| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:MYO5B-HTATIP2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: MYO5B-HTATIP2 | FusionPDB ID: 56727 | FusionGDB2.0 ID: 56727 | Hgene | Tgene | Gene symbol | MYO5B | HTATIP2 | Gene ID | 4645 | 10553 |
| Gene name | myosin VB | HIV-1 Tat interactive protein 2 | |
| Synonyms | - | CC3|SDR44U1|TIP30 | |
| Cytomap | 18q21.1 | 11p15.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | unconventional myosin-VbMYO5B variant proteinmyosin-Vb | oxidoreductase HTATIP230 kDa HIV-1 TAT-interacting proteinHIV-1 TAT-interactive protein 2HIV-1 Tat interactive protein 2, 30kDaTat-interacting protein (30kD)short chain dehydrogenase/reductase family 44U, member 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9ULV0 | Q9BUP3 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000285039, ENST00000324581, ENST00000592688, ENST00000587895, | ENST00000530266, ENST00000532081, ENST00000532505, ENST00000419348, ENST00000421577, ENST00000443524, ENST00000451739, ENST00000531058, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 18 X 15 X 7=1890 | 4 X 3 X 3=36 |
| # samples | 21 | 4 | |
| ** MAII score | log2(21/1890*10)=-3.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/36*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: MYO5B [Title/Abstract] AND HTATIP2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | MYO5B(47361713)-HTATIP2(20388719), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | MYO5B-HTATIP2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MYO5B-HTATIP2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. MYO5B-HTATIP2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MYO5B-HTATIP2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. MYO5B-HTATIP2 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. MYO5B-HTATIP2 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | HTATIP2 | GO:0043068 | positive regulation of programmed cell death | 10698937 |
| Tgene | HTATIP2 | GO:0045765 | regulation of angiogenesis | 11313954 |
| Tgene | HTATIP2 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 10698937 |
| Tgene | HTATIP2 | GO:0046777 | protein autophosphorylation | 10698937 |
| Tgene | HTATIP2 | GO:0051170 | import into nucleus | 15282309 |
 Fusion gene breakpoints across MYO5B (5'-gene) Fusion gene breakpoints across MYO5B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
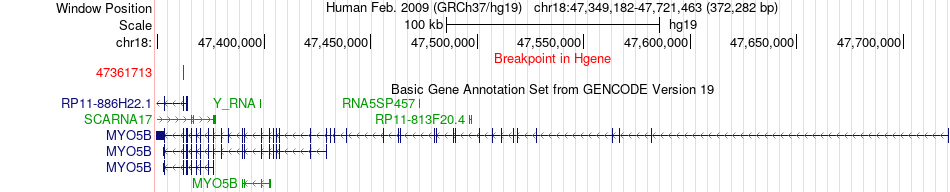 |
 Fusion gene breakpoints across HTATIP2 (3'-gene) Fusion gene breakpoints across HTATIP2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
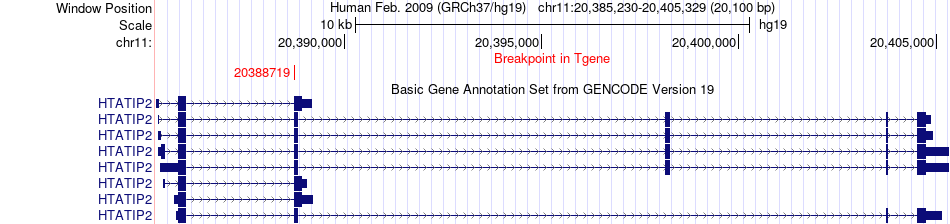 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-CD-A48C | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000285039 | MYO5B | chr18 | 47361713 | - | ENST00000421577 | HTATIP2 | chr11 | 20388719 | + | 6357 | 5694 | 300 | 6227 | 1975 |
| ENST00000285039 | MYO5B | chr18 | 47361713 | - | ENST00000443524 | HTATIP2 | chr11 | 20388719 | + | 6391 | 5694 | 300 | 6227 | 1975 |
| ENST00000285039 | MYO5B | chr18 | 47361713 | - | ENST00000419348 | HTATIP2 | chr11 | 20388719 | + | 6805 | 5694 | 300 | 6227 | 1975 |
| ENST00000285039 | MYO5B | chr18 | 47361713 | - | ENST00000451739 | HTATIP2 | chr11 | 20388719 | + | 6806 | 5694 | 300 | 6227 | 1975 |
| ENST00000285039 | MYO5B | chr18 | 47361713 | - | ENST00000531058 | HTATIP2 | chr11 | 20388719 | + | 6501 | 5694 | 300 | 6089 | 1929 |
| ENST00000324581 | MYO5B | chr18 | 47361713 | - | ENST00000421577 | HTATIP2 | chr11 | 20388719 | + | 3410 | 2747 | 8 | 3280 | 1090 |
| ENST00000324581 | MYO5B | chr18 | 47361713 | - | ENST00000443524 | HTATIP2 | chr11 | 20388719 | + | 3444 | 2747 | 8 | 3280 | 1090 |
| ENST00000324581 | MYO5B | chr18 | 47361713 | - | ENST00000419348 | HTATIP2 | chr11 | 20388719 | + | 3858 | 2747 | 8 | 3280 | 1090 |
