| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:MYO10-MAST1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: MYO10-MAST1 | FusionPDB ID: 56552 | FusionGDB2.0 ID: 56552 | Hgene | Tgene | Gene symbol | MYO10 | MAST1 | Gene ID | 4651 | 23332 |
| Gene name | myosin X | cytoplasmic linker associated protein 1 | |
| Synonyms | - | MAST1 | |
| Cytomap | 5p15.1 | 2q14.2-q14.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | unconventional myosin-Xunconventional myosin-10unconventionnal myosin-X | CLIP-associating protein 1multiple asters 1multiple asters homolog 1protein Orbit homolog 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9HD67 | Q9Y2H9 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000274203, ENST00000515803, ENST00000427430, ENST00000505695, ENST00000513610, ENST00000507288, ENST00000512061, | ENST00000251472, ENST00000591495, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 20 X 14 X 10=2800 | 7 X 10 X 6=420 |
| # samples | 25 | 11 | |
| ** MAII score | log2(25/2800*10)=-3.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/420*10)=-1.93288580414146 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: MYO10 [Title/Abstract] AND MAST1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MYO10(16685846)-MAST1(12969175), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MYO10-MAST1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MYO10-MAST1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MYO10-MAST1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MYO10-MAST1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MYO10-MAST1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. MYO10-MAST1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. MYO10-MAST1 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. MYO10-MAST1 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | MAST1 | GO:0051294 | establishment of spindle orientation | 21822276 |
| Tgene | MAST1 | GO:0051301 | cell division | 21822276 |
 Fusion gene breakpoints across MYO10 (5'-gene) Fusion gene breakpoints across MYO10 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
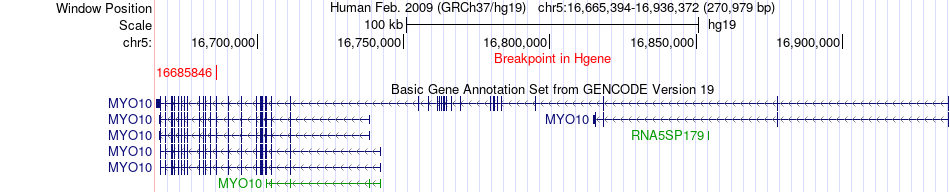 |
 Fusion gene breakpoints across MAST1 (3'-gene) Fusion gene breakpoints across MAST1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
