | chr15 | 89444781 | - | ENST00000338797 | CRTC1 | chr19 | 18882251 | + | 6359 | 962 | 47 | 1546 | 499 |
| ENST00000268150 | MFGE8 | chr15 | 89444781 | - | ENST00000594658 | CRTC1 | chr19 | 18882251 | + | 2117 | 962 | 47 | 1546 | 499 |
| ENST00000542878 | MFGE8 | chr15 | 89444781 | - | ENST00000321949 | CRTC1 | chr19 | 18882251 | + | 1953 | 798 | 15 | 1382 | 455 |
| ENST00000542878 | MFGE8 | chr15 | 89444781 | - | ENST00000338797 | CRTC1 | chr19 | 18882251 | + | 6195 | 798 | 15 | 1382 | 455 |
| ENST00000542878 | MFGE8 | chr15 | 89444781 | - | ENST00000594658 | CRTC1 | chr19 | 18882251 | + | 1953 | 798 | 15 | 1382 | 455 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000268151 | ENST00000321949 | MFGE8 | chr15 | 89444781 | - | CRTC1 | chr19 | 18882251 | + | 0.017202448 | 0.9827976 |
| ENST00000268151 | ENST00000338797 | MFGE8 | chr15 | 89444781 | - | CRTC1 | chr19 | 18882251 | + | 0.003235545 | 0.9967644 |
| ENST00000268151 | ENST00000594658 | MFGE8 | chr15 | 89444781 | - | CRTC1 | chr19 | 18882251 | + | 0.017202448 | 0.9827976 |
| ENST00000268150 | ENST00000321949 | MFGE8 | chr15 | 89444781 | - | CRTC1 | chr19 | 18882251 | + | 0.01638786 | 0.9836121 |
| ENST00000268150 | ENST00000338797 | MFGE8 | chr15 | 89444781 | - | CRTC1 | chr19 | 18882251 | + | 0.003197067 | 0.9968029 |
| ENST00000268150 | ENST00000594658 | MFGE8 | chr15 | 89444781 | - | CRTC1 | chr19 | 18882251 | + | 0.01638786 | 0.9836121 |
| ENST00000542878 | ENST00000321949 | MFGE8 | chr15 | 89444781 | - | CRTC1 | chr19 | 18882251 | + | 0.013821748 | 0.9861783 |
| ENST00000542878 | ENST00000338797 | MFGE8 | chr15 | 89444781 | - | CRTC1 | chr19 | 18882251 | + | 0.002659698 | 0.99734026 |
| ENST00000542878 | ENST00000594658 | MFGE8 | chr15 | 89444781 | - | CRTC1 | chr19 | 18882251 | + | 0.013821748 | 0.9861783 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >53177_53177_1_MFGE8-CRTC1_MFGE8_chr15_89444781_ENST00000268150_CRTC1_chr19_18882251_ENST00000321949_length(amino acids)=499AA_BP=304 MSSPACPFQRPRPRSMPRPRLLAALCGALLCAPSLLVALDICSKNPCHNGGLCEEISQEVRGDVFPSYTCTCLKGYAGNHCETKCVEPLG LENGNIANSQIAASSVRVTFLGLQHWVPELARLNRAGMVNAWTPSSNDDNPWIQVNLLRRMWVTGVVTQGASRLASHEYLKAFKVAYSLN GHEFDFIHDVNKKHKEFVGNWNKNAVHVNLFETPVEAQYVRLYPTSCHTACTLRFELLGCELNGCANPLGLKNNSIPDKQITASSSYKTW GLHLFSWNPSYARLDKQGNFNAWVAGSYGNDQWLQAPALQQYRTSAGSPANQSPTSPVSNQGFSPGSSPQHTSTLGSVFGDAYYEQQMAA RQANALSHQLEQFNMMENAISSSSLYSPGSTLNYSQAAMMGLTGSHGSLPDSQQLGYASHSGIPNIILTVTGESPPSLSKELTSSLAGVG -------------------------------------------------------------- >53177_53177_2_MFGE8-CRTC1_MFGE8_chr15_89444781_ENST00000268150_CRTC1_chr19_18882251_ENST00000338797_length(amino acids)=499AA_BP=304 MSSPACPFQRPRPRSMPRPRLLAALCGALLCAPSLLVALDICSKNPCHNGGLCEEISQEVRGDVFPSYTCTCLKGYAGNHCETKCVEPLG LENGNIANSQIAASSVRVTFLGLQHWVPELARLNRAGMVNAWTPSSNDDNPWIQVNLLRRMWVTGVVTQGASRLASHEYLKAFKVAYSLN GHEFDFIHDVNKKHKEFVGNWNKNAVHVNLFETPVEAQYVRLYPTSCHTACTLRFELLGCELNGCANPLGLKNNSIPDKQITASSSYKTW GLHLFSWNPSYARLDKQGNFNAWVAGSYGNDQWLQAPALQQYRTSAGSPANQSPTSPVSNQGFSPGSSPQHTSTLGSVFGDAYYEQQMAA RQANALSHQLEQFNMMENAISSSSLYSPGSTLNYSQAAMMGLTGSHGSLPDSQQLGYASHSGIPNIILTVTGESPPSLSKELTSSLAGVG -------------------------------------------------------------- >53177_53177_3_MFGE8-CRTC1_MFGE8_chr15_89444781_ENST00000268150_CRTC1_chr19_18882251_ENST00000594658_length(amino acids)=499AA_BP=304 MSSPACPFQRPRPRSMPRPRLLAALCGALLCAPSLLVALDICSKNPCHNGGLCEEISQEVRGDVFPSYTCTCLKGYAGNHCETKCVEPLG LENGNIANSQIAASSVRVTFLGLQHWVPELARLNRAGMVNAWTPSSNDDNPWIQVNLLRRMWVTGVVTQGASRLASHEYLKAFKVAYSLN GHEFDFIHDVNKKHKEFVGNWNKNAVHVNLFETPVEAQYVRLYPTSCHTACTLRFELLGCELNGCANPLGLKNNSIPDKQITASSSYKTW GLHLFSWNPSYARLDKQGNFNAWVAGSYGNDQWLQAPALQQYRTSAGSPANQSPTSPVSNQGFSPGSSPQHTSTLGSVFGDAYYEQQMAA RQANALSHQLEQFNMMENAISSSSLYSPGSTLNYSQAAMMGLTGSHGSLPDSQQLGYASHSGIPNIILTVTGESPPSLSKELTSSLAGVG -------------------------------------------------------------- >53177_53177_4_MFGE8-CRTC1_MFGE8_chr15_89444781_ENST00000268151_CRTC1_chr19_18882251_ENST00000321949_length(amino acids)=499AA_BP=304 MSSPACPFQRPRPRSMPRPRLLAALCGALLCAPSLLVALDICSKNPCHNGGLCEEISQEVRGDVFPSYTCTCLKGYAGNHCETKCVEPLG LENGNIANSQIAASSVRVTFLGLQHWVPELARLNRAGMVNAWTPSSNDDNPWIQVNLLRRMWVTGVVTQGASRLASHEYLKAFKVAYSLN GHEFDFIHDVNKKHKEFVGNWNKNAVHVNLFETPVEAQYVRLYPTSCHTACTLRFELLGCELNGCANPLGLKNNSIPDKQITASSSYKTW GLHLFSWNPSYARLDKQGNFNAWVAGSYGNDQWLQAPALQQYRTSAGSPANQSPTSPVSNQGFSPGSSPQHTSTLGSVFGDAYYEQQMAA RQANALSHQLEQFNMMENAISSSSLYSPGSTLNYSQAAMMGLTGSHGSLPDSQQLGYASHSGIPNIILTVTGESPPSLSKELTSSLAGVG -------------------------------------------------------------- >53177_53177_5_MFGE8-CRTC1_MFGE8_chr15_89444781_ENST00000268151_CRTC1_chr19_18882251_ENST00000338797_length(amino acids)=499AA_BP=304 MSSPACPFQRPRPRSMPRPRLLAALCGALLCAPSLLVALDICSKNPCHNGGLCEEISQEVRGDVFPSYTCTCLKGYAGNHCETKCVEPLG LENGNIANSQIAASSVRVTFLGLQHWVPELARLNRAGMVNAWTPSSNDDNPWIQVNLLRRMWVTGVVTQGASRLASHEYLKAFKVAYSLN GHEFDFIHDVNKKHKEFVGNWNKNAVHVNLFETPVEAQYVRLYPTSCHTACTLRFELLGCELNGCANPLGLKNNSIPDKQITASSSYKTW GLHLFSWNPSYARLDKQGNFNAWVAGSYGNDQWLQAPALQQYRTSAGSPANQSPTSPVSNQGFSPGSSPQHTSTLGSVFGDAYYEQQMAA RQANALSHQLEQFNMMENAISSSSLYSPGSTLNYSQAAMMGLTGSHGSLPDSQQLGYASHSGIPNIILTVTGESPPSLSKELTSSLAGVG -------------------------------------------------------------- >53177_53177_6_MFGE8-CRTC1_MFGE8_chr15_89444781_ENST00000268151_CRTC1_chr19_18882251_ENST00000594658_length(amino acids)=499AA_BP=304 MSSPACPFQRPRPRSMPRPRLLAALCGALLCAPSLLVALDICSKNPCHNGGLCEEISQEVRGDVFPSYTCTCLKGYAGNHCETKCVEPLG LENGNIANSQIAASSVRVTFLGLQHWVPELARLNRAGMVNAWTPSSNDDNPWIQVNLLRRMWVTGVVTQGASRLASHEYLKAFKVAYSLN GHEFDFIHDVNKKHKEFVGNWNKNAVHVNLFETPVEAQYVRLYPTSCHTACTLRFELLGCELNGCANPLGLKNNSIPDKQITASSSYKTW GLHLFSWNPSYARLDKQGNFNAWVAGSYGNDQWLQAPALQQYRTSAGSPANQSPTSPVSNQGFSPGSSPQHTSTLGSVFGDAYYEQQMAA RQANALSHQLEQFNMMENAISSSSLYSPGSTLNYSQAAMMGLTGSHGSLPDSQQLGYASHSGIPNIILTVTGESPPSLSKELTSSLAGVG -------------------------------------------------------------- >53177_53177_7_MFGE8-CRTC1_MFGE8_chr15_89444781_ENST00000542878_CRTC1_chr19_18882251_ENST00000321949_length(amino acids)=455AA_BP=260 MSSPACPFQRPRPRSMPRPRLLAALCGALLCAPSLLVALECVEPLGLENGNIANSQIAASSVRVTFLGLQHWVPELARLNRAGMVNAWTP SSNDDNPWIQVNLLRRMWVTGVVTQGASRLASHEYLKAFKVAYSLNGHEFDFIHDVNKKHKEFVGNWNKNAVHVNLFETPVEAQYVRLYP TSCHTACTLRFELLGCELNGCANPLGLKNNSIPDKQITASSSYKTWGLHLFSWNPSYARLDKQGNFNAWVAGSYGNDQWLQAPALQQYRT SAGSPANQSPTSPVSNQGFSPGSSPQHTSTLGSVFGDAYYEQQMAARQANALSHQLEQFNMMENAISSSSLYSPGSTLNYSQAAMMGLTG SHGSLPDSQQLGYASHSGIPNIILTVTGESPPSLSKELTSSLAGVGDVSFDSDSQFPLDELKIDPLTLDGLHMLNDPDMVLADPATEDTF -------------------------------------------------------------- >53177_53177_8_MFGE8-CRTC1_MFGE8_chr15_89444781_ENST00000542878_CRTC1_chr19_18882251_ENST00000338797_length(amino acids)=455AA_BP=260 MSSPACPFQRPRPRSMPRPRLLAALCGALLCAPSLLVALECVEPLGLENGNIANSQIAASSVRVTFLGLQHWVPELARLNRAGMVNAWTP SSNDDNPWIQVNLLRRMWVTGVVTQGASRLASHEYLKAFKVAYSLNGHEFDFIHDVNKKHKEFVGNWNKNAVHVNLFETPVEAQYVRLYP TSCHTACTLRFELLGCELNGCANPLGLKNNSIPDKQITASSSYKTWGLHLFSWNPSYARLDKQGNFNAWVAGSYGNDQWLQAPALQQYRT SAGSPANQSPTSPVSNQGFSPGSSPQHTSTLGSVFGDAYYEQQMAARQANALSHQLEQFNMMENAISSSSLYSPGSTLNYSQAAMMGLTG SHGSLPDSQQLGYASHSGIPNIILTVTGESPPSLSKELTSSLAGVGDVSFDSDSQFPLDELKIDPLTLDGLHMLNDPDMVLADPATEDTF -------------------------------------------------------------- >53177_53177_9_MFGE8-CRTC1_MFGE8_chr15_89444781_ENST00000542878_CRTC1_chr19_18882251_ENST00000594658_length(amino acids)=455AA_BP=260 MSSPACPFQRPRPRSMPRPRLLAALCGALLCAPSLLVALECVEPLGLENGNIANSQIAASSVRVTFLGLQHWVPELARLNRAGMVNAWTP SSNDDNPWIQVNLLRRMWVTGVVTQGASRLASHEYLKAFKVAYSLNGHEFDFIHDVNKKHKEFVGNWNKNAVHVNLFETPVEAQYVRLYP TSCHTACTLRFELLGCELNGCANPLGLKNNSIPDKQITASSSYKTWGLHLFSWNPSYARLDKQGNFNAWVAGSYGNDQWLQAPALQQYRT SAGSPANQSPTSPVSNQGFSPGSSPQHTSTLGSVFGDAYYEQQMAARQANALSHQLEQFNMMENAISSSSLYSPGSTLNYSQAAMMGLTG SHGSLPDSQQLGYASHSGIPNIILTVTGESPPSLSKELTSSLAGVGDVSFDSDSQFPLDELKIDPLTLDGLHMLNDPDMVLADPATEDTF -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:89444781/chr19:18882251) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MFGE8 | CRTC1 |
| FUNCTION: Plays an important role in the maintenance of intestinal epithelial homeostasis and the promotion of mucosal healing. Promotes VEGF-dependent neovascularization (By similarity). Contributes to phagocytic removal of apoptotic cells in many tissues. Specific ligand for the alpha-v/beta-3 and alpha-v/beta-5 receptors. Also binds to phosphatidylserine-enriched cell surfaces in a receptor-independent manner. Zona pellucida-binding protein which may play a role in gamete interaction. {ECO:0000250, ECO:0000269|PubMed:19204935}.; FUNCTION: [Medin]: Main constituent of aortic medial amyloid. {ECO:0000269|PubMed:19204935}. | FUNCTION: Transcriptional coactivator for CREB1 which activates transcription through both consensus and variant cAMP response element (CRE) sites. Acts as a coactivator, in the SIK/TORC signaling pathway, being