| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:MFN1-KMT2C |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: MFN1-KMT2C | FusionPDB ID: 53215 | FusionGDB2.0 ID: 53215 | Hgene | Tgene | Gene symbol | MFN1 | KMT2C | Gene ID | 55669 | 58508 |
| Gene name | mitofusin 1 | lysine methyltransferase 2C | |
| Synonyms | hfzo1|hfzo2 | HALR|KLEFS2|MLL3 | |
| Cytomap | 3q26.33 | 7q36.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | mitofusin-1fzo homologmitochondrial transmembrane GTPase FZO-2mitochondrial transmembrane GTPase Fzo-1putative transmembrane GTPasetransmembrane GTPase MFN1 | histone-lysine N-methyltransferase 2CALR-like proteinhistone-lysine N-methyltransferase MLL3histone-lysine N-methyltransferase, H3 lysine-4 specifichomologous to ALR proteinlysine (K)-specific methyltransferase 2Cmyeloid/lymphoid or mixed-lineage le | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q8IWA4 | Q8NEZ4 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000263969, ENST00000280653, ENST00000471841, | ENST00000485241, ENST00000485655, ENST00000262189, ENST00000355193, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 3 X 2 X 3=18 | 12 X 17 X 8=1632 |
| # samples | 3 | 17 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(17/1632*10)=-3.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: MFN1 [Title/Abstract] AND KMT2C [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MFN1(179069823)-KMT2C(151921264), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MFN1-KMT2C seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MFN1-KMT2C seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MFN1-KMT2C seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MFN1-KMT2C seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | MFN1 | GO:0008053 | mitochondrial fusion | 20436456 |
| Hgene | MFN1 | GO:0046039 | GTP metabolic process | 27920125 |
| Tgene | KMT2C | GO:0097692 | histone H3-K4 monomethylation | 26324722 |
 Fusion gene breakpoints across MFN1 (5'-gene) Fusion gene breakpoints across MFN1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
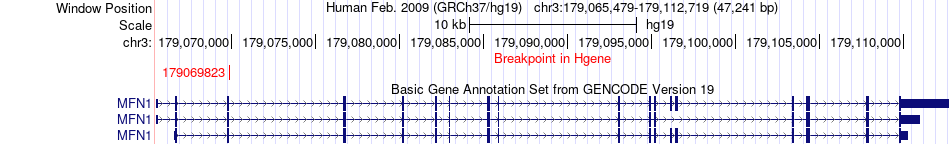 |
 Fusion gene breakpoints across KMT2C (3'-gene) Fusion gene breakpoints across KMT2C (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
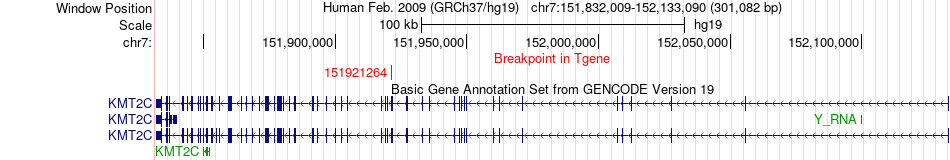 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUSC | TCGA-L3-A524-01A | MFN1 | chr3 | 179069823 | + | KMT2C | chr7 | 151921264 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000280653 | MFN1 | chr3 | 179069823 | + | ENST00000262189 | KMT2C | chr7 | 151921264 | - | 13859 | 374 | 105 | 11951 | 3948 |
| ENST00000280653 | MFN1 | chr3 | 179069823 | + | ENST00000355193 | KMT2C | chr7 | 151921264 | - | 14026 | 374 | 105 | 12122 | 4005 |
| ENST00000471841 | MFN1 | chr3 | 179069823 | + | ENST00000262189 | KMT2C | chr7 | 151921264 | - | 13859 | 374 | 105 | 11951 | 3948 |
| ENST00000471841 | MFN1 | chr3 | 179069823 | + | ENST00000355193 | KMT2C | chr7 | 151921264 | - | 14026 | 374 | 105 | 12122 | 4005 |
| ENST00000263969 | MFN1 | chr3 | 179069823 | + | ENST00000262189 | KMT2C | chr7 | 151921264 | - | 13824 | 339 | 7 | 11916 | 3969 |
| ENST00000263969 | MFN1 | chr3 | 179069823 | + | ENST00000355193 | KMT2C | chr7 | 151921264 | - | 13991 | 339 | 7 | 12087 | 4026 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000280653 | ENST00000262189 | MFN1 | chr3 | 179069823 | + | KMT2C | chr7 | 151921264 | - | 0.000700028 | 0.9993 |
| ENST00000280653 | ENST00000355193 | MFN1 | chr3 | 179069823 | + | KMT2C | chr7 | 151921264 | - | 0.000718067 | 0.99928194 |
| ENST00000471841 | ENST00000262189 | MFN1 | chr3 | 179069823 | + | KMT2C | chr7 | 151921264 | - | 0.000700028 | 0.9993 |
| ENST00000471841 | ENST00000355193 | MFN1 | chr3 | 179069823 | + | KMT2C | chr7 | 151921264 | - | 0.000718067 | 0.99928194 |
| ENST00000263969 | ENST00000262189 | MFN1 | chr3 | 179069823 | + | KMT2C | chr7 | 151921264 | - | 0.000603809 | 0.9993962 |
| ENST00000263969 | ENST00000355193 | MFN1 | chr3 | 179069823 | + | KMT2C | chr7 | 151921264 | - | 0.000618791 | 0.9993812 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >53215_53215_1_MFN1-KMT2C_MFN1_chr3_179069823_ENST00000263969_KMT2C_chr7_151921264_ENST00000262189_length(amino acids)=3969AA_BP=110 MSLVCHQFACLSSTLVGAFLTLSPSSSIMAEPVSPLKHFVLAKKAITAIFDQLLEFVTEGSHFVEATYKNPELDRIATEDDLVEMQGYKD KLSIIGEVLSRRHMKVAFFGRCVWCRHCGATSAGLRCEWQNNYTQCAPCASLSSCPVCYRNYREEDLILQCRQCDRWMHAVCQNLNTEEE VENVADIGFDCSMCRPYMPASNVPSSDCCESSLVAQIVTKVKELDPPKTYTQDGVCLTESGMTQLQSLTVTVPRRKRSKPKLKLKIINQN SVAVLQTPPDIQSEHSRDGEMDDSREGELMDCDGKSESSPEREAVDDETKGVEGTDGVKKRKRKPYRPGIGGFMVRQRSRTGQGKTKRSV IRKDSSGSISEQLPCRDDGWSEQLPDTLVDESVSVTESTEKIKKRYRKRKNKLEETFPAYLQEAFFGKDLLDTSRQSKISLDNLSEDGAQ LLYKTNMNTGFLDPSLDPLLSSSSAPTKSGTHGPADDPLADISEVLNTDDDILGIISDDLAKSVDHSDIGPVTDDPSSLPQPNVNQSSRP LSEEQLDGILSPELDKMVTDGAILGKLYKIPELGGKDVEDLFTAVLSPANTQPTPLPQPPPPTQLLPIHNQDAFSRMPLMNGLIGSSPHL PHNSLPPGSGLGTFSAIAQSSYPDARDKNSAFNPMASDPNNSWTSSAPTVEGENDTMSNAQRSTLKWEKEEALGEMATVAPVLYTNINFP NLKEEFPDWTTRVKQIAKLWRKASSQERAPYVQKARDNRAALRINKVQMSNDSMKRQQQQDSIDPSSRIDSELFKDPLKQRESEHEQEWK FRQQMRQKSKQQAKIEATQKLEQVKNEQQQQQQQQFGSQHLLVQSGSDTPSSGIQSPLTPQPGNGNMSPAQSFHKELFTKQPPSTPTSTS SDDVFVKPQAPPPPPAPSRIPIQDSLSQAQTSQPPSPQVFSPGSSNSRPPSPMDPYAKMVGTPRPPPVGHSFSRRNSAAPVENCTPLSSV SRPLQMNETTANRPSPVRDLCSSSTTNNDPYAKPPDTPRPVMTDQFPKSLGLSRSPVVSEQTAKGPIAAGTSDHFTKPSPRADVFQRQRI PDSYARPLLTPAPLDSGPGPFKTPMQPPPSSQDPYGSVSQASRRLSVDPYERPALTPRPIDNFSHNQSNDPYSQPPLTPHPAVNESFAHP