| ENST00000324581 | MYO5B | chr18 | 47361713 | - | ENST00000451739 | HTATIP2 | chr11 | 20388719 | + | 3859 | 2747 | 8 | 3280 | 1090 |
| ENST00000324581 | MYO5B | chr18 | 47361713 | - | ENST00000531058 | HTATIP2 | chr11 | 20388719 | + | 3554 | 2747 | 8 | 3142 | 1044 |
| ENST00000592688 | MYO5B | chr18 | 47361713 | - | ENST00000421577 | HTATIP2 | chr11 | 20388719 | + | 2022 | 1359 | 156 | 1892 | 578 |
| ENST00000592688 | MYO5B | chr18 | 47361713 | - | ENST00000443524 | HTATIP2 | chr11 | 20388719 | + | 2056 | 1359 | 156 | 1892 | 578 |
| ENST00000592688 | MYO5B | chr18 | 47361713 | - | ENST00000419348 | HTATIP2 | chr11 | 20388719 | + | 2470 | 1359 | 156 | 1892 | 578 |
| ENST00000592688 | MYO5B | chr18 | 47361713 | - | ENST00000451739 | HTATIP2 | chr11 | 20388719 | + | 2471 | 1359 | 156 | 1892 | 578 |
| ENST00000592688 | MYO5B | chr18 | 47361713 | - | ENST00000531058 | HTATIP2 | chr11 | 20388719 | + | 2166 | 1359 | 156 | 1754 | 532 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000285039 | ENST00000421577 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.006818909 | 0.99318105 |
| ENST00000285039 | ENST00000443524 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.006832735 | 0.99316734 |
| ENST00000285039 | ENST00000419348 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.004484423 | 0.9955155 |
| ENST00000285039 | ENST00000451739 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.004496655 | 0.99550337 |
| ENST00000285039 | ENST00000531058 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.006485765 | 0.9935142 |
| ENST00000324581 | ENST00000421577 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.010694211 | 0.9893058 |
| ENST00000324581 | ENST00000443524 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.010762624 | 0.98923737 |
| ENST00000324581 | ENST00000419348 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.005019364 | 0.99498063 |
| ENST00000324581 | ENST00000451739 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.005044685 | 0.99495536 |
| ENST00000324581 | ENST00000531058 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.006610879 | 0.9933891 |
| ENST00000592688 | ENST00000421577 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.002365268 | 0.99763477 |
| ENST00000592688 | ENST00000443524 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.002390266 | 0.99760973 |
| ENST00000592688 | ENST00000419348 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.001078336 | 0.99892163 |
| ENST00000592688 | ENST00000451739 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.001075622 | 0.9989243 |
| ENST00000592688 | ENST00000531058 | MYO5B | chr18 | 47361713 | - | HTATIP2 | chr11 | 20388719 | + | 0.002084099 | 0.9979159 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >56727_56727_1_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000285039_HTATIP2_chr11_20388719_ENST00000419348_length(amino acids)=1975AA_BP=1798 MSVGELYSQCTRVWIPDPDEVWRSAELTKDYKEGDKSLQLRLEDETILEYPIDVQRNQLPFLRNPDILVGENDLTALSYLHEPAVLHNLK VRFLESNHIYTYCGIVLVAINPYEQLPIYGQDVIYTYSGQNMGDMDPHIFAVAEEAYKQMARDEKNQSIIVSGESGAGKTVSAKYAMRYF ATVGGSASETNIEEKVLASSPIMEAIGNAKTTRNDNSSRFGKYIQIGFDKRYHIIGANMRTYLLEKSRVVFQADDERNYHIFYQLCAAAG LPEFKELALTSAEDFFYTSQGGDTSIEGVDDAEDFEKTRQAFTLLGVKESHQMSIFKIIASILHLGSVAIQAERDGDSCSISPQDVYLSN FCRLLGVEHSQMEHWLCHRKLVTTSETYVKTMSLQQVINARNALAKHIYAQLFGWIVEHINKALHTSLKQHSFIGVLDIYGFETFEVNSF EQFCINYANEKLQQQFNSHVFKLEQEEYMKEQIPWTLIDFYDNQPCIDLIEAKLGILDLLDEECKVPKGTDQNWAQKLYDRHSSSQHFQK PRMSNTAFIIVHFADKVEYLSDGFLEKNRDTVYEEQINILKASKFPLVADLFHDDKDPVPATTPGKGSSSKISVRSARPPMKVSNKEHKK TVGHQFRTSLHLLMETLNATTPHYVRCIKPNDEKLPFHFDPKRAVQQLRACGVLETIRISAAGYPSRWAYHDFFNRYRVLVKKRELANTD KKAICRSVLENLIKDPDKFQFGRTKIFFRAGQVAYLEKLRADKFRTATIMIQKTVRGWLQKVKYHRLKGATLTLQRYCRGHLARRLAEHL RRIRAAVVLQKHYRMQRARQAYQRVRRAAVVIQAFTRAMFVRRTYRQVLMEHKATTIQKHVRGWMARRHFQRLRDAAIVIQCAFRMLKAR