active when dephosphorylated and acts independently of CREB1 'Ser-133' phosphorylation. Enhances the interaction of CREB1 with TAF4. Regulates the expression of specific CREB-activated genes such as the steroidogenic gene, StAR. Potent coactivator of PGC1alpha and inducer of mitochondrial biogenesis in muscle cells. In the hippocampus, involved in late-phase long-term potentiation (L-LTP) maintenance at the Schaffer collateral-CA1 synapses. May be required for dendritic growth of developing cortical neurons (By similarity). In concert with SIK1, regulates the light-induced entrainment of the circadian clock. In response to light stimulus, coactivates the CREB-mediated transcription of PER1 which plays an important role in the photic entrainment of the circadian clock. {ECO:0000250|UniProtKB:Q157S1, ECO:0000250|UniProtKB:Q68ED7, ECO:0000269|PubMed:23699513}.; FUNCTION: (Microbial infection) Plays a role of coactivator for TAX activation of the human T-cell leukemia virus type 1 (HTLV-1) long terminal repeats (LTR). {ECO:0000269|PubMed:16809310}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MFGE8 | chr15:89444781 | chr19:18882251 | ENST00000268150 | - | 6 | 8 | 24_67 | 290.0 | 388.0 | Domain | EGF-like |
| Hgene | MFGE8 | chr15:89444781 | chr19:18882251 | ENST00000566497 | - | 6 | 9 | 24_67 | 290.0 | 499.3333333333333 | Domain | EGF-like |
| Hgene | MFGE8 | chr15:89444781 | chr19:18882251 | ENST00000268150 | - | 6 | 8 | 46_48 | 290.0 | 388.0 | Motif | Note=Cell attachment site |
| Hgene | MFGE8 | chr15:89444781 | chr19:18882251 | ENST00000566497 | - | 6 | 9 | 46_48 | 290.0 | 499.3333333333333 | Motif | Note=Cell attachment site |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CRTC1 | chr15:89444781 | chr19:18882251 | ENST00000321949 | 9 | 14 | 299_366 | 440.0 | 635.0 | Compositional bias | Note=Ser-rich | |
| Tgene | CRTC1 | chr15:89444781 | chr19:18882251 | ENST00000338797 | 10 | 15 | 299_366 | 456.0 | 651.0 | Compositional bias | Note=Ser-rich | |
| Tgene | CRTC1 | chr15:89444781 | chr19:18882251 | ENST00000321949 | 9 | 14 | 242_258 | 440.0 | 635.0 | Motif | Nuclear export signal | |
| Tgene | CRTC1 | chr15:89444781 | chr19:18882251 | ENST00000338797 | 10 | 15 | 242_258 | 456.0 | 651.0 | Motif | Nuclear export signal |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| CRTC1 | CREB1, MAP3K1, MEIS1, YWHAE, CREBBP, EP300, SRPK2, XPO1, KCTD3, KIF13B, CBY1, BICD2, nsp16, ESR1, TBK1, TFG, TRAF3, VASP, YWHAG, YWHAB, YWHAZ, YWHAH, YWHAQ, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| MFGE8 | |
| CRTC1 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to MFGE8-CRTC1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to MFGE8-CRTC1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | CRTC1 | C0279628 | Adenocarcinoma Of Esophagus | 1 | CTD_human |
| Tgene | CRTC1 | C0525045 | Mood Disorders | 1 | PSYGENET |