SRAFSQPGTISRPTSQDPYSQPPGTPRPVVDSYSQSSGTARSNTDPYSQPPGTPRPTTVDPYSQQPQTPRPSTQTDLFVTPVTNQRHSDP YAHPPGTPRPGISVPYSQPPATPRPRISEGFTRSSMTRPVLMPNQDPFLQAAQNRGPALPGPLVRPPDTCSQTPRPPGPGLSDTFSRVSP SAARDPYDQSPMTPRSQSDSFGTSQTAHDVADQPRPGSEGSFCASSNSPMHSQGQQFSGVSQLPGPVPTSGVTDTQNTVNMAQADTEKLR QRQKLREIILQQQQQKKIAGRQEKGSQDSPAVPHPGPLQHWQPENVNQAFTRPPPPYPGNIRSPVAPPLGPRYAVFPKDQRGPYPPDVAS MGMRPHGFRFGFPGGSHGTMPSQERFLVPPQQIQGSGVSPQLRRSVSVDMPRPLNNSQMNNPVGLPQHFSPQSLPVQQHNILGQAYIELR HRAPDGRQRLPFSAPPGSVVEASSNLRHGNFIPRPDFPGPRHTDPMRRPPQGLPNQLPVHPDLEQVPPSQQEQGHSVHSSSMVMRTLNHP LGGEFSEAPLSTSVPSETTSDNLQITTQPSDGLEEKLDSDDPSVKELDVKDLEGVEVKDLDDEDLENLNLDTEDGKVVELDTLDNLETND PNLDDLLRSGEFDIIAYTDPELDMGDKKSMFNEELDLPIDDKLDNQCVSVEPKKKEQENKTLVLSDKHSPQKKSTVTNEVKTEVLSPNSK VESKCETEKNDENKDNVDTPCSQASAHSDLNDGEKTSLHPCDPDLFEKRTNRETAGPSANVIQASTQLPAQDVINSCGITGSTPVLSSLL ANEKSDNSDIRPSGSPPPPTLPASPSNHVSSLPPFIAPPGRVLDNAMNSNVTVVSRVNHVFSQGVQVNPGLIPGQSTVNHSLGTGKPATQ TGPQTSQSGTSSMSGPQQLMIPQTLAQQNRERPLLLEEQPLLLQDLLDQERQEQQQQRQMQAMIRQRSEPFFPNIDFDAITDPIMKAKMV ALKGINKVMAQNNLGMPPMVMSRFPFMGQVVTGTQNSEGQNLGPQAIPQDGSITHQISRPNPPNFGPGFVNDSQRKQYEEWLQETQQLLQ MQQKYLEEQIGAHRKSKKALSAKQRTAKKAGREFPEEDAEQLKHVTEQQSMVQKQLEQIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMP SVQPQPPLIPGATPPTMSQPTFPMVPQQLQHQQHTTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSN ANPQSGPPPRVEFDDNNPFSESFQERERKERLREQQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDF MQPLGPLQQSPQHQQQMGQVLQQQNIQQGSINSPSTQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPS FTPALPAAPPVANSSLPCGQDSTITHGHSYPGSTQSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTS TPAVSTPSELPQQADQESVEPVGPSTPNMAAGQLCTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQE EPKLEEQNGSKVEGNAVACPVSSAQSPPHSAGAPAAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQT LGAQMQGGFGCGNQLPKTDGGSETKKQRSKRTQRTGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQQNNLSNPPTPPASLPPTPPPMA CQKMANGFATTEELAGKAGVLVSHEVTKTLGPKPFQLPFRPQDDLLARALAQGPKTVDVPASLPTPPHNNQEELRIQDHCGDRDTPDSFV PSSSPESVVGVEVSRYPDLSLVKEEPPEPVPSPIIPILPSTAGKSSESRRNDIKTEPGTLYFASPFGPSPNGPRSGLISVAITLHPTAAE NISSVVAAFSDLLHVRIPNSYEVSSAPDVPSMGLVSSHRINPGLEYRQHLLLRGPPPGSANPPRLVSSYRLKQPNVPFPPTSNGLSGYKD SSHGIAESAALRPQWCCHCKVVILGSGVRKSFKDLTLLNKDSRESTKRVEKDIVFCSNNCFILYSSTAQAKNSENKESIPSLPQSPMRET PSKAFHQYSNNISTLDVHCLPQLPEKASPPASPPIAFPPAFEAAQVEAKPDELKVTVKLKPRLRAVHGGFEDCRPLNKKWRGMKWKKWSI HIVIPKGTFKPPCEDEIDEFLKKLGTSLKPDPVPKDYRKCCFCHEEGDGLTDGPARLLNLDLDLWVHLNCALWSTEVYETQAGALINVEL ALRRGLQMKCVFCHKTGATSGCHRFRCTNIYHFTCAIKAQCMFFKDKTMLCPMHKPKGIHEQELSYFAVFRRVYVQRDEVRQIASIVQRG ERDHTFRVGSLIFHTIGQLLPQQMQAFHSPKALFPVGYEASRLYWSTRYANRRCRYLCSIEEKDGRPVFVIRIVEQGHEDLVLSDISPKG VWDKILEPVACVRKKSEMLQLFPAYLKGEDLFGLTVSAVARIAESLPGVEACENYTFRYGRNPLMELPLAVNPTGCARSEPKMSAHVKRF VLRPHTLNSTSTSKSFQSTVTGELNAPYSKQFVHSKSSQYRKMKTEWKSNVYLARSRIQGLGLYAARDIEKHTMVIEYIGTIIRNEVANR KEKLYESQNRGVYMFRMDNDHVIDATLTGGPARYINHSCAPNCVAEVVTFERGHKIIISSSRRIQKGEELCYDYKFDFEDDQHKIPCHCG -------------------------------------------------------------- >53215_53215_2_MFN1-KMT2C_MFN1_chr3_179069823_ENST00000263969_KMT2C_chr7_151921264_ENST00000355193_length(amino acids)=4026AA_BP=110 MSLVCHQFACLSSTLVGAFLTLSPSSSIMAEPVSPLKHFVLAKKAITAIFDQLLEFVTEGSHFVEATYKNPELDRIATEDDLVEMQGYKD KLSIIGEVLSRRHMKVAFFGRCVWCRHCGATSAGLRCEWQNNYTQCAPCASLSSCPVCYRNYREEDLILQCRQCDRWMHAVCQNLNTEEE VENVADIGFDCSMCRPYMPASNVPSSDCCESSLVAQIVTKVKELDPPKTYTQDGVCLTESGMTQLQSLTVTVPRRKRSKPKLKLKIINQN SVAVLQTPPDIQSEHSRDGEMDDSREGELMDCDGKSESSPEREAVDDETKGVEGTDGVKKRKRKPYRPGIGGFMVRQRSRTGQGKTKRSV IRKDSSGSISEQLPCRDDGWSEQLPDTLVDESVSVTESTEKIKKRYRKRKNKLEETFPAYLQEAFFGKDLLDTSRQSKISLDNLSEDGAQ LLYKTNMNTGFLDPSLDPLLSSSSAPTKSGTHGPADDPLADISEVLNTDDDILGIISDDLAKSVDHSDIGPVTDDPSSLPQPNVNQSSRP LSEEQLDGILSPELDKMVTDGAILGKLYKIPELGGKDVEDLFTAVLSPANTQPTPLPQPPPPTQLLPIHNQDAFSRMPLMNGLIGSSPHL PHNSLPPGSGLGTFSAIAQSSYPDARDKNSAFNPMASDPNNSWTSSAPTVEGENDTMSNAQRSTLKWEKEEALGEMATVAPVLYTNINFP NLKEEFPDWTTRVKQIAKLWRKASSQERAPYVQKARDNRAALRINKVQMSNDSMKRQQQQDSIDPSSRIDSELFKDPLKQRESEHEQEWK FRQQMRQKSKQQAKIEATQKLEQVKNEQQQQQQQQFGSQHLLVQSGSDTPSSGIQSPLTPQPGNGNMSPAQSFHKELFTKQPPSTPTSTS SDDVFVKPQAPPPPPAPSRIPIQDSLSQAQTSQPPSPQVFSPGSSNSRPPSPMDPYAKMVGTPRPPPVGHSFSRRNSAAPVENCTPLSSV SRPLQMNETTANRPSPVRDLCSSSTTNNDPYAKPPDTPRPVMTDQFPKSLGLSRSPVVSEQTAKGPIAAGTSDHFTKPSPRADVFQRQRI PDSYARPLLTPAPLDSGPGPFKTPMQPPPSSQDPYGSVSQASRRLSVDPYERPALTPRPIDNFSHNQSNDPYSQPPLTPHPAVNESFAHP SRAFSQPGTISRPTSQDPYSQPPGTPRPVVDSYSQSSGTARSNTDPYSQPPGTPRPTTVDPYSQQPQTPRPSTQTDLFVTPVTNQRHSDP YAHPPGTPRPGISVPYSQPPATPRPRISEGFTRSSMTRPVLMPNQDPFLQAAQNRGPALPGPLVRPPDTCSQTPRPPGPGLSDTFSRVSP SAARDPYDQSPMTPRSQSDSFGTSQTAHDVADQPRPGSEGSFCASSNSPMHSQGQQFSGVSQLPGPVPTSGVTDTQNTVNMAQADTEKLR QRQKLREIILQQQQQKKIAGRQEKGSQDSPAVPHPGPLQHWQPENVNQAFTRPPPPYPGNIRSPVAPPLGPRYAVFPKDQRGPYPPDVAS MGMRPHGFRFGFPGGSHGTMPSQERFLVPPQQIQGSGVSPQLRRSVSVDMPRPLNNSQMNNPVGLPQHFSPQSLPVQQHNILGQAYIELR HRAPDGRQRLPFSAPPGSVVEASSNLRHGNFIPRPDFPGPRHTDPMRRPPQGLPNQLPVHPDLEQVPPSQQEQGHSVHSSSMVMRTLNHP LGGEFSEAPLSTSVPSETTSDNLQITTQPSDGLEEKLDSDDPSVKELDVKDLEGVEVKDLDDEDLENLNLDTEDGKVVELDTLDNLETND PNLDDLLRSGEFDIIAYTDPELDMGDKKSMFNEELDLPIDDKLDNQCVSVEPKKKEQENKTLVLSDKHSPQKKSTVTNEVKTEVLSPNSK VESKCETEKNDENKDNVDTPCSQASAHSDLNDGEKTSLHPCDPDLFEKRTNRETAGPSANVIQASTQLPAQDVINSCGITGSTPVLSSLL ANEKSDNSDIRPSGSPPPPTLPASPSNHVSSLPPFIAPPGRVLDNAMNSNVTVVSRVNHVFSQGVQVNPGLIPGQSTVNHSLGTGKPATQ TGPQTSQSGTSSMSGPQQLMIPQTLAQQNRERPLLLEEQPLLLQDLLDQERQEQQQQRQMQAMIRQRSEPFFPNIDFDAITDPIMKAKMV ALKGINKVMAQNNLGMPPMVMSRFPFMGQVVTGTQNSEGQNLGPQAIPQDGSITHQISRPNPPNFGPGFVNDSQRKQYEEWLQETQQLLQ MQQKYLEEQIGAHRKSKKALSAKQRTAKKAGREFPEEDAEQLKHVTEQQSMVQKQLEQIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMP SVQPQPPLIPGATPPTMSQPTFPMVPQQLQHQQHTTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSN ANPQSGPPPRVEFDDNNPFSESFQERERKERLREQQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDF MQPLGPLQQSPQHQQQMGQVLQQQNIQQGSINSPSTQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPS FTPALPAAPPVANSSLPCGQDSTITHGHSYPGSTQSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTS TPAVSTPSELPQQADQESVEPVGPSTPNMAAGQLCTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQE EPKLEEQNGSKVEGNAVACPVSSAQSPPHSAGAPAAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQT LGAQMQGGFGCGNQLPKTDGGSETKKQRSKRTQRTGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQVRQLSLLPLMEPIIGVNFAHFL PYGSGQFNSGNRLLGTFGSATLEGVSDYYSQLIYKQNNLSNPPTPPASLPPTPPPMACQKMANGFATTEELAGKAGVLVSHEVTKTLGPK PFQLPFRPQDDLLARALAQGPKTVDVPASLPTPPHNNQEELRIQDHCGDRDTPDSFVPSSSPESVVGVEVSRYPDLSLVKEEPPEPVPSP IIPILPSTAGKSSESRRNDIKTEPGTLYFASPFGPSPNGPRSGLISVAITLHPTAAENISSVVAAFSDLLHVRIPNSYEVSSAPDVPSMG LVSSHRINPGLEYRQHLLLRGPPPGSANPPRLVSSYRLKQPNVPFPPTSNGLSGYKDSSHGIAESAALRPQWCCHCKVVILGSGVRKSFK DLTLLNKDSRESTKRVEKDIVFCSNNCFILYSSTAQAKNSENKESIPSLPQSPMRETPSKAFHQYSNNISTLDVHCLPQLPEKASPPASP PIAFPPAFEAAQVEAKPDELKVTVKLKPRLRAVHGGFEDCRPLNKKWRGMKWKKWSIHIVIPKGTFKPPCEDEIDEFLKKLGTSLKPDPV PKDYRKCCFCHEEGDGLTDGPARLLNLDLDLWVHLNCALWSTEVYETQAGALINVELALRRGLQMKCVFCHKTGATSGCHRFRCTNIYHF TCAIKAQCMFFKDKTMLCPMHKPKGIHEQELSYFAVFRRVYVQRDEVRQIASIVQRGERDHTFRVGSLIFHTIGQLLPQQMQAFHSPKAL FPVGYEASRLYWSTRYANRRCRYLCSIEEKDGRPVFVIRIVEQGHEDLVLSDISPKGVWDKILEPVACVRKKSEMLQLFPAYLKGEDLFG LTVSAVARIAESLPGVEACENYTFRYGRNPLMELPLAVNPTGCARSEPKMSAHVKRFVLRPHTLNSTSTSKSFQSTVTGELNAPYSKQFV HSKSSQYRKMKTEWKSNVYLARSRIQGLGLYAARDIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDHVIDATLTGGPAR -------------------------------------------------------------- >53215_53215_3_MFN1-KMT2C_MFN1_chr3_179069823_ENST00000280653_KMT2C_chr7_151921264_ENST00000262189_length(amino acids)=3948AA_BP=89 MERRLSIMAEPVSPLKHFVLAKKAITAIFDQLLEFVTEGSHFVEATYKNPELDRIATEDDLVEMQGYKDKLSIIGEVLSRRHMKVAFFGR CVWCRHCGATSAGLRCEWQNNYTQCAPCASLSSCPVCYRNYREEDLILQCRQCDRWMHAVCQNLNTEEEVENVADIGFDCSMCRPYMPAS NVPSSDCCESSLVAQIVTKVKELDPPKTYTQDGVCLTESGMTQLQSLTVTVPRRKRSKPKLKLKIINQNSVAVLQTPPDIQSEHSRDGEM DDSREGELMDCDGKSESSPEREAVDDETKGVEGTDGVKKRKRKPYRPGIGGFMVRQRSRTGQGKTKRSVIRKDSSGSISEQLPCRDDGWS EQLPDTLVDESVSVTESTEKIKKRYRKRKNKLEETFPAYLQEAFFGKDLLDTSRQSKISLDNLSEDGAQLLYKTNMNTGFLDPSLDPLLS SSSAPTKSGTHGPADDPLADISEVLNTDDDILGIISDDLAKSVDHSDIGPVTDDPSSLPQPNVNQSSRPLSEEQLDGILSPELDKMVTDG AILGKLYKIPELGGKDVEDLFTAVLSPANTQPTPLPQPPPPTQLLPIHNQDAFSRMPLMNGLIGSSPHLPHNSLPPGSGLGTFSAIAQSS YPDARDKNSAFNPMASDPNNSWTSSAPTVEGENDTMSNAQRSTLKWEKEEALGEMATVAPVLYTNINFPNLKEEFPDWTTRVKQIAKLWR KASSQERAPYVQKARDNRAALRINKVQMSNDSMKRQQQQDSIDPSSRIDSELFKDPLKQRESEHEQEWKFRQQMRQKSKQQAKIEATQKL EQVKNEQQQQQQQQFGSQHLLVQSGSDTPSSGIQSPLTPQPGNGNMSPAQSFHKELFTKQPPSTPTSTSSDDVFVKPQAPPPPPAPSRIP IQDSLSQAQTSQPPSPQVFSPGSSNSRPPSPMDPYAKMVGTPRPPPVGHSFSRRNSAAPVENCTPLSSVSRPLQMNETTANRPSPVRDLC SSSTTNNDPYAKPPDTPRPVMTDQFPKSLGLSRSPVVSEQTAKGPIAAGTSDHFTKPSPRADVFQRQRIPDSYARPLLTPAPLDSGPGPF KTPMQPPPSSQDPYGSVSQASRRLSVDPYERPALTPRPIDNFSHNQSNDPYSQPPLTPHPAVNESFAHPSRAFSQPGTISRPTSQDPYSQ PPGTPRPVVDSYSQSSGTARSNTDPYSQPPGTPRPTTVDPYSQQPQTPRPSTQTDLFVTPVTNQRHSDPYAHPPGTPRPGISVPYSQPPA TPRPRISEGFTRSSMTRPVLMPNQDPFLQAAQNRGPALPGPLVRPPDTCSQTPRPPGPGLSDTFSRVSPSAARDPYDQSPMTPRSQSDSF GTSQTAHDVADQPRPGSEGSFCASSNSPMHSQGQQFSGVSQLPGPVPTSGVTDTQNTVNMAQADTEKLRQRQKLREIILQQQQQKKIAGR QEKGSQDSPAVPHPGPLQHWQPENVNQAFTRPPPPYPGNIRSPVAPPLGPRYAVFPKDQRGPYPPDVASMGMRPHGFRFGFPGGSHGTMP SQERFLVPPQQIQGSGVSPQLRRSVSVDMPRPLNNSQMNNPVGLPQHFSPQSLPVQQHNILGQAYIELRHRAPDGRQRLPFSAPPGSVVE ASSNLRHGNFIPRPDFPGPRHTDPMRRPPQGLPNQLPVHPDLEQVPPSQQEQGHSVHSSSMVMRTLNHPLGGEFSEAPLSTSVPSETTSD NLQITTQPSDGLEEKLDSDDPSVKELDVKDLEGVEVKDLDDEDLENLNLDTEDGKVVELDTLDNLETNDPNLDDLLRSGEFDIIAYTDPE LDMGDKKSMFNEELDLPIDDKLDNQCVSVEPKKKEQENKTLVLSDKHSPQKKSTVTNEVKTEVLSPNSKVESKCETEKNDENKDNVDTPC SQASAHSDLNDGEKTSLHPCDPDLFEKRTNRETAGPSANVIQASTQLPAQDVINSCGITGSTPVLSSLLANEKSDNSDIRPSGSPPPPTL PASPSNHVSSLPPFIAPPGRVLDNAMNSNVTVVSRVNHVFSQGVQVNPGLIPGQSTVNHSLGTGKPATQTGPQTSQSGTSSMSGPQQLMI PQTLAQQNRERPLLLEEQPLLLQDLLDQERQEQQQQRQMQAMIRQRSEPFFPNIDFDAITDPIMKAKMVALKGINKVMAQNNLGMPPMVM SRFPFMGQVVTGTQNSEGQNLGPQAIPQDGSITHQISRPNPPNFGPGFVNDSQRKQYEEWLQETQQLLQMQQKYLEEQIGAHRKSKKALS AKQRTAKKAGREFPEEDAEQLKHVTEQQSMVQKQLEQIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMPSVQPQPPLIPGATPPTMSQPT FPMVPQQLQHQQHTTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSNANPQSGPPPRVEFDDNNPFSE SFQERERKERLREQQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDFMQPLGPLQQSPQHQQQMGQVL QQQNIQQGSINSPSTQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPSFTPALPAAPPVANSSLPCGQD STITHGHSYPGSTQSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTSTPAVSTPSELPQQADQESVEP VGPSTPNMAAGQLCTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQEEPKLEEQNGSKVEGNAVACPV SSAQSPPHSAGAPAAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQTLGAQMQGGFGCGNQLPKTDGG SETKKQRSKRTQRTGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQQNNLSNPPTPPASLPPTPPPMACQKMANGFATTEELAGKAGVL VSHEVTKTLGPKPFQLPFRPQDDLLARALAQGPKTVDVPASLPTPPHNNQEELRIQDHCGDRDTPDSFVPSSSPESVVGVEVSRYPDLSL VKEEPPEPVPSPIIPILPSTAGKSSESRRNDIKTEPGTLYFASPFGPSPNGPRSGLISVAITLHPTAAENISSVVAAFSDLLHVRIPNSY EVSSAPDVPSMGLVSSHRINPGLEYRQHLLLRGPPPGSANPPRLVSSYRLKQPNVPFPPTSNGLSGYKDSSHGIAESAALRPQWCCHCKV VILGSGVRKSFKDLTLLNKDSRESTKRVEKDIVFCSNNCFILYSSTAQAKNSENKESIPSLPQSPMRETPSKAFHQYSNNISTLDVHCLP QLPEKASPPASPPIAFPPAFEAAQVEAKPDELKVTVKLKPRLRAVHGGFEDCRPLNKKWRGMKWKKWSIHIVIPKGTFKPPCEDEIDEFL KKLGTSLKPDPVPKDYRKCCFCHEEGDGLTDGPARLLNLDLDLWVHLNCALWSTEVYETQAGALINVELALRRGLQMKCVFCHKTGATSG CHRFRCTNIYHFTCAIKAQCMFFKDKTMLCPMHKPKGIHEQELSYFAVFRRVYVQRDEVRQIASIVQRGERDHTFRVGSLIFHTIGQLLP