| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:NFASC-ATP2B4 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: NFASC-ATP2B4 | FusionPDB ID: 58738 | FusionGDB2.0 ID: 58738 | Hgene | Tgene | Gene symbol | NFASC | ATP2B4 | Gene ID | 23114 | 493 |
| Gene name | neurofascin | ATPase plasma membrane Ca2+ transporting 4 | |
| Synonyms | NEDCPMD|NF|NRCAML | ATP2B2|MXRA1|PMCA4|PMCA4b|PMCA4x | |
| Cytomap | 1q32.1 | 1q32.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | neurofascinneurofascin homologneurofascin isoform 140 | plasma membrane calcium-transporting ATPase 4ATPase, Ca++ transporting, plasma membrane 4matrix-remodeling-associated protein 1sarcolemmal calcium pump | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | O94856 | P23634 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000495396, ENST00000338515, ENST00000338586, ENST00000339876, ENST00000360049, ENST00000367169, ENST00000367170, ENST00000367171, ENST00000367172, ENST00000401399, ENST00000404076, ENST00000404907, ENST00000513543, ENST00000539706, ENST00000403080, | ENST00000341360, ENST00000357681, ENST00000367218, ENST00000367219, ENST00000391954, ENST00000466407, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 16 X 10 X 4=640 | 13 X 15 X 8=1560 |
| # samples | 21 | 16 | |
| ** MAII score | log2(21/640*10)=-1.60768257722124 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/1560*10)=-3.28540221886225 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: NFASC [Title/Abstract] AND ATP2B4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | NFASC(204797910)-ATP2B4(203667285), # samples:2 ATP2B4(203596347)-NFASC(204889760), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | NFASC-ATP2B4 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NFASC-ATP2B4 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NFASC-ATP2B4 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NFASC-ATP2B4 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NFASC-ATP2B4 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. NFASC-ATP2B4 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ATP2B4 | GO:0006874 | cellular calcium ion homeostasis | 19278978|25798335 |
| Tgene | ATP2B4 | GO:0010629 | negative regulation of gene expression | 25147342 |
| Tgene | ATP2B4 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction | 19278978 |
| Tgene | ATP2B4 | GO:0016525 | negative regulation of angiogenesis | 25147342 |
| Tgene | ATP2B4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation | 19278978 |
| Tgene | ATP2B4 | GO:0043537 | negative regulation of blood vessel endothelial cell migration | 25147342 |
| Tgene | ATP2B4 | GO:0045019 | negative regulation of nitric oxide biosynthetic process | 11591728|19278978 |
| Tgene | ATP2B4 | GO:0051001 | negative regulation of nitric-oxide synthase activity | 11591728|17242280|19278978|19287093 |
| Tgene | ATP2B4 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade | 19287093|25147342 |
| Tgene | ATP2B4 | GO:0071872 | cellular response to epinephrine stimulus | 19278978 |
| Tgene | ATP2B4 | GO:0098736 | negative regulation of the force of heart contraction | 17242280 |
| Tgene | ATP2B4 | GO:0140199 | negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway involved in heart process | 17242280 |
| Tgene | ATP2B4 | GO:1900082 | negative regulation of arginine catabolic process | 19278978 |
| Tgene | ATP2B4 | GO:1901660 | calcium ion export | 25798335 |
| Tgene | ATP2B4 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 25147342 |
| Tgene | ATP2B4 | GO:1903078 | positive regulation of protein localization to plasma membrane | 15955804 |
| Tgene | ATP2B4 | GO:1903249 | negative regulation of citrulline biosynthetic process | 19278978 |
| Tgene | ATP2B4 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity | 19278978 |
 Fusion gene breakpoints across NFASC (5'-gene) Fusion gene breakpoints across NFASC (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