RELKALRIEARSAEHLKRLNVGMENKVVQLQRKIDEQNKEFKTLSEQLSVTTSTYTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRT ELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQILCQSKDEFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQR YDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQQVEEIGLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKR EQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNELRKAVADQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEE VLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHGVCQTNSKTEDWGYLNEDGELGLAYQGLKQVARLLEAQLQAQ SLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAA QALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTIN GIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSA MLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGM QLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEFEERVTVAFIRTIQNQ EVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLKSAELAKAGGCKHFNLLSSKGADKSSNFLYLQVKGEVEAKVEEL -------------------------------------------------------------- >56727_56727_2_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000285039_HTATIP2_chr11_20388719_ENST00000421577_length(amino acids)=1975AA_BP=1798 MSVGELYSQCTRVWIPDPDEVWRSAELTKDYKEGDKSLQLRLEDETILEYPIDVQRNQLPFLRNPDILVGENDLTALSYLHEPAVLHNLK VRFLESNHIYTYCGIVLVAINPYEQLPIYGQDVIYTYSGQNMGDMDPHIFAVAEEAYKQMARDEKNQSIIVSGESGAGKTVSAKYAMRYF ATVGGSASETNIEEKVLASSPIMEAIGNAKTTRNDNSSRFGKYIQIGFDKRYHIIGANMRTYLLEKSRVVFQADDERNYHIFYQLCAAAG LPEFKELALTSAEDFFYTSQGGDTSIEGVDDAEDFEKTRQAFTLLGVKESHQMSIFKIIASILHLGSVAIQAERDGDSCSISPQDVYLSN FCRLLGVEHSQMEHWLCHRKLVTTSETYVKTMSLQQVINARNALAKHIYAQLFGWIVEHINKALHTSLKQHSFIGVLDIYGFETFEVNSF EQFCINYANEKLQQQFNSHVFKLEQEEYMKEQIPWTLIDFYDNQPCIDLIEAKLGILDLLDEECKVPKGTDQNWAQKLYDRHSSSQHFQK PRMSNTAFIIVHFADKVEYLSDGFLEKNRDTVYEEQINILKASKFPLVADLFHDDKDPVPATTPGKGSSSKISVRSARPPMKVSNKEHKK TVGHQFRTSLHLLMETLNATTPHYVRCIKPNDEKLPFHFDPKRAVQQLRACGVLETIRISAAGYPSRWAYHDFFNRYRVLVKKRELANTD KKAICRSVLENLIKDPDKFQFGRTKIFFRAGQVAYLEKLRADKFRTATIMIQKTVRGWLQKVKYHRLKGATLTLQRYCRGHLARRLAEHL RRIRAAVVLQKHYRMQRARQAYQRVRRAAVVIQAFTRAMFVRRTYRQVLMEHKATTIQKHVRGWMARRHFQRLRDAAIVIQCAFRMLKAR RELKALRIEARSAEHLKRLNVGMENKVVQLQRKIDEQNKEFKTLSEQLSVTTSTYTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRT ELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQILCQSKDEFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQR YDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQQVEEIGLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKR EQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNELRKAVADQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEE VLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHGVCQTNSKTEDWGYLNEDGELGLAYQGLKQVARLLEAQLQAQ SLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAA QALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTIN GIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSA MLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGM QLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEFEERVTVAFIRTIQNQ EVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLKSAELAKAGGCKHFNLLSSKGADKSSNFLYLQVKGEVEAKVEEL -------------------------------------------------------------- >56727_56727_3_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000285039_HTATIP2_chr11_20388719_ENST00000443524_length(amino acids)=1975AA_BP=1798 MSVGELYSQCTRVWIPDPDEVWRSAELTKDYKEGDKSLQLRLEDETILEYPIDVQRNQLPFLRNPDILVGENDLTALSYLHEPAVLHNLK VRFLESNHIYTYCGIVLVAINPYEQLPIYGQDVIYTYSGQNMGDMDPHIFAVAEEAYKQMARDEKNQSIIVSGESGAGKTVSAKYAMRYF ATVGGSASETNIEEKVLASSPIMEAIGNAKTTRNDNSSRFGKYIQIGFDKRYHIIGANMRTYLLEKSRVVFQADDERNYHIFYQLCAAAG LPEFKELALTSAEDFFYTSQGGDTSIEGVDDAEDFEKTRQAFTLLGVKESHQMSIFKIIASILHLGSVAIQAERDGDSCSISPQDVYLSN FCRLLGVEHSQMEHWLCHRKLVTTSETYVKTMSLQQVINARNALAKHIYAQLFGWIVEHINKALHTSLKQHSFIGVLDIYGFETFEVNSF EQFCINYANEKLQQQFNSHVFKLEQEEYMKEQIPWTLIDFYDNQPCIDLIEAKLGILDLLDEECKVPKGTDQNWAQKLYDRHSSSQHFQK