QQMQAFHSPKALFPVGYEASRLYWSTRYANRRCRYLCSIEEKDGRPVFVIRIVEQGHEDLVLSDISPKGVWDKILEPVACVRKKSEMLQL FPAYLKGEDLFGLTVSAVARIAESLPGVEACENYTFRYGRNPLMELPLAVNPTGCARSEPKMSAHVKRFVLRPHTLNSTSTSKSFQSTVT GELNAPYSKQFVHSKSSQYRKMKTEWKSNVYLARSRIQGLGLYAARDIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDH -------------------------------------------------------------- >53215_53215_4_MFN1-KMT2C_MFN1_chr3_179069823_ENST00000280653_KMT2C_chr7_151921264_ENST00000355193_length(amino acids)=4005AA_BP=89 MERRLSIMAEPVSPLKHFVLAKKAITAIFDQLLEFVTEGSHFVEATYKNPELDRIATEDDLVEMQGYKDKLSIIGEVLSRRHMKVAFFGR CVWCRHCGATSAGLRCEWQNNYTQCAPCASLSSCPVCYRNYREEDLILQCRQCDRWMHAVCQNLNTEEEVENVADIGFDCSMCRPYMPAS NVPSSDCCESSLVAQIVTKVKELDPPKTYTQDGVCLTESGMTQLQSLTVTVPRRKRSKPKLKLKIINQNSVAVLQTPPDIQSEHSRDGEM DDSREGELMDCDGKSESSPEREAVDDETKGVEGTDGVKKRKRKPYRPGIGGFMVRQRSRTGQGKTKRSVIRKDSSGSISEQLPCRDDGWS EQLPDTLVDESVSVTESTEKIKKRYRKRKNKLEETFPAYLQEAFFGKDLLDTSRQSKISLDNLSEDGAQLLYKTNMNTGFLDPSLDPLLS SSSAPTKSGTHGPADDPLADISEVLNTDDDILGIISDDLAKSVDHSDIGPVTDDPSSLPQPNVNQSSRPLSEEQLDGILSPELDKMVTDG AILGKLYKIPELGGKDVEDLFTAVLSPANTQPTPLPQPPPPTQLLPIHNQDAFSRMPLMNGLIGSSPHLPHNSLPPGSGLGTFSAIAQSS YPDARDKNSAFNPMASDPNNSWTSSAPTVEGENDTMSNAQRSTLKWEKEEALGEMATVAPVLYTNINFPNLKEEFPDWTTRVKQIAKLWR KASSQERAPYVQKARDNRAALRINKVQMSNDSMKRQQQQDSIDPSSRIDSELFKDPLKQRESEHEQEWKFRQQMRQKSKQQAKIEATQKL EQVKNEQQQQQQQQFGSQHLLVQSGSDTPSSGIQSPLTPQPGNGNMSPAQSFHKELFTKQPPSTPTSTSSDDVFVKPQAPPPPPAPSRIP IQDSLSQAQTSQPPSPQVFSPGSSNSRPPSPMDPYAKMVGTPRPPPVGHSFSRRNSAAPVENCTPLSSVSRPLQMNETTANRPSPVRDLC SSSTTNNDPYAKPPDTPRPVMTDQFPKSLGLSRSPVVSEQTAKGPIAAGTSDHFTKPSPRADVFQRQRIPDSYARPLLTPAPLDSGPGPF KTPMQPPPSSQDPYGSVSQASRRLSVDPYERPALTPRPIDNFSHNQSNDPYSQPPLTPHPAVNESFAHPSRAFSQPGTISRPTSQDPYSQ PPGTPRPVVDSYSQSSGTARSNTDPYSQPPGTPRPTTVDPYSQQPQTPRPSTQTDLFVTPVTNQRHSDPYAHPPGTPRPGISVPYSQPPA TPRPRISEGFTRSSMTRPVLMPNQDPFLQAAQNRGPALPGPLVRPPDTCSQTPRPPGPGLSDTFSRVSPSAARDPYDQSPMTPRSQSDSF GTSQTAHDVADQPRPGSEGSFCASSNSPMHSQGQQFSGVSQLPGPVPTSGVTDTQNTVNMAQADTEKLRQRQKLREIILQQQQQKKIAGR QEKGSQDSPAVPHPGPLQHWQPENVNQAFTRPPPPYPGNIRSPVAPPLGPRYAVFPKDQRGPYPPDVASMGMRPHGFRFGFPGGSHGTMP SQERFLVPPQQIQGSGVSPQLRRSVSVDMPRPLNNSQMNNPVGLPQHFSPQSLPVQQHNILGQAYIELRHRAPDGRQRLPFSAPPGSVVE ASSNLRHGNFIPRPDFPGPRHTDPMRRPPQGLPNQLPVHPDLEQVPPSQQEQGHSVHSSSMVMRTLNHPLGGEFSEAPLSTSVPSETTSD NLQITTQPSDGLEEKLDSDDPSVKELDVKDLEGVEVKDLDDEDLENLNLDTEDGKVVELDTLDNLETNDPNLDDLLRSGEFDIIAYTDPE LDMGDKKSMFNEELDLPIDDKLDNQCVSVEPKKKEQENKTLVLSDKHSPQKKSTVTNEVKTEVLSPNSKVESKCETEKNDENKDNVDTPC SQASAHSDLNDGEKTSLHPCDPDLFEKRTNRETAGPSANVIQASTQLPAQDVINSCGITGSTPVLSSLLANEKSDNSDIRPSGSPPPPTL PASPSNHVSSLPPFIAPPGRVLDNAMNSNVTVVSRVNHVFSQGVQVNPGLIPGQSTVNHSLGTGKPATQTGPQTSQSGTSSMSGPQQLMI PQTLAQQNRERPLLLEEQPLLLQDLLDQERQEQQQQRQMQAMIRQRSEPFFPNIDFDAITDPIMKAKMVALKGINKVMAQNNLGMPPMVM SRFPFMGQVVTGTQNSEGQNLGPQAIPQDGSITHQISRPNPPNFGPGFVNDSQRKQYEEWLQETQQLLQMQQKYLEEQIGAHRKSKKALS AKQRTAKKAGREFPEEDAEQLKHVTEQQSMVQKQLEQIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMPSVQPQPPLIPGATPPTMSQPT FPMVPQQLQHQQHTTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSNANPQSGPPPRVEFDDNNPFSE SFQERERKERLREQQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDFMQPLGPLQQSPQHQQQMGQVL QQQNIQQGSINSPSTQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPSFTPALPAAPPVANSSLPCGQD STITHGHSYPGSTQSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTSTPAVSTPSELPQQADQESVEP VGPSTPNMAAGQLCTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQEEPKLEEQNGSKVEGNAVACPV SSAQSPPHSAGAPAAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQTLGAQMQGGFGCGNQLPKTDGG SETKKQRSKRTQRTGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQVRQLSLLPLMEPIIGVNFAHFLPYGSGQFNSGNRLLGTFGSAT LEGVSDYYSQLIYKQNNLSNPPTPPASLPPTPPPMACQKMANGFATTEELAGKAGVLVSHEVTKTLGPKPFQLPFRPQDDLLARALAQGP KTVDVPASLPTPPHNNQEELRIQDHCGDRDTPDSFVPSSSPESVVGVEVSRYPDLSLVKEEPPEPVPSPIIPILPSTAGKSSESRRNDIK TEPGTLYFASPFGPSPNGPRSGLISVAITLHPTAAENISSVVAAFSDLLHVRIPNSYEVSSAPDVPSMGLVSSHRINPGLEYRQHLLLRG PPPGSANPPRLVSSYRLKQPNVPFPPTSNGLSGYKDSSHGIAESAALRPQWCCHCKVVILGSGVRKSFKDLTLLNKDSRESTKRVEKDIV FCSNNCFILYSSTAQAKNSENKESIPSLPQSPMRETPSKAFHQYSNNISTLDVHCLPQLPEKASPPASPPIAFPPAFEAAQVEAKPDELK VTVKLKPRLRAVHGGFEDCRPLNKKWRGMKWKKWSIHIVIPKGTFKPPCEDEIDEFLKKLGTSLKPDPVPKDYRKCCFCHEEGDGLTDGP ARLLNLDLDLWVHLNCALWSTEVYETQAGALINVELALRRGLQMKCVFCHKTGATSGCHRFRCTNIYHFTCAIKAQCMFFKDKTMLCPMH KPKGIHEQELSYFAVFRRVYVQRDEVRQIASIVQRGERDHTFRVGSLIFHTIGQLLPQQMQAFHSPKALFPVGYEASRLYWSTRYANRRC RYLCSIEEKDGRPVFVIRIVEQGHEDLVLSDISPKGVWDKILEPVACVRKKSEMLQLFPAYLKGEDLFGLTVSAVARIAESLPGVEACEN YTFRYGRNPLMELPLAVNPTGCARSEPKMSAHVKRFVLRPHTLNSTSTSKSFQSTVTGELNAPYSKQFVHSKSSQYRKMKTEWKSNVYLA RSRIQGLGLYAARDIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDHVIDATLTGGPARYINHSCAPNCVAEVVTFERGH -------------------------------------------------------------- >53215_53215_5_MFN1-KMT2C_MFN1_chr3_179069823_ENST00000471841_KMT2C_chr7_151921264_ENST00000262189_length(amino acids)=3948AA_BP=89 MERRLSIMAEPVSPLKHFVLAKKAITAIFDQLLEFVTEGSHFVEATYKNPELDRIATEDDLVEMQGYKDKLSIIGEVLSRRHMKVAFFGR CVWCRHCGATSAGLRCEWQNNYTQCAPCASLSSCPVCYRNYREEDLILQCRQCDRWMHAVCQNLNTEEEVENVADIGFDCSMCRPYMPAS NVPSSDCCESSLVAQIVTKVKELDPPKTYTQDGVCLTESGMTQLQSLTVTVPRRKRSKPKLKLKIINQNSVAVLQTPPDIQSEHSRDGEM DDSREGELMDCDGKSESSPEREAVDDETKGVEGTDGVKKRKRKPYRPGIGGFMVRQRSRTGQGKTKRSVIRKDSSGSISEQLPCRDDGWS EQLPDTLVDESVSVTESTEKIKKRYRKRKNKLEETFPAYLQEAFFGKDLLDTSRQSKISLDNLSEDGAQLLYKTNMNTGFLDPSLDPLLS SSSAPTKSGTHGPADDPLADISEVLNTDDDILGIISDDLAKSVDHSDIGPVTDDPSSLPQPNVNQSSRPLSEEQLDGILSPELDKMVTDG AILGKLYKIPELGGKDVEDLFTAVLSPANTQPTPLPQPPPPTQLLPIHNQDAFSRMPLMNGLIGSSPHLPHNSLPPGSGLGTFSAIAQSS YPDARDKNSAFNPMASDPNNSWTSSAPTVEGENDTMSNAQRSTLKWEKEEALGEMATVAPVLYTNINFPNLKEEFPDWTTRVKQIAKLWR