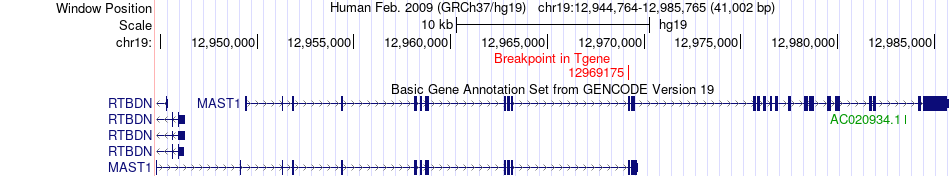 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-23-1023 | MYO10 | chr5 | 16685846 | - | MAST1 | chr19 | 12969175 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000513610 | MYO10 | chr5 | 16685846 | - | ENST00000251472 | MAST1 | chr19 | 12969175 | + | 8162 | 4445 | 401 | 8080 | 2559 |
| ENST00000513610 | MYO10 | chr5 | 16685846 | - | ENST00000591495 | MAST1 | chr19 | 12969175 | + | 4859 | 4445 | 401 | 4807 | 1468 |
| ENST00000427430 | MYO10 | chr5 | 16685846 | - | ENST00000251472 | MAST1 | chr19 | 12969175 | + | 6203 | 2486 | 425 | 6121 | 1898 |
| ENST00000427430 | MYO10 | chr5 | 16685846 | - | ENST00000591495 | MAST1 | chr19 | 12969175 | + | 2900 | 2486 | 425 | 2848 | 807 |
| ENST00000505695 | MYO10 | chr5 | 16685846 | - | ENST00000251472 | MAST1 | chr19 | 12969175 | + | 6203 | 2486 | 425 | 6121 | 1898 |
| ENST00000505695 | MYO10 | chr5 | 16685846 | - | ENST00000591495 | MAST1 | chr19 | 12969175 | + | 2900 | 2486 | 425 | 2848 | 807 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000513610 | ENST00000251472 | MYO10 | chr5 | 16685846 | - | MAST1 | chr19 | 12969175 | + | 0.004438419 | 0.9955616 |
| ENST00000513610 | ENST00000591495 | MYO10 | chr5 | 16685846 | - | MAST1 | chr19 | 12969175 | + | 0.004815842 | 0.9951841 |
| ENST00000427430 | ENST00000251472 | MYO10 | chr5 | 16685846 | - | MAST1 | chr19 | 12969175 | + | 0.004701676 | 0.9952983 |
| ENST00000427430 | ENST00000591495 | MYO10 | chr5 | 16685846 | - | MAST1 | chr19 | 12969175 | + | 0.011205519 | 0.98879445 |
| ENST00000505695 | ENST00000251472 | MYO10 | chr5 | 16685846 | - | MAST1 | chr19 | 12969175 | + | 0.004701676 | 0.9952983 |
| ENST00000505695 | ENST00000591495 | MYO10 | chr5 | 16685846 | - | MAST1 | chr19 | 12969175 | + | 0.011205519 | 0.98879445 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >56552_56552_1_MYO10-MAST1_MYO10_chr5_16685846_ENST00000427430_MAST1_chr19_12969175_ENST00000251472_length(amino acids)=1898AA_BP=687 MPDQFDQAVVLNQLRYSGMLETVRIRKAGYAVRRPFQDFYKRYKVLMRNLALPEDVRGKCTSLLQLYDASNSEWQLGKTKVFLRESLEQK LEKRREEEVSHAAMVIRAHVLGFLARKQYRKVLYCVVIIQKNYRAFLLRRRFLHLKKAAIVFQKQLRGQIARRVYRQLLAEKREQEEKKK QEEEEKKKREEEERERERERREAELRAQQEEETRKQQELEALQKSQKEAELTRELEKQKENKQVEEILRLEKEIEDLQRMKEQQELSLTE