PRMSNTAFIIVHFADKVEYLSDGFLEKNRDTVYEEQINILKASKFPLVADLFHDDKDPVPATTPGKGSSSKISVRSARPPMKVSNKEHKK TVGHQFRTSLHLLMETLNATTPHYVRCIKPNDEKLPFHFDPKRAVQQLRACGVLETIRISAAGYPSRWAYHDFFNRYRVLVKKRELANTD KKAICRSVLENLIKDPDKFQFGRTKIFFRAGQVAYLEKLRADKFRTATIMIQKTVRGWLQKVKYHRLKGATLTLQRYCRGHLARRLAEHL RRIRAAVVLQKHYRMQRARQAYQRVRRAAVVIQAFTRAMFVRRTYRQVLMEHKATTIQKHVRGWMARRHFQRLRDAAIVIQCAFRMLKAR RELKALRIEARSAEHLKRLNVGMENKVVQLQRKIDEQNKEFKTLSEQLSVTTSTYTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRT ELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQILCQSKDEFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQR YDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQQVEEIGLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKR EQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNELRKAVADQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEE VLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHGVCQTNSKTEDWGYLNEDGELGLAYQGLKQVARLLEAQLQAQ SLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAA QALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTIN GIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSA MLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGM QLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEFEERVTVAFIRTIQNQ EVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLKSAELAKAGGCKHFNLLSSKGADKSSNFLYLQVKGEVEAKVEEL -------------------------------------------------------------- >56727_56727_4_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000285039_HTATIP2_chr11_20388719_ENST00000451739_length(amino acids)=1975AA_BP=1798 MSVGELYSQCTRVWIPDPDEVWRSAELTKDYKEGDKSLQLRLEDETILEYPIDVQRNQLPFLRNPDILVGENDLTALSYLHEPAVLHNLK VRFLESNHIYTYCGIVLVAINPYEQLPIYGQDVIYTYSGQNMGDMDPHIFAVAEEAYKQMARDEKNQSIIVSGESGAGKTVSAKYAMRYF ATVGGSASETNIEEKVLASSPIMEAIGNAKTTRNDNSSRFGKYIQIGFDKRYHIIGANMRTYLLEKSRVVFQADDERNYHIFYQLCAAAG LPEFKELALTSAEDFFYTSQGGDTSIEGVDDAEDFEKTRQAFTLLGVKESHQMSIFKIIASILHLGSVAIQAERDGDSCSISPQDVYLSN FCRLLGVEHSQMEHWLCHRKLVTTSETYVKTMSLQQVINARNALAKHIYAQLFGWIVEHINKALHTSLKQHSFIGVLDIYGFETFEVNSF EQFCINYANEKLQQQFNSHVFKLEQEEYMKEQIPWTLIDFYDNQPCIDLIEAKLGILDLLDEECKVPKGTDQNWAQKLYDRHSSSQHFQK PRMSNTAFIIVHFADKVEYLSDGFLEKNRDTVYEEQINILKASKFPLVADLFHDDKDPVPATTPGKGSSSKISVRSARPPMKVSNKEHKK TVGHQFRTSLHLLMETLNATTPHYVRCIKPNDEKLPFHFDPKRAVQQLRACGVLETIRISAAGYPSRWAYHDFFNRYRVLVKKRELANTD KKAICRSVLENLIKDPDKFQFGRTKIFFRAGQVAYLEKLRADKFRTATIMIQKTVRGWLQKVKYHRLKGATLTLQRYCRGHLARRLAEHL RRIRAAVVLQKHYRMQRARQAYQRVRRAAVVIQAFTRAMFVRRTYRQVLMEHKATTIQKHVRGWMARRHFQRLRDAAIVIQCAFRMLKAR RELKALRIEARSAEHLKRLNVGMENKVVQLQRKIDEQNKEFKTLSEQLSVTTSTYTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRT ELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQILCQSKDEFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQR YDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQQVEEIGLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKR EQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNELRKAVADQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEE VLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHGVCQTNSKTEDWGYLNEDGELGLAYQGLKQVARLLEAQLQAQ SLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAA QALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTIN GIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSA MLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGM QLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEFEERVTVAFIRTIQNQ EVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLKSAELAKAGGCKHFNLLSSKGADKSSNFLYLQVKGEVEAKVEEL -------------------------------------------------------------- >56727_56727_5_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000285039_HTATIP2_chr11_20388719_ENST00000531058_length(amino acids)=1929AA_BP=1798 MSVGELYSQCTRVWIPDPDEVWRSAELTKDYKEGDKSLQLRLEDETILEYPIDVQRNQLPFLRNPDILVGENDLTALSYLHEPAVLHNLK VRFLESNHIYTYCGIVLVAINPYEQLPIYGQDVIYTYSGQNMGDMDPHIFAVAEEAYKQMARDEKNQSIIVSGESGAGKTVSAKYAMRYF