KASSQERAPYVQKARDNRAALRINKVQMSNDSMKRQQQQDSIDPSSRIDSELFKDPLKQRESEHEQEWKFRQQMRQKSKQQAKIEATQKL EQVKNEQQQQQQQQFGSQHLLVQSGSDTPSSGIQSPLTPQPGNGNMSPAQSFHKELFTKQPPSTPTSTSSDDVFVKPQAPPPPPAPSRIP IQDSLSQAQTSQPPSPQVFSPGSSNSRPPSPMDPYAKMVGTPRPPPVGHSFSRRNSAAPVENCTPLSSVSRPLQMNETTANRPSPVRDLC SSSTTNNDPYAKPPDTPRPVMTDQFPKSLGLSRSPVVSEQTAKGPIAAGTSDHFTKPSPRADVFQRQRIPDSYARPLLTPAPLDSGPGPF KTPMQPPPSSQDPYGSVSQASRRLSVDPYERPALTPRPIDNFSHNQSNDPYSQPPLTPHPAVNESFAHPSRAFSQPGTISRPTSQDPYSQ PPGTPRPVVDSYSQSSGTARSNTDPYSQPPGTPRPTTVDPYSQQPQTPRPSTQTDLFVTPVTNQRHSDPYAHPPGTPRPGISVPYSQPPA TPRPRISEGFTRSSMTRPVLMPNQDPFLQAAQNRGPALPGPLVRPPDTCSQTPRPPGPGLSDTFSRVSPSAARDPYDQSPMTPRSQSDSF GTSQTAHDVADQPRPGSEGSFCASSNSPMHSQGQQFSGVSQLPGPVPTSGVTDTQNTVNMAQADTEKLRQRQKLREIILQQQQQKKIAGR QEKGSQDSPAVPHPGPLQHWQPENVNQAFTRPPPPYPGNIRSPVAPPLGPRYAVFPKDQRGPYPPDVASMGMRPHGFRFGFPGGSHGTMP SQERFLVPPQQIQGSGVSPQLRRSVSVDMPRPLNNSQMNNPVGLPQHFSPQSLPVQQHNILGQAYIELRHRAPDGRQRLPFSAPPGSVVE ASSNLRHGNFIPRPDFPGPRHTDPMRRPPQGLPNQLPVHPDLEQVPPSQQEQGHSVHSSSMVMRTLNHPLGGEFSEAPLSTSVPSETTSD NLQITTQPSDGLEEKLDSDDPSVKELDVKDLEGVEVKDLDDEDLENLNLDTEDGKVVELDTLDNLETNDPNLDDLLRSGEFDIIAYTDPE LDMGDKKSMFNEELDLPIDDKLDNQCVSVEPKKKEQENKTLVLSDKHSPQKKSTVTNEVKTEVLSPNSKVESKCETEKNDENKDNVDTPC SQASAHSDLNDGEKTSLHPCDPDLFEKRTNRETAGPSANVIQASTQLPAQDVINSCGITGSTPVLSSLLANEKSDNSDIRPSGSPPPPTL PASPSNHVSSLPPFIAPPGRVLDNAMNSNVTVVSRVNHVFSQGVQVNPGLIPGQSTVNHSLGTGKPATQTGPQTSQSGTSSMSGPQQLMI PQTLAQQNRERPLLLEEQPLLLQDLLDQERQEQQQQRQMQAMIRQRSEPFFPNIDFDAITDPIMKAKMVALKGINKVMAQNNLGMPPMVM SRFPFMGQVVTGTQNSEGQNLGPQAIPQDGSITHQISRPNPPNFGPGFVNDSQRKQYEEWLQETQQLLQMQQKYLEEQIGAHRKSKKALS AKQRTAKKAGREFPEEDAEQLKHVTEQQSMVQKQLEQIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMPSVQPQPPLIPGATPPTMSQPT FPMVPQQLQHQQHTTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSNANPQSGPPPRVEFDDNNPFSE SFQERERKERLREQQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDFMQPLGPLQQSPQHQQQMGQVL QQQNIQQGSINSPSTQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPSFTPALPAAPPVANSSLPCGQD STITHGHSYPGSTQSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTSTPAVSTPSELPQQADQESVEP VGPSTPNMAAGQLCTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQEEPKLEEQNGSKVEGNAVACPV SSAQSPPHSAGAPAAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQTLGAQMQGGFGCGNQLPKTDGG SETKKQRSKRTQRTGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQQNNLSNPPTPPASLPPTPPPMACQKMANGFATTEELAGKAGVL VSHEVTKTLGPKPFQLPFRPQDDLLARALAQGPKTVDVPASLPTPPHNNQEELRIQDHCGDRDTPDSFVPSSSPESVVGVEVSRYPDLSL VKEEPPEPVPSPIIPILPSTAGKSSESRRNDIKTEPGTLYFASPFGPSPNGPRSGLISVAITLHPTAAENISSVVAAFSDLLHVRIPNSY EVSSAPDVPSMGLVSSHRINPGLEYRQHLLLRGPPPGSANPPRLVSSYRLKQPNVPFPPTSNGLSGYKDSSHGIAESAALRPQWCCHCKV VILGSGVRKSFKDLTLLNKDSRESTKRVEKDIVFCSNNCFILYSSTAQAKNSENKESIPSLPQSPMRETPSKAFHQYSNNISTLDVHCLP QLPEKASPPASPPIAFPPAFEAAQVEAKPDELKVTVKLKPRLRAVHGGFEDCRPLNKKWRGMKWKKWSIHIVIPKGTFKPPCEDEIDEFL KKLGTSLKPDPVPKDYRKCCFCHEEGDGLTDGPARLLNLDLDLWVHLNCALWSTEVYETQAGALINVELALRRGLQMKCVFCHKTGATSG CHRFRCTNIYHFTCAIKAQCMFFKDKTMLCPMHKPKGIHEQELSYFAVFRRVYVQRDEVRQIASIVQRGERDHTFRVGSLIFHTIGQLLP QQMQAFHSPKALFPVGYEASRLYWSTRYANRRCRYLCSIEEKDGRPVFVIRIVEQGHEDLVLSDISPKGVWDKILEPVACVRKKSEMLQL FPAYLKGEDLFGLTVSAVARIAESLPGVEACENYTFRYGRNPLMELPLAVNPTGCARSEPKMSAHVKRFVLRPHTLNSTSTSKSFQSTVT GELNAPYSKQFVHSKSSQYRKMKTEWKSNVYLARSRIQGLGLYAARDIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDH -------------------------------------------------------------- >53215_53215_6_MFN1-KMT2C_MFN1_chr3_179069823_ENST00000471841_KMT2C_chr7_151921264_ENST00000355193_length(amino acids)=4005AA_BP=89 MERRLSIMAEPVSPLKHFVLAKKAITAIFDQLLEFVTEGSHFVEATYKNPELDRIATEDDLVEMQGYKDKLSIIGEVLSRRHMKVAFFGR CVWCRHCGATSAGLRCEWQNNYTQCAPCASLSSCPVCYRNYREEDLILQCRQCDRWMHAVCQNLNTEEEVENVADIGFDCSMCRPYMPAS NVPSSDCCESSLVAQIVTKVKELDPPKTYTQDGVCLTESGMTQLQSLTVTVPRRKRSKPKLKLKIINQNSVAVLQTPPDIQSEHSRDGEM DDSREGELMDCDGKSESSPEREAVDDETKGVEGTDGVKKRKRKPYRPGIGGFMVRQRSRTGQGKTKRSVIRKDSSGSISEQLPCRDDGWS EQLPDTLVDESVSVTESTEKIKKRYRKRKNKLEETFPAYLQEAFFGKDLLDTSRQSKISLDNLSEDGAQLLYKTNMNTGFLDPSLDPLLS SSSAPTKSGTHGPADDPLADISEVLNTDDDILGIISDDLAKSVDHSDIGPVTDDPSSLPQPNVNQSSRPLSEEQLDGILSPELDKMVTDG AILGKLYKIPELGGKDVEDLFTAVLSPANTQPTPLPQPPPPTQLLPIHNQDAFSRMPLMNGLIGSSPHLPHNSLPPGSGLGTFSAIAQSS YPDARDKNSAFNPMASDPNNSWTSSAPTVEGENDTMSNAQRSTLKWEKEEALGEMATVAPVLYTNINFPNLKEEFPDWTTRVKQIAKLWR KASSQERAPYVQKARDNRAALRINKVQMSNDSMKRQQQQDSIDPSSRIDSELFKDPLKQRESEHEQEWKFRQQMRQKSKQQAKIEATQKL EQVKNEQQQQQQQQFGSQHLLVQSGSDTPSSGIQSPLTPQPGNGNMSPAQSFHKELFTKQPPSTPTSTSSDDVFVKPQAPPPPPAPSRIP IQDSLSQAQTSQPPSPQVFSPGSSNSRPPSPMDPYAKMVGTPRPPPVGHSFSRRNSAAPVENCTPLSSVSRPLQMNETTANRPSPVRDLC SSSTTNNDPYAKPPDTPRPVMTDQFPKSLGLSRSPVVSEQTAKGPIAAGTSDHFTKPSPRADVFQRQRIPDSYARPLLTPAPLDSGPGPF KTPMQPPPSSQDPYGSVSQASRRLSVDPYERPALTPRPIDNFSHNQSNDPYSQPPLTPHPAVNESFAHPSRAFSQPGTISRPTSQDPYSQ PPGTPRPVVDSYSQSSGTARSNTDPYSQPPGTPRPTTVDPYSQQPQTPRPSTQTDLFVTPVTNQRHSDPYAHPPGTPRPGISVPYSQPPA TPRPRISEGFTRSSMTRPVLMPNQDPFLQAAQNRGPALPGPLVRPPDTCSQTPRPPGPGLSDTFSRVSPSAARDPYDQSPMTPRSQSDSF GTSQTAHDVADQPRPGSEGSFCASSNSPMHSQGQQFSGVSQLPGPVPTSGVTDTQNTVNMAQADTEKLRQRQKLREIILQQQQQKKIAGR QEKGSQDSPAVPHPGPLQHWQPENVNQAFTRPPPPYPGNIRSPVAPPLGPRYAVFPKDQRGPYPPDVASMGMRPHGFRFGFPGGSHGTMP SQERFLVPPQQIQGSGVSPQLRRSVSVDMPRPLNNSQMNNPVGLPQHFSPQSLPVQQHNILGQAYIELRHRAPDGRQRLPFSAPPGSVVE ASSNLRHGNFIPRPDFPGPRHTDPMRRPPQGLPNQLPVHPDLEQVPPSQQEQGHSVHSSSMVMRTLNHPLGGEFSEAPLSTSVPSETTSD NLQITTQPSDGLEEKLDSDDPSVKELDVKDLEGVEVKDLDDEDLENLNLDTEDGKVVELDTLDNLETNDPNLDDLLRSGEFDIIAYTDPE LDMGDKKSMFNEELDLPIDDKLDNQCVSVEPKKKEQENKTLVLSDKHSPQKKSTVTNEVKTEVLSPNSKVESKCETEKNDENKDNVDTPC SQASAHSDLNDGEKTSLHPCDPDLFEKRTNRETAGPSANVIQASTQLPAQDVINSCGITGSTPVLSSLLANEKSDNSDIRPSGSPPPPTL