ASLQKLQERRDQELRRLEEEACRAAQEFLESLNFDEIDECVRNIERSLSVGSEFSSELAESACEEKPNFNFSQPYPEEEVDEGFEADDDA FKDSPNPSEHGHSDQRTSGIRTSDDSSEEDPYMNDTVVPTSPSADSTVLLAPSVQDSGSLHNSSSGESTYCMPQNAGDLPSPDGDYDYDQ DDYEDGAITSGSSVTFSNSYGSQWSPDYRCSVGTYNSSGAYRFSSEGAQSSFEDSEEDFDSRFDTDDELSYRRDSVYSCVTLPYFHSFLY MKGGLMNSWKRRWCVLKDETFLWFRSKQEALKQGWLHKKGGGSSTLSRRNWKKRWFVLRQSKLMYFENDSEEKLKGTVEVRTAKEIIDNT TKENGIDIIMADRTFHLIAESPEDASQWFSVLSQVHASTDQEIQEMHDEQANPQNAVGRSSKAKKPPGENDFDTIKLISNGAYGAVYLVR HRDTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVGMFCSFETRRHLCMVMEYVEGGDCATLLKNIGALPVEMARMYFAETV LALEYLHNYGIVHRDLKPDNLLITSMGHIKLTDFGLSKMGLMSLTTNLYEGHIEKDAREFLDKQVCGTPEYIAPEVILRQGYGKPVDWWA MGIILYEFLVGCVPFFGDTPEELFGQVISDDILWPEGDEALPTEAQLLISSLLQTNPLVRLGAGGAFEVKQHSFFRDLDWTGLLRQKAEF IPHLESEDDTSYFDTRSDRYHHVNSYDEDDTTEEEPVEIRQFSSCSPRFSKVYSSMEQLSQHEPKTPVAAAGSSKREPSTKGPEEKVAGK REGLGGLTLREKTWRGGSPEIKRFSASEASFLEGEASPPLGARRRFSALLEPSRFSAPQEDEDEARLRRPPRPSSDPAGSLDARAPKEET QGEGTSSAGDSEATDRPRPGDLCPPSKDGDASGPRATNDLVLRRARHQQMSGDVAVEKRPSRTGGKVIKSASATALSVMIPAVDPHGSSP LASPMSPRSLSSNPSSRDSSPSRDYSPAVSGLRSPITIQRSGKKYGFTLRAIRVYMGDTDVYSVHHIVWHVEEGGPAQEAGLCAGDLITH VNGEPVHGMVHPEVVELILKSGNKVAVTTTPFENTSIRIGPARRSSYKAKMARRNKRPSAKEGQESKKRSSLFRKITKQSNLLHTSRSLS SLNRSLSSSDSLPGSPTHGLPARSPTHSYRSTPDSAYLGASSQSSSPASSTPNSPASSASHHIRPSTLHGLSPKLHRQYRSARCKSAGNI PLSPLAHTPSPTQASPPPLPGHTVGSSHTTQSFPAKLHSSPPVVRPRPKSAEPPRSPLLKRVQSAEKLGASLSADKKGALRKHSLEVGHP DFRKDFHGELALHSLAESDGETPPVEGLGAPRQVAVRRLGRQESPLSLGADPLLPEGASRPPVSSKEKESPGGAEACTPPRATTPGGRTL ERDVGCTRHQSVQTEDGTGGMARAVAKAALSPVQEHETGRRSSSGEAGTPLVPIVVEPARPGAKAVVPQPLGADSKGLQEPAPLAPSVPE APRGRERWVLEVVEERTTLSGPRSKPASPKLSPEPQTPSLAPAKCSAPSSAVTPVPPASLLGSGTKPQVGLTSRCPAEAVPPAGLTKKGV -------------------------------------------------------------- >56552_56552_2_MYO10-MAST1_MYO10_chr5_16685846_ENST00000427430_MAST1_chr19_12969175_ENST00000591495_length(amino acids)=807AA_BP=687 MPDQFDQAVVLNQLRYSGMLETVRIRKAGYAVRRPFQDFYKRYKVLMRNLALPEDVRGKCTSLLQLYDASNSEWQLGKTKVFLRESLEQK LEKRREEEVSHAAMVIRAHVLGFLARKQYRKVLYCVVIIQKNYRAFLLRRRFLHLKKAAIVFQKQLRGQIARRVYRQLLAEKREQEEKKK QEEEEKKKREEEERERERERREAELRAQQEEETRKQQELEALQKSQKEAELTRELEKQKENKQVEEILRLEKEIEDLQRMKEQQELSLTE ASLQKLQERRDQELRRLEEEACRAAQEFLESLNFDEIDECVRNIERSLSVGSEFSSELAESACEEKPNFNFSQPYPEEEVDEGFEADDDA FKDSPNPSEHGHSDQRTSGIRTSDDSSEEDPYMNDTVVPTSPSADSTVLLAPSVQDSGSLHNSSSGESTYCMPQNAGDLPSPDGDYDYDQ DDYEDGAITSGSSVTFSNSYGSQWSPDYRCSVGTYNSSGAYRFSSEGAQSSFEDSEEDFDSRFDTDDELSYRRDSVYSCVTLPYFHSFLY MKGGLMNSWKRRWCVLKDETFLWFRSKQEALKQGWLHKKGGGSSTLSRRNWKKRWFVLRQSKLMYFENDSEEKLKGTVEVRTAKEIIDNT TKENGIDIIMADRTFHLIAESPEDASQWFSVLSQVHASTDQEIQEMHDEQANPQNAVGRSSKAKKPPGENDFDTIKLISNGAYGAVYLVR -------------------------------------------------------------- >56552_56552_3_MYO10-MAST1_MYO10_chr5_16685846_ENST00000505695_MAST1_chr19_12969175_ENST00000251472_length(amino acids)=1898AA_BP=687 MPDQFDQAVVLNQLRYSGMLETVRIRKAGYAVRRPFQDFYKRYKVLMRNLALPEDVRGKCTSLLQLYDASNSEWQLGKTKVFLRESLEQK LEKRREEEVSHAAMVIRAHVLGFLARKQYRKVLYCVVIIQKNYRAFLLRRRFLHLKKAAIVFQKQLRGQIARRVYRQLLAEKREQEEKKK QEEEEKKKREEEERERERERREAELRAQQEEETRKQQELEALQKSQKEAELTRELEKQKENKQVEEILRLEKEIEDLQRMKEQQELSLTE ASLQKLQERRDQELRRLEEEACRAAQEFLESLNFDEIDECVRNIERSLSVGSEFSSELAESACEEKPNFNFSQPYPEEEVDEGFEADDDA FKDSPNPSEHGHSDQRTSGIRTSDDSSEEDPYMNDTVVPTSPSADSTVLLAPSVQDSGSLHNSSSGESTYCMPQNAGDLPSPDGDYDYDQ DDYEDGAITSGSSVTFSNSYGSQWSPDYRCSVGTYNSSGAYRFSSEGAQSSFEDSEEDFDSRFDTDDELSYRRDSVYSCVTLPYFHSFLY MKGGLMNSWKRRWCVLKDETFLWFRSKQEALKQGWLHKKGGGSSTLSRRNWKKRWFVLRQSKLMYFENDSEEKLKGTVEVRTAKEIIDNT TKENGIDIIMADRTFHLIAESPEDASQWFSVLSQVHASTDQEIQEMHDEQANPQNAVGRSSKAKKPPGENDFDTIKLISNGAYGAVYLVR HRDTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVGMFCSFETRRHLCMVMEYVEGGDCATLLKNIGALPVEMARMYFAETV LALEYLHNYGIVHRDLKPDNLLITSMGHIKLTDFGLSKMGLMSLTTNLYEGHIEKDAREFLDKQVCGTPEYIAPEVILRQGYGKPVDWWA MGIILYEFLVGCVPFFGDTPEELFGQVISDDILWPEGDEALPTEAQLLISSLLQTNPLVRLGAGGAFEVKQHSFFRDLDWTGLLRQKAEF IPHLESEDDTSYFDTRSDRYHHVNSYDEDDTTEEEPVEIRQFSSCSPRFSKVYSSMEQLSQHEPKTPVAAAGSSKREPSTKGPEEKVAGK REGLGGLTLREKTWRGGSPEIKRFSASEASFLEGEASPPLGARRRFSALLEPSRFSAPQEDEDEARLRRPPRPSSDPAGSLDARAPKEET QGEGTSSAGDSEATDRPRPGDLCPPSKDGDASGPRATNDLVLRRARHQQMSGDVAVEKRPSRTGGKVIKSASATALSVMIPAVDPHGSSP LASPMSPRSLSSNPSSRDSSPSRDYSPAVSGLRSPITIQRSGKKYGFTLRAIRVYMGDTDVYSVHHIVWHVEEGGPAQEAGLCAGDLITH VNGEPVHGMVHPEVVELILKSGNKVAVTTTPFENTSIRIGPARRSSYKAKMARRNKRPSAKEGQESKKRSSLFRKITKQSNLLHTSRSLS SLNRSLSSSDSLPGSPTHGLPARSPTHSYRSTPDSAYLGASSQSSSPASSTPNSPASSASHHIRPSTLHGLSPKLHRQYRSARCKSAGNI PLSPLAHTPSPTQASPPPLPGHTVGSSHTTQSFPAKLHSSPPVVRPRPKSAEPPRSPLLKRVQSAEKLGASLSADKKGALRKHSLEVGHP DFRKDFHGELALHSLAESDGETPPVEGLGAPRQVAVRRLGRQESPLSLGADPLLPEGASRPPVSSKEKESPGGAEACTPPRATTPGGRTL ERDVGCTRHQSVQTEDGTGGMARAVAKAALSPVQEHETGRRSSSGEAGTPLVPIVVEPARPGAKAVVPQPLGADSKGLQEPAPLAPSVPE