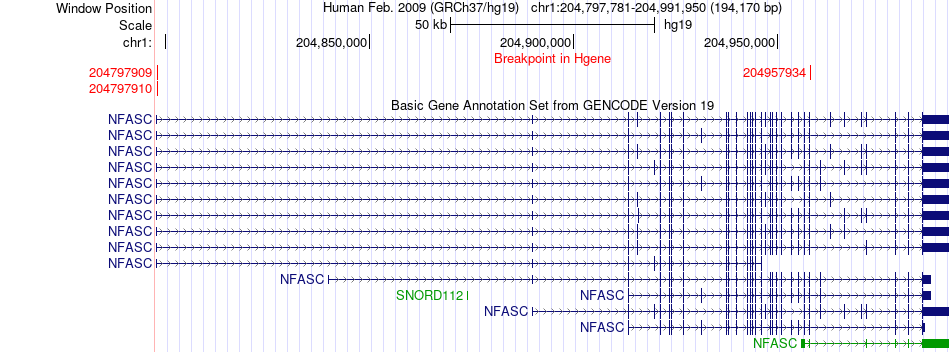 |
 Fusion gene breakpoints across ATP2B4 (3'-gene) Fusion gene breakpoints across ATP2B4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
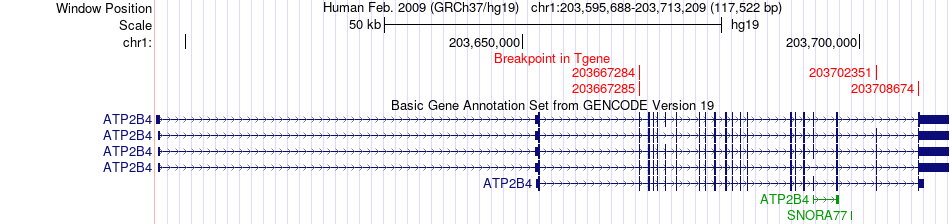 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LGG | TCGA-DU-7292 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203702351 | + |
| ChimerDB4 | LGG | TCGA-DU-7292 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + |
| ChimerDB4 | LGG | TCGA-DU-A7TD-01A | NFASC | chr1 | 204797909 | + | ATP2B4 | chr1 | 203667284 | + |
| ChimerDB4 | LGG | TCGA-DU-A7TD-01A | NFASC | chr1 | 204797910 | - | ATP2B4 | chr1 | 203667285 | + |
| ChimerDB4 | LGG | TCGA-DU-A7TD | NFASC | chr1 | 204797910 | + | ATP2B4 | chr1 | 203667284 | + |
| ChimerDB4 | LGG | TCGA-DU-A7TD | NFASC | chr1 | 204797910 | + | ATP2B4 | chr1 | 203667285 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000338586 | NFASC | chr1 | 204957934 | + | ENST00000391954 | ATP2B4 | chr1 | 203708674 | + | 7950 | 3416 | 7 | 3441 | 1144 |
| ENST00000338586 | NFASC | chr1 | 204957934 | + | ENST00000367218 | ATP2B4 | chr1 | 203708674 | + | 7950 | 3416 | 7 | 3441 | 1144 |
| ENST00000338586 | NFASC | chr1 | 204957934 | + | ENST00000367219 | ATP2B4 | chr1 | 203708674 | + | 7950 | 3416 | 7 | 3441 | 1144 |
| ENST00000338515 | NFASC | chr1 | 204957934 | + | ENST00000341360 | ATP2B4 | chr1 | 203708674 | + | 4194 | 3420 | 7 | 3504 | 1165 |
| ENST00000367171 | NFASC | chr1 | 204957934 | + | ENST00000341360 | ATP2B4 | chr1 | 203708674 | + | 4011 | 3237 | 7 | 3321 | 1104 |
| ENST00000367169 | NFASC | chr1 | 204957934 | + | ENST00000341360 | ATP2B4 | chr1 | 203708674 | + | 3873 | 3099 | 7 | 3183 | 1058 |
| ENST00000539706 | NFASC | chr1 | 204957934 | + | ENST00000391954 | ATP2B4 | chr1 | 203708674 | + | 7938 | 3404 | 7 | 3429 | 1140 |
| ENST00000539706 | NFASC | chr1 | 204957934 | + | ENST00000367218 | ATP2B4 | chr1 | 203708674 | + | 7938 | 3404 | 7 | 3429 | 1140 |
| ENST00000539706 | NFASC | chr1 | 204957934 | + | ENST00000367219 | ATP2B4 | chr1 | 203708674 | + | 7938 | 3404 | 7 | 3429 | 1140 |
| ENST00000339876 | NFASC | chr1 | 204957934 | + | ENST00000391954 | ATP2B4 | chr1 | 203708674 | + | 7629 | 3095 | 7 | 3120 | 1037 |
| ENST00000339876 | NFASC | chr1 | 204957934 | + | ENST00000367218 | ATP2B4 | chr1 | 203708674 | + | 7629 | 3095 | 7 | 3120 | 1037 |
| ENST00000339876 | NFASC | chr1 | 204957934 | + | ENST00000367219 | ATP2B4 | chr1 | 203708674 | + | 7629 | 3095 | 7 | 3120 | 1037 |
| ENST00000367170 | NFASC | chr1 | 204957934 | + | ENST00000341360 | ATP2B4 | chr1 | 203708674 | + | 4194 | 3420 | 7 | 3504 | 1165 |
| ENST00000360049 | NFASC | chr1 | 204957934 | + | ENST00000391954 | ATP2B4 | chr1 | 203708674 | + | 7938 | 3404 | 7 | 3429 | 1140 |
| ENST00000360049 | NFASC | chr1 | 204957934 | + | ENST00000367218 | ATP2B4 | chr1 | 203708674 | + | 7938 | 3404 | 7 | 3429 | 1140 |
| ENST00000360049 | NFASC | chr1 | 204957934 | + | ENST00000367219 | ATP2B4 | chr1 | 203708674 | + | 7938 | 3404 | 7 | 3429 | 1140 |
| ENST00000367172 | NFASC | chr1 | 204957934 | + | ENST00000341360 | ATP2B4 | chr1 | 203708674 | + | 4194 | 3420 | 7 | 3504 | 1165 |
| ENST00000404076 | NFASC | chr1 | 204957934 | + | ENST00000391954 | ATP2B4 | chr1 | 203708674 | + | 7981 | 3447 | 224 | 3472 | 1082 |
| ENST00000404076 | NFASC | chr1 | 204957934 | + | ENST00000367218 | ATP2B4 | chr1 | 203708674 | + | 7981 | 3447 | 224 | 3472 | 1082 |
| ENST00000404076 | NFASC | chr1 | 204957934 | + | ENST00000367219 | ATP2B4 | chr1 | 203708674 | + | 7981 | 3447 | 224 | 3472 | 1082 |
| ENST00000401399 | NFASC | chr1 | 204957934 | + | ENST00000391954 | ATP2B4 | chr1 | 203708674 | + | 7500 | 2966 | 1 | 2991 | 996 |
| ENST00000401399 | NFASC | chr1 | 204957934 | + | ENST00000367218 | ATP2B4 | chr1 | 203708674 | + | 7500 | 2966 | 1 | 2991 | 996 |