PASPSNHVSSLPPFIAPPGRVLDNAMNSNVTVVSRVNHVFSQGVQVNPGLIPGQSTVNHSLGTGKPATQTGPQTSQSGTSSMSGPQQLMI PQTLAQQNRERPLLLEEQPLLLQDLLDQERQEQQQQRQMQAMIRQRSEPFFPNIDFDAITDPIMKAKMVALKGINKVMAQNNLGMPPMVM SRFPFMGQVVTGTQNSEGQNLGPQAIPQDGSITHQISRPNPPNFGPGFVNDSQRKQYEEWLQETQQLLQMQQKYLEEQIGAHRKSKKALS AKQRTAKKAGREFPEEDAEQLKHVTEQQSMVQKQLEQIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMPSVQPQPPLIPGATPPTMSQPT FPMVPQQLQHQQHTTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSNANPQSGPPPRVEFDDNNPFSE SFQERERKERLREQQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDFMQPLGPLQQSPQHQQQMGQVL QQQNIQQGSINSPSTQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPSFTPALPAAPPVANSSLPCGQD STITHGHSYPGSTQSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTSTPAVSTPSELPQQADQESVEP VGPSTPNMAAGQLCTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQEEPKLEEQNGSKVEGNAVACPV SSAQSPPHSAGAPAAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQTLGAQMQGGFGCGNQLPKTDGG SETKKQRSKRTQRTGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQVRQLSLLPLMEPIIGVNFAHFLPYGSGQFNSGNRLLGTFGSAT LEGVSDYYSQLIYKQNNLSNPPTPPASLPPTPPPMACQKMANGFATTEELAGKAGVLVSHEVTKTLGPKPFQLPFRPQDDLLARALAQGP KTVDVPASLPTPPHNNQEELRIQDHCGDRDTPDSFVPSSSPESVVGVEVSRYPDLSLVKEEPPEPVPSPIIPILPSTAGKSSESRRNDIK TEPGTLYFASPFGPSPNGPRSGLISVAITLHPTAAENISSVVAAFSDLLHVRIPNSYEVSSAPDVPSMGLVSSHRINPGLEYRQHLLLRG PPPGSANPPRLVSSYRLKQPNVPFPPTSNGLSGYKDSSHGIAESAALRPQWCCHCKVVILGSGVRKSFKDLTLLNKDSRESTKRVEKDIV FCSNNCFILYSSTAQAKNSENKESIPSLPQSPMRETPSKAFHQYSNNISTLDVHCLPQLPEKASPPASPPIAFPPAFEAAQVEAKPDELK VTVKLKPRLRAVHGGFEDCRPLNKKWRGMKWKKWSIHIVIPKGTFKPPCEDEIDEFLKKLGTSLKPDPVPKDYRKCCFCHEEGDGLTDGP ARLLNLDLDLWVHLNCALWSTEVYETQAGALINVELALRRGLQMKCVFCHKTGATSGCHRFRCTNIYHFTCAIKAQCMFFKDKTMLCPMH KPKGIHEQELSYFAVFRRVYVQRDEVRQIASIVQRGERDHTFRVGSLIFHTIGQLLPQQMQAFHSPKALFPVGYEASRLYWSTRYANRRC RYLCSIEEKDGRPVFVIRIVEQGHEDLVLSDISPKGVWDKILEPVACVRKKSEMLQLFPAYLKGEDLFGLTVSAVARIAESLPGVEACEN YTFRYGRNPLMELPLAVNPTGCARSEPKMSAHVKRFVLRPHTLNSTSTSKSFQSTVTGELNAPYSKQFVHSKSSQYRKMKTEWKSNVYLA RSRIQGLGLYAARDIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDHVIDATLTGGPARYINHSCAPNCVAEVVTFERGH -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:179069823/chr7:151921264) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MFN1 | KMT2C |
| FUNCTION: Mitochondrial outer membrane GTPase that mediates mitochondrial clustering and fusion (PubMed:12475957, PubMed:12759376, PubMed:27920125, PubMed:28114303). Membrane clustering requires GTPase activity (PubMed:27920125). It may involve a major rearrangement of the coiled coil domains (PubMed:27920125, PubMed:28114303). Mitochondria are highly dynamic organelles, and their morphology is determined by the equilibrium between mitochondrial fusion and fission events (PubMed:12475957, PubMed:12759376). Overexpression induces the formation of mitochondrial networks (in vitro) (PubMed:12759376). Has low GTPase activity (PubMed:27920125, PubMed:28114303). {ECO:0000269|PubMed:12475957, ECO:0000269|PubMed:12759376, ECO:0000269|PubMed:27920125, ECO:0000269|PubMed:28114303}. | FUNCTION: Histone methyltransferase that methylates 'Lys-4' of histone H3 (PubMed:22266653). H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation. Central component of the MLL2/3 complex, a coactivator complex of nuclear receptors, involved in transcriptional coactivation. KMT2C/MLL3 may be a catalytic subunit of this complex. May be involved in leukemogenesis and developmental disorder. {ECO:0000269|PubMed:17500065, ECO:0000269|PubMed:22266653}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 9_73 | 82.66666666666667 | 742.0 | Region | Part of a helix bundle domain%2C formed by helices from N-terminal and C-terminal regions |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 9_73 | 82.66666666666667 | 631.0 | Region | Part of a helix bundle domain%2C formed by helices from N-terminal and C-terminal regions |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 9_73 | 82.66666666666667 | 742.0 | Region | Part of a helix bundle domain%2C formed by helices from N-terminal and C-terminal regions |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 1338_1366 | 1052.6666666666667 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 1754_1787 | 1052.6666666666667 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 3054_3081 | 1052.6666666666667 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 3173_3272 | 1052.6666666666667 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 3391_3433 | 1052.6666666666667 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 1338_1366 | 1052.6666666666667 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 1754_1787 | 1052.6666666666667 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 3054_3081 | 1052.6666666666667 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 3173_3272 | 1052.6666666666667 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 3391_3433 | 1052.6666666666667 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 1719_1796 | 1052.6666666666667 | 4912.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 1834_2281 | 1052.6666666666667 | 4912.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 2412_2630 | 1052.6666666666667 | 4912.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 2690_2786 | 1052.6666666666667 | 4912.0 | Compositional bias | Note=Asp-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 3012_3509 | 1052.6666666666667 | 4912.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 3277_3381 | 1052.6666666666667 | 4912.