APRGRERWVLEVVEERTTLSGPRSKPASPKLSPEPQTPSLAPAKCSAPSSAVTPVPPASLLGSGTKPQVGLTSRCPAEAVPPAGLTKKGV -------------------------------------------------------------- >56552_56552_4_MYO10-MAST1_MYO10_chr5_16685846_ENST00000505695_MAST1_chr19_12969175_ENST00000591495_length(amino acids)=807AA_BP=687 MPDQFDQAVVLNQLRYSGMLETVRIRKAGYAVRRPFQDFYKRYKVLMRNLALPEDVRGKCTSLLQLYDASNSEWQLGKTKVFLRESLEQK LEKRREEEVSHAAMVIRAHVLGFLARKQYRKVLYCVVIIQKNYRAFLLRRRFLHLKKAAIVFQKQLRGQIARRVYRQLLAEKREQEEKKK QEEEEKKKREEEERERERERREAELRAQQEEETRKQQELEALQKSQKEAELTRELEKQKENKQVEEILRLEKEIEDLQRMKEQQELSLTE ASLQKLQERRDQELRRLEEEACRAAQEFLESLNFDEIDECVRNIERSLSVGSEFSSELAESACEEKPNFNFSQPYPEEEVDEGFEADDDA FKDSPNPSEHGHSDQRTSGIRTSDDSSEEDPYMNDTVVPTSPSADSTVLLAPSVQDSGSLHNSSSGESTYCMPQNAGDLPSPDGDYDYDQ DDYEDGAITSGSSVTFSNSYGSQWSPDYRCSVGTYNSSGAYRFSSEGAQSSFEDSEEDFDSRFDTDDELSYRRDSVYSCVTLPYFHSFLY MKGGLMNSWKRRWCVLKDETFLWFRSKQEALKQGWLHKKGGGSSTLSRRNWKKRWFVLRQSKLMYFENDSEEKLKGTVEVRTAKEIIDNT TKENGIDIIMADRTFHLIAESPEDASQWFSVLSQVHASTDQEIQEMHDEQANPQNAVGRSSKAKKPPGENDFDTIKLISNGAYGAVYLVR -------------------------------------------------------------- >56552_56552_5_MYO10-MAST1_MYO10_chr5_16685846_ENST00000513610_MAST1_chr19_12969175_ENST00000251472_length(amino acids)=2559AA_BP=1348 MTSASRSGTRRVRDCAGTMDNFFTEGTRVWLRENGQHFPSTVNSCAEGIVVFRTDYGQVFTYKQSTITHQKVTAMHPTNEEGVDDMASLT ELHGGSIMYNLFQRYKRNQIYTYIGSILASVNPYQPIAGLYEPATMEQYSRRHLGELPPHIFAIANECYRCLWKRHDNQCILISGESGAG KTESTKLILKFLSVISQQSLELSLKEKTSCVERAILESSPIMEAFGNAKTVYNNNSSRFGKFVQLNICQKGNIQGGRIVDYLLEKNRVVR QNPGERNYHIFYALLAGLEHEEREEFYLSTPENYHYLNQSGCVEDKTISDQESFREVITAMDVMQFSKEEVREVSRLLAGILHLGNIEFI TAGGAQVSFKTALGRSAELLGLDPTQLTDALTQRSMFLRGEEILTPLNVQQAVDSRDSLAMALYACCFEWVIKKINSRIKGNEDFKSIGI LDIFGFENFEVNHFEQFNINYANEKLQEYFNKHIFSLEQLEYSREGLVWEDIDWIDNGECLDLIEKKLGLLALINEESHFPQATDSTLLE KLHSQHANNHFYVKPRVAVNNFGVKHYAGEVQYDVRGILEKNRDTFRDDLLNLLRESRFDFIYDLFEHVSSRNNQDTLKCGSKHRRPTVS SQFKDSLHSLMATLSSSNPFFVRCIKPNMQKMPDQFDQAVVLNQLRYSGMLETVRIRKAGYAVRRPFQDFYKRYKVLMRNLALPEDVRGK CTSLLQLYDASNSEWQLGKTKVFLRESLEQKLEKRREEEVSHAAMVIRAHVLGFLARKQYRKVLYCVVIIQKNYRAFLLRRRFLHLKKAA IVFQKQLRGQIARRVYRQLLAEKREQEEKKKQEEEEKKKREEEERERERERREAELRAQQEEETRKQQELEALQKSQKEAELTRELEKQK ENKQVEEILRLEKEIEDLQRMKEQQELSLTEASLQKLQERRDQELRRLEEEACRAAQEFLESLNFDEIDECVRNIERSLSVGSEFSSELA ESACEEKPNFNFSQPYPEEEVDEGFEADDDAFKDSPNPSEHGHSDQRTSGIRTSDDSSEEDPYMNDTVVPTSPSADSTVLLAPSVQDSGS