ATVGGSASETNIEEKVLASSPIMEAIGNAKTTRNDNSSRFGKYIQIGFDKRYHIIGANMRTYLLEKSRVVFQADDERNYHIFYQLCAAAG LPEFKELALTSAEDFFYTSQGGDTSIEGVDDAEDFEKTRQAFTLLGVKESHQMSIFKIIASILHLGSVAIQAERDGDSCSISPQDVYLSN FCRLLGVEHSQMEHWLCHRKLVTTSETYVKTMSLQQVINARNALAKHIYAQLFGWIVEHINKALHTSLKQHSFIGVLDIYGFETFEVNSF EQFCINYANEKLQQQFNSHVFKLEQEEYMKEQIPWTLIDFYDNQPCIDLIEAKLGILDLLDEECKVPKGTDQNWAQKLYDRHSSSQHFQK PRMSNTAFIIVHFADKVEYLSDGFLEKNRDTVYEEQINILKASKFPLVADLFHDDKDPVPATTPGKGSSSKISVRSARPPMKVSNKEHKK TVGHQFRTSLHLLMETLNATTPHYVRCIKPNDEKLPFHFDPKRAVQQLRACGVLETIRISAAGYPSRWAYHDFFNRYRVLVKKRELANTD KKAICRSVLENLIKDPDKFQFGRTKIFFRAGQVAYLEKLRADKFRTATIMIQKTVRGWLQKVKYHRLKGATLTLQRYCRGHLARRLAEHL RRIRAAVVLQKHYRMQRARQAYQRVRRAAVVIQAFTRAMFVRRTYRQVLMEHKATTIQKHVRGWMARRHFQRLRDAAIVIQCAFRMLKAR RELKALRIEARSAEHLKRLNVGMENKVVQLQRKIDEQNKEFKTLSEQLSVTTSTYTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRT ELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQILCQSKDEFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQR YDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQQVEEIGLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKR EQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNELRKAVADQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEE VLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHGVCQTNSKTEDWGYLNEDGELGLAYQGLKQVARLLEAQLQAQ SLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAA QALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTIN GIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSA MLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGM QLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEFEERVTVAFIRTIQNQ EVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPGEWLVRKFFGSLPDSWASGHSVPV -------------------------------------------------------------- >56727_56727_6_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000324581_HTATIP2_chr11_20388719_ENST00000419348_length(amino acids)=1090AA_BP=913 MEHKATTIQKHVRGWMARRHFQRLRDAAIVIQCAFRMLKARRELKALRIEARSAEHLKRLNVGMENKVVQLQRKIDEQNKEFKTLSEQLS VTTSTYTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRTELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQIL CQSKDEFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQRYDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQ QVEEIGLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKREQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNEL RKAVADQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEEVLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHG VCQTNRLLEAQLQAQSLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKK QLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYT NDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQL IKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLN NLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEF EERVTVAFIRTIQNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLKSAELAKAGGCKHFNLLSSKGADKSSNFL YLQVKGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPGEWLVRKFFGSLPDSWASGHSVPVVTVVRAMLNNVVRPRDKQMELLENKAIHD -------------------------------------------------------------- >56727_56727_7_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000324581_HTATIP2_chr11_20388719_ENST00000421577_length(amino acids)=1090AA_BP=913 MEHKATTIQKHVRGWMARRHFQRLRDAAIVIQCAFRMLKARRELKALRIEARSAEHLKRLNVGMENKVVQLQRKIDEQNKEFKTLSEQLS VTTSTYTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRTELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQIL CQSKDEFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQRYDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQ QVEEIGLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKREQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNEL RKAVADQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEEVLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHG VCQTNRLLEAQLQAQSLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKK QLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYT NDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQL IKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLN NLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEF EERVTVAFIRTIQNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLKSAELAKAGGCKHFNLLSSKGADKSSNFL YLQVKGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPGEWLVRKFFGSLPDSWASGHSVPVVTVVRAMLNNVVRPRDKQMELLENKAIHD -------------------------------------------------------------- >56727_56727_8_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000324581_HTATIP2_chr11_20388719_ENST00000443524_length(amino acids)=1090AA_BP=913 MEHKATTIQKHVRGWMARRHFQRLRDAAIVIQCAFRMLKARRELKALRIEARSAEHLKRLNVGMENKVVQLQRKIDEQNKEFKTLSEQLS VTTSTYTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRTELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQIL CQSKDEFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQRYDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQ QVEEIGLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKREQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNEL RKAVADQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEEVLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHG VCQTNRLLEAQLQAQSLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKK QLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYT NDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQL IKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLN NLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEF EERVTVAFIRTIQNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLKSAELAKAGGCKHFNLLSSKGADKSSNFL YLQVKGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPGEWLVRKFFGSLPDSWASGHSVPVVTVVRAMLNNVVRPRDKQMELLENKAIHD -------------------------------------------------------------- >56727_56727_9_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000324581_HTATIP2_chr11_20388719_ENST00000451739_length(amino acids)=1090AA_BP=913 MEHKATTIQKHVRGWMARRHFQRLRDAAIVIQCAFRMLKARRELKALRIEARSAEHLKRLNVGMENKVVQLQRKIDEQNKEFKTLSEQLS VTTSTYTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRTELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQIL CQSKDEFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQRYDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQ QVEEIGLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKREQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNEL RKAVADQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEEVLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHG VCQTNRLLEAQLQAQSLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKK QLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYT NDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQL IKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLN NLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEF EERVTVAFIRTIQNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLKSAELAKAGGCKHFNLLSSKGADKSSNFL YLQVKGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPGEWLVRKFFGSLPDSWASGHSVPVVTVVRAMLNNVVRPRDKQMELLENKAIHD -------------------------------------------------------------- >56727_56727_10_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000324581_HTATIP2_chr11_20388719_ENST00000531058_length(amino acids)=1044AA_BP=913 MEHKATTIQKHVRGWMARRHFQRLRDAAIVIQCAFRMLKARRELKALRIEARSAEHLKRLNVGMENKVVQLQRKIDEQNKEFKTLSEQLS VTTSTYTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRTELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQIL CQSKDEFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQRYDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQ QVEEIGLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKREQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNEL RKAVADQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEEVLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHG VCQTNRLLEAQLQAQSLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKK QLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYT NDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQL IKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLN NLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEF EERVTVAFIRTIQNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPGEWLVRKFF -------------------------------------------------------------- >56727_56727_11_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000592688_HTATIP2_chr11_20388719_ENST00000419348_length(amino acids)=578AA_BP=401 MFSHCFLCFQDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTD LKPQMLSGTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQ NEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTV MCDQGLDPEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDA EAICSLCTSLSTQQIVKILNLYTPLNEFEERVTVAFIRTIQNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLK SAELAKAGGCKHFNLLSSKGADKSSNFLYLQVKGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPGEWLVRKFFGSLPDSWASGHSVPVV -------------------------------------------------------------- >56727_56727_12_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000592688_HTATIP2_chr11_20388719_ENST00000421577_length(amino acids)=578AA_BP=401 MFSHCFLCFQDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTD LKPQMLSGTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQ NEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTV MCDQGLDPEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDA EAICSLCTSLSTQQIVKILNLYTPLNEFEERVTVAFIRTIQNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLK SAELAKAGGCKHFNLLSSKGADKSSNFLYLQVKGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPGEWLVRKFFGSLPDSWASGHSVPVV -------------------------------------------------------------- >56727_56727_13_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000592688_HTATIP2_chr11_20388719_ENST00000443524_length(amino acids)=578AA_BP=401 MFSHCFLCFQDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTD LKPQMLSGTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQ NEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTV MCDQGLDPEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDA EAICSLCTSLSTQQIVKILNLYTPLNEFEERVTVAFIRTIQNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLK SAELAKAGGCKHFNLLSSKGADKSSNFLYLQVKGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPGEWLVRKFFGSLPDSWASGHSVPVV -------------------------------------------------------------- >56727_56727_14_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000592688_HTATIP2_chr11_20388719_ENST00000451739_length(amino acids)=578AA_BP=401 MFSHCFLCFQDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTD LKPQMLSGTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQ NEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTV MCDQGLDPEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDA EAICSLCTSLSTQQIVKILNLYTPLNEFEERVTVAFIRTIQNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAEGFVRVDRDYVLK SAELAKAGGCKHFNLLSSKGADKSSNFLYLQVKGEVEAKVEELKFDRYSVFRPGVLLCDRQESRPGEWLVRKFFGSLPDSWASGHSVPVV -------------------------------------------------------------- >56727_56727_15_MYO5B-HTATIP2_MYO5B_chr18_47361713_ENST00000592688_HTATIP2_chr11_20388719_ENST00000531058_length(amino acids)=532AA_BP=401 MFSHCFLCFQDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTD LKPQMLSGTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQ NEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTV MCDQGLDPEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDA EAICSLCTSLSTQQIVKILNLYTPLNEFEERVTVAFIRTIQNQEVVDFEKLDDYASAFQGHDVGFCCLGTTRGKAGAGEVEAKVEELKFD -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr18:47361713/chr11:20388719) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| MYO5B | HTATIP2 |