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 1719_1796 | 1052.6666666666667 | 4969.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 1834_2281 | 1052.6666666666667 | 4969.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 2412_2630 | 1052.6666666666667 | 4969.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 2690_2786 | 1052.6666666666667 | 4969.0 | Compositional bias | Note=Asp-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 3012_3509 | 1052.6666666666667 | 4969.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 3277_3381 | 1052.6666666666667 | 4969.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 4545_4605 | 1052.6666666666667 | 4912.0 | Domain | FYR N-terminal | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 4606_4691 | 1052.6666666666667 | 4912.0 | Domain | FYR C-terminal | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 4771_4887 | 1052.6666666666667 | 4912.0 | Domain | SET | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 4895_4911 | 1052.6666666666667 | 4912.0 | Domain | Post-SET | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 4545_4605 | 1052.6666666666667 | 4969.0 | Domain | FYR N-terminal | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 4606_4691 | 1052.6666666666667 | 4969.0 | Domain | FYR C-terminal | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 4771_4887 | 1052.6666666666667 | 4969.0 | Domain | SET | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 4895_4911 | 1052.6666666666667 | 4969.0 | Domain | Post-SET | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 4848_4849 | 1052.6666666666667 | 4912.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 4848_4849 | 1052.6666666666667 | 4969.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 1084_1139 | 1052.6666666666667 | 4912.0 | Zinc finger | PHD-type 7 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 4399_4439 | 1052.6666666666667 | 4912.0 | Zinc finger | C2HC pre-PHD-type 2 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 4460_4507 | 1052.6666666666667 | 4912.0 | Zinc finger | PHD-type 8 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 1084_1139 | 1052.6666666666667 | 4969.0 | Zinc finger | PHD-type 7 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 4399_4439 | 1052.6666666666667 | 4969.0 | Zinc finger | C2HC pre-PHD-type 2 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 4460_4507 | 1052.6666666666667 | 4969.0 | Zinc finger | PHD-type 8 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 371_408 | 82.66666666666667 | 742.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 679_734 | 82.66666666666667 | 742.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 371_408 | 82.66666666666667 | 631.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 679_734 | 82.66666666666667 | 631.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 371_408 | 82.66666666666667 | 742.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 679_734 | 82.66666666666667 | 742.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 72_321 | 82.66666666666667 | 742.0 | Domain | Dynamin-type G |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 72_321 | 82.66666666666667 | 631.0 | Domain | Dynamin-type G |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 72_321 | 82.66666666666667 | 742.0 | Domain | Dynamin-type G |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 85_90 | 82.66666666666667 | 742.0 | Nucleotide binding | GTP |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 85_90 | 82.66666666666667 | 631.0 | Nucleotide binding | GTP |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 85_90 | 82.66666666666667 | 742.0 | Nucleotide binding | GTP |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 108_109 | 82.66666666666667 | 742.0 | Region | G2 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 178_181 | 82.66666666666667 | 742.0 | Region | G3 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 237_240 | 82.66666666666667 | 742.0 | Region | G4 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 266_266 | 82.66666666666667 | 742.0 | Region | G5 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 338_364 | 82.66666666666667 | 742.0 | Region | Part of a helix bundle domain%2C formed by helices from N-terminal and C-terminal regions |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 703_734 | 82.66666666666667 | 742.0 | Region | Part of a helix bundle domain%2C formed by helices from N-terminal and C-terminal regions |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 82_89 | 82.66666666666667 | 742.0 | Region | G1 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 108_109 | 82.66666666666667 | 631.0 | Region | G2 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 178_181 | 82.66666666666667 | 631.0 | Region | G3 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 237_240 | 82.66666666666667 | 631.0 | Region | G4 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 266_266 | 82.66666666666667 | 631.0 | Region | G5 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 338_364 | 82.66666666666667 | 631.0 | Region | Part of a helix bundle domain%2C formed by helices from N-terminal and C-terminal regions |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 703_734 | 82.66666666666667 | 631.0 | Region | Part of a helix bundle domain%2C formed by helices from N-terminal and C-terminal regions |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 82_89 | 82.66666666666667 | 631.0 | Region | G1 