LHNSSSGESTYCMPQNAGDLPSPDGDYDYDQDDYEDGAITSGSSVTFSNSYGSQWSPDYRCSVGTYNSSGAYRFSSEGAQSSFEDSEEDF DSRFDTDDELSYRRDSVYSCVTLPYFHSFLYMKGGLMNSWKRRWCVLKDETFLWFRSKQEALKQGWLHKKGGGSSTLSRRNWKKRWFVLR QSKLMYFENDSEEKLKGTVEVRTAKEIIDNTTKENGIDIIMADRTFHLIAESPEDASQWFSVLSQVHASTDQEIQEMHDEQANPQNAVGR SSKAKKPPGENDFDTIKLISNGAYGAVYLVRHRDTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVGMFCSFETRRHLCMVM EYVEGGDCATLLKNIGALPVEMARMYFAETVLALEYLHNYGIVHRDLKPDNLLITSMGHIKLTDFGLSKMGLMSLTTNLYEGHIEKDARE FLDKQVCGTPEYIAPEVILRQGYGKPVDWWAMGIILYEFLVGCVPFFGDTPEELFGQVISDDILWPEGDEALPTEAQLLISSLLQTNPLV RLGAGGAFEVKQHSFFRDLDWTGLLRQKAEFIPHLESEDDTSYFDTRSDRYHHVNSYDEDDTTEEEPVEIRQFSSCSPRFSKVYSSMEQL SQHEPKTPVAAAGSSKREPSTKGPEEKVAGKREGLGGLTLREKTWRGGSPEIKRFSASEASFLEGEASPPLGARRRFSALLEPSRFSAPQ EDEDEARLRRPPRPSSDPAGSLDARAPKEETQGEGTSSAGDSEATDRPRPGDLCPPSKDGDASGPRATNDLVLRRARHQQMSGDVAVEKR PSRTGGKVIKSASATALSVMIPAVDPHGSSPLASPMSPRSLSSNPSSRDSSPSRDYSPAVSGLRSPITIQRSGKKYGFTLRAIRVYMGDT DVYSVHHIVWHVEEGGPAQEAGLCAGDLITHVNGEPVHGMVHPEVVELILKSGNKVAVTTTPFENTSIRIGPARRSSYKAKMARRNKRPS AKEGQESKKRSSLFRKITKQSNLLHTSRSLSSLNRSLSSSDSLPGSPTHGLPARSPTHSYRSTPDSAYLGASSQSSSPASSTPNSPASSA SHHIRPSTLHGLSPKLHRQYRSARCKSAGNIPLSPLAHTPSPTQASPPPLPGHTVGSSHTTQSFPAKLHSSPPVVRPRPKSAEPPRSPLL KRVQSAEKLGASLSADKKGALRKHSLEVGHPDFRKDFHGELALHSLAESDGETPPVEGLGAPRQVAVRRLGRQESPLSLGADPLLPEGAS RPPVSSKEKESPGGAEACTPPRATTPGGRTLERDVGCTRHQSVQTEDGTGGMARAVAKAALSPVQEHETGRRSSSGEAGTPLVPIVVEPA RPGAKAVVPQPLGADSKGLQEPAPLAPSVPEAPRGRERWVLEVVEERTTLSGPRSKPASPKLSPEPQTPSLAPAKCSAPSSAVTPVPPAS -------------------------------------------------------------- >56552_56552_6_MYO10-MAST1_MYO10_chr5_16685846_ENST00000513610_MAST1_chr19_12969175_ENST00000591495_length(amino acids)=1468AA_BP=1348 MTSASRSGTRRVRDCAGTMDNFFTEGTRVWLRENGQHFPSTVNSCAEGIVVFRTDYGQVFTYKQSTITHQKVTAMHPTNEEGVDDMASLT ELHGGSIMYNLFQRYKRNQIYTYIGSILASVNPYQPIAGLYEPATMEQYSRRHLGELPPHIFAIANECYRCLWKRHDNQCILISGESGAG KTESTKLILKFLSVISQQSLELSLKEKTSCVERAILESSPIMEAFGNAKTVYNNNSSRFGKFVQLNICQKGNIQGGRIVDYLLEKNRVVR QNPGERNYHIFYALLAGLEHEEREEFYLSTPENYHYLNQSGCVEDKTISDQESFREVITAMDVMQFSKEEVREVSRLLAGILHLGNIEFI TAGGAQVSFKTALGRSAELLGLDPTQLTDALTQRSMFLRGEEILTPLNVQQAVDSRDSLAMALYACCFEWVIKKINSRIKGNEDFKSIGI LDIFGFENFEVNHFEQFNINYANEKLQEYFNKHIFSLEQLEYSREGLVWEDIDWIDNGECLDLIEKKLGLLALINEESHFPQATDSTLLE KLHSQHANNHFYVKPRVAVNNFGVKHYAGEVQYDVRGILEKNRDTFRDDLLNLLRESRFDFIYDLFEHVSSRNNQDTLKCGSKHRRPTVS