| FUNCTION: May be involved in vesicular trafficking via its association with the CART complex. The CART complex is necessary for efficient transferrin receptor recycling but not for EGFR degradation. Required in a complex with RAB11A and RAB11FIP2 for the transport of NPC1L1 to the plasma membrane. Together with RAB11A participates in CFTR trafficking to the plasma membrane and TF (transferrin) recycling in nonpolarized cells. Together with RAB11A and RAB8A participates in epithelial cell polarization. Together with RAB25 regulates transcytosis. {ECO:0000269|PubMed:21206382, ECO:0000269|PubMed:21282656}. | FUNCTION: Oxidoreductase required for tumor suppression. NAPDH-bound form inhibits nuclear import by competing with nuclear import substrates for binding to a subset of nuclear transport receptors. May act as a redox sensor linked to transcription through regulation of nuclear import. Isoform 1 is a metastasis suppressor with proapoptotic as well as antiangiogenic properties. Isoform 2 has an antiapoptotic effect. {ECO:0000269|PubMed:10611237, ECO:0000269|PubMed:11313954, ECO:0000269|PubMed:15282309, ECO:0000269|PubMed:9174052}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 1341_1471 | 1798.0 | 1849.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 899_1266 | 1798.0 | 1849.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 801_916 | 1798.0 | 1849.0 | Compositional bias | Note=Arg-rich |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 69_761 | 1798.0 | 1849.0 | Domain | Myosin motor |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 769_798 | 1798.0 | 1849.0 | Domain | IQ 1 |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 792_821 | 1798.0 | 1849.0 | Domain | IQ 2 |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 817_848 | 1798.0 | 1849.0 | Domain | IQ 3 |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 840_869 | 1798.0 | 1849.0 | Domain | IQ 4 |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 865_896 | 1798.0 | 1849.0 | Domain | IQ 5 |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 888_917 | 1798.0 | 1849.0 | Domain | IQ 6 |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 8_60 | 1798.0 | 1849.0 | Domain | Myosin N-terminal SH3-like |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 163_170 | 1798.0 | 1849.0 | Nucleotide binding | ATP |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 640_662 | 1798.0 | 1849.0 | Region | Actin-binding |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | MYO5B | chr18:47361713 | chr11:20388719 | ENST00000285039 | - | 39 | 40 | 1526_1803 | 1798.0 | 1849.0 | Domain | Dilute |
| Tgene | HTATIP2 | chr18:47361713 | chr11:20388719 | ENST00000419348 | 1 | 6 | 19_52 | 99.0 | 277.0 | Nucleotide binding | NADP | |
| Tgene | HTATIP2 | chr18:47361713 | chr11:20388719 | ENST00000421577 | 1 | 6 | 19_52 | 65.0 | 243.0 | Nucleotide binding | NADP | |
| Tgene | HTATIP2 | chr18:47361713 | chr11:20388719 | ENST00000443524 | 1 | 6 | 19_52 | 65.0 | 243.0 | Nucleotide binding | NADP | |
| Tgene | HTATIP2 | chr18:47361713 | chr11:20388719 | ENST00000451739 | 0 | 5 | 19_52 | 65.0 | 243.0 | Nucleotide binding | NADP | |
| Tgene | HTATIP2 | chr18:47361713 | chr11:20388719 | ENST00000530266 | 1 | 3 | 19_52 | 65.0 | 134.0 | Nucleotide binding | NADP | |
| Tgene | HTATIP2 | chr18:47361713 | chr11:20388719 | ENST00000532081 | 0 | 2 | 19_52 | 65.0 | 134.0 | Nucleotide binding | NADP | |
| Tgene | HTATIP2 | chr18:47361713 | chr11:20388719 | ENST00000532505 | 1 | 3 | 19_52 | 65.0 | 134.0 | Nucleotide binding | NADP |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| MYO5B | |
| HTATIP2 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to MYO5B-HTATIP2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to MYO5B-HTATIP2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