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 108_109 | 82.66666666666667 | 742.0 | Region | G2 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 178_181 | 82.66666666666667 | 742.0 | Region | G3 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 237_240 | 82.66666666666667 | 742.0 | Region | G4 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 266_266 | 82.66666666666667 | 742.0 | Region | G5 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 338_364 | 82.66666666666667 | 742.0 | Region | Part of a helix bundle domain%2C formed by helices from N-terminal and C-terminal regions |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 703_734 | 82.66666666666667 | 742.0 | Region | Part of a helix bundle domain%2C formed by helices from N-terminal and C-terminal regions |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 82_89 | 82.66666666666667 | 742.0 | Region | G1 motif |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 1_584 | 82.66666666666667 | 742.0 | Topological domain | Cytoplasmic |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 606_608 | 82.66666666666667 | 742.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 630_741 | 82.66666666666667 | 742.0 | Topological domain | Cytoplasmic |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 1_584 | 82.66666666666667 | 631.0 | Topological domain | Cytoplasmic |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 606_608 | 82.66666666666667 | 631.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 630_741 | 82.66666666666667 | 631.0 | Topological domain | Cytoplasmic |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 1_584 | 82.66666666666667 | 742.0 | Topological domain | Cytoplasmic |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 606_608 | 82.66666666666667 | 742.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 630_741 | 82.66666666666667 | 742.0 | Topological domain | Cytoplasmic |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 585_605 | 82.66666666666667 | 742.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000263969 | + | 2 | 17 | 609_629 | 82.66666666666667 | 742.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 585_605 | 82.66666666666667 | 631.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000280653 | + | 3 | 16 | 609_629 | 82.66666666666667 | 631.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 585_605 | 82.66666666666667 | 742.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | MFN1 | chr3:179069823 | chr7:151921264 | ENST00000471841 | + | 3 | 18 | 609_629 | 82.66666666666667 | 742.0 | Transmembrane | Helical%3B Name%3D2 |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 644_672 | 1052.6666666666667 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 92_112 | 1052.6666666666667 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 644_672 | 1052.6666666666667 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 92_112 | 1052.6666666666667 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 34_46 | 1052.6666666666667 | 4912.0 | DNA binding | Note=A.T hook | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 34_46 | 1052.6666666666667 | 4969.0 | DNA binding | Note=A.T hook | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 436_489 | 1052.6666666666667 | 4912.0 | Domain | DHHC | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 436_489 | 1052.6666666666667 | 4969.0 | Domain | DHHC | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 1007_1057 | 1052.6666666666667 | 4912.0 | Zinc finger | PHD-type 6 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 227_262 | 1052.6666666666667 | 4912.0 | Zinc finger | C2HC pre-PHD-type 1%3B degenerate | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 283_331 | 1052.6666666666667 | 4912.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 341_391 | 1052.6666666666667 | 4912.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 344_389 | 1052.6666666666667 | 4912.0 | Zinc finger | RING-type | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 388_438 | 1052.6666666666667 | 4912.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 464_520 | 1052.6666666666667 | 4912.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000262189 | 18 | 59 | 957_1010 | 1052.6666666666667 | 4912.0 | Zinc finger | PHD-type 5 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 1007_1057 | 1052.6666666666667 | 4969.0 | Zinc finger | PHD-type 6 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 227_262 | 1052.6666666666667 | 4969.0 | Zinc finger | C2HC pre-PHD-type 1%3B degenerate | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 283_331 | 1052.6666666666667 | 4969.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 341_391 | 1052.6666666666667 | 4969.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 344_389 | 1052.6666666666667 | 4969.0 | Zinc finger | RING-type | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 388_438 | 1052.6666666666667 | 4969.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 464_520 | 1052.6666666666667 | 4969.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2C | chr3:179069823 | chr7:151921264 | ENST00000355193 | 18 | 60 | 957_1010 | 1052.6666666666667 | 4969.0 | Zinc finger | PHD-type 5 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| MFN1 | |
| KMT2C |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to MFN1-KMT2C |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to MFN1-KMT2C |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