SQFKDSLHSLMATLSSSNPFFVRCIKPNMQKMPDQFDQAVVLNQLRYSGMLETVRIRKAGYAVRRPFQDFYKRYKVLMRNLALPEDVRGK CTSLLQLYDASNSEWQLGKTKVFLRESLEQKLEKRREEEVSHAAMVIRAHVLGFLARKQYRKVLYCVVIIQKNYRAFLLRRRFLHLKKAA IVFQKQLRGQIARRVYRQLLAEKREQEEKKKQEEEEKKKREEEERERERERREAELRAQQEEETRKQQELEALQKSQKEAELTRELEKQK ENKQVEEILRLEKEIEDLQRMKEQQELSLTEASLQKLQERRDQELRRLEEEACRAAQEFLESLNFDEIDECVRNIERSLSVGSEFSSELA ESACEEKPNFNFSQPYPEEEVDEGFEADDDAFKDSPNPSEHGHSDQRTSGIRTSDDSSEEDPYMNDTVVPTSPSADSTVLLAPSVQDSGS LHNSSSGESTYCMPQNAGDLPSPDGDYDYDQDDYEDGAITSGSSVTFSNSYGSQWSPDYRCSVGTYNSSGAYRFSSEGAQSSFEDSEEDF DSRFDTDDELSYRRDSVYSCVTLPYFHSFLYMKGGLMNSWKRRWCVLKDETFLWFRSKQEALKQGWLHKKGGGSSTLSRRNWKKRWFVLR QSKLMYFENDSEEKLKGTVEVRTAKEIIDNTTKENGIDIIMADRTFHLIAESPEDASQWFSVLSQVHASTDQEIQEMHDEQANPQNAVGR SSKAKKPPGENDFDTIKLISNGAYGAVYLVRHRDTRQRFAMKKINKQNLILRNQIQQAFVERDILTFAENPFVVGMFCSFETRRHLCMVM -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:16685846/chr19:12969175) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MYO10 | MAST1 |
| FUNCTION: Myosins are actin-based motor molecules with ATPase activity. Unconventional myosins serve in intracellular movements. MYO10 binds to actin filaments and actin bundles and functions as plus end-directed motor. The tail domain binds to membranous compartments containing phosphatidylinositol 3,4,5-trisphosphate or integrins, and mediates cargo transport along actin filaments. Regulates cell shape, cell spreading and cell adhesion. Stimulates the formation and elongation of filopodia. May play a role in neurite outgrowth and axon guidance. In hippocampal neurons it induces the formation of dendritic filopodia by trafficking the actin-remodeling protein VASP to the tips of filopodia, where it promotes actin elongation. Plays a role in formation of the podosome belt in osteoclasts. {ECO:0000269|PubMed:16894163, ECO:0000269|PubMed:18570893}.; FUNCTION: [Isoform Headless]: Functions as a dominant-negative regulator of isoform 1, suppressing its filopodia-inducing and axon outgrowth-promoting activities. In hippocampal neurons, it increases VASP retention in spine heads to induce spine formation and spine head expansion (By similarity). {ECO:0000250}. | FUNCTION: Microtubule-associated protein essential for correct brain development (PubMed:30449657). Appears to link the dystrophin/utrophin network with microtubule filaments via the syntrophins. Phosphorylation of DMD or UTRN may modulate their affinities for associated proteins (By similarity). {ECO:0000250|UniProtKB:Q9R1L5, ECO:0000269|PubMed:30449657}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 1212_1310 | 1330.0 | 2059.0 | Domain | PH 1 |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 63_739 | 1330.0 | 2059.0 | Domain | Myosin motor |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 742_763 | 1330.0 | 2059.0 | Domain | IQ 1 |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 764_787 | 1330.0 | 2059.0 | Domain | IQ 2 |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 788_817 | 1330.0 | 2059.0 | Domain | IQ 3 |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 157_164 | 1330.0 | 2059.0 | Nucleotide binding | ATP |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 619_641 | 1330.0 | 2059.0 | Region | Actin-binding |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 815_908 | 1330.0 | 2059.0 | Region | SAH |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 883_933 | 1330.0 | 2059.0 | Region | Mediates antiparallel dimerization |
| Tgene | MAST1 | chr5:16685846 | chr19:12969175 | ENST00000251472 | 9 | 26 | 374_647 | 359.0 | 1571.0 | Domain | Protein kinase | |
| Tgene | MAST1 | chr5:16685846 | chr19:12969175 | ENST00000251472 | 9 | 26 | 648_719 | 359.0 | 1571.0 | Domain | AGC-kinase C-terminal | |
| Tgene | MAST1 | chr5:16685846 | chr19:12969175 | ENST00000251472 | 9 | 26 | 967_1055 | 359.0 | 1571.0 | Domain | PDZ | |
| Tgene | MAST1 | chr5:16685846 | chr19:12969175 | ENST00000251472 | 9 | 26 | 380_388 | 359.0 | 1571.0 | Nucleotide binding | ATP |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 1392_1497 | 1330.0 | 2059.0 | Domain | PH 2 |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 1547_1695 | 1330.0 | 2059.0 | Domain | MyTH4 |
| Hgene | MYO10 | chr5:16685846 | chr19:12969175 | ENST00000513610 | - | 29 | 41 | 1700_2044 | 1330.0 | 2059.0 | Domain | FERM |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| MYO10 | |
| MAST1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to MYO10-MAST1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to MYO10-MAST1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