| ENST00000401399 | NFASC | chr1 | 204957934 | + | ENST00000367219 | ATP2B4 | chr1 | 203708674 | + | 7500 | 2966 | 1 | 2991 | 996 |
| ENST00000404907 | NFASC | chr1 | 204957934 | + | ENST00000391954 | ATP2B4 | chr1 | 203708674 | + | 7700 | 3166 | 0 | 3191 | 1063 |
| ENST00000404907 | NFASC | chr1 | 204957934 | + | ENST00000367218 | ATP2B4 | chr1 | 203708674 | + | 7700 | 3166 | 0 | 3191 | 1063 |
| ENST00000404907 | NFASC | chr1 | 204957934 | + | ENST00000367219 | ATP2B4 | chr1 | 203708674 | + | 7700 | 3166 | 0 | 3191 | 1063 |
| ENST00000513543 | NFASC | chr1 | 204957934 | + | ENST00000391954 | ATP2B4 | chr1 | 203708674 | + | 7675 | 3141 | 32 | 3166 | 1044 |
| ENST00000513543 | NFASC | chr1 | 204957934 | + | ENST00000367218 | ATP2B4 | chr1 | 203708674 | + | 7675 | 3141 | 32 | 3166 | 1044 |
| ENST00000513543 | NFASC | chr1 | 204957934 | + | ENST00000367219 | ATP2B4 | chr1 | 203708674 | + | 7675 | 3141 | 32 | 3166 | 1044 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000338586 | ENST00000391954 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000160884 | 0.99983907 |
| ENST00000338586 | ENST00000367218 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000160884 | 0.99983907 |
| ENST00000338586 | ENST00000367219 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000160884 | 0.99983907 |
| ENST00000338515 | ENST00000341360 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000937634 | 0.99906236 |
| ENST00000367171 | ENST00000341360 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.001196274 | 0.99880373 |
| ENST00000367169 | ENST00000341360 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.001378938 | 0.9986211 |
| ENST00000539706 | ENST00000391954 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000151455 | 0.9998485 |
| ENST00000539706 | ENST00000367218 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000151455 | 0.9998485 |
| ENST00000539706 | ENST00000367219 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000151455 | 0.9998485 |
| ENST00000339876 | ENST00000391954 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000157755 | 0.99984217 |
| ENST00000339876 | ENST00000367218 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000157755 | 0.99984217 |
| ENST00000339876 | ENST00000367219 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000157755 | 0.99984217 |
| ENST00000367170 | ENST00000341360 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000937634 | 0.99906236 |
| ENST00000360049 | ENST00000391954 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000151455 | 0.9998485 |
| ENST00000360049 | ENST00000367218 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000151455 | 0.9998485 |
| ENST00000360049 | ENST00000367219 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000151455 | 0.9998485 |
| ENST00000367172 | ENST00000341360 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000937634 | 0.99906236 |
| ENST00000404076 | ENST00000391954 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000401423 | 0.99959856 |
| ENST00000404076 | ENST00000367218 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000401423 | 0.99959856 |
| ENST00000404076 | ENST00000367219 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000401423 | 0.99959856 |
| ENST00000401399 | ENST00000391954 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000162759 | 0.99983716 |
| ENST00000401399 | ENST00000367218 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000162759 | 0.99983716 |
| ENST00000401399 | ENST00000367219 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000162759 | 0.99983716 |
| ENST00000404907 | ENST00000391954 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000147137 | 0.9998529 |
| ENST00000404907 | ENST00000367218 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000147137 | 0.9998529 |
| ENST00000404907 | ENST00000367219 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000147137 | 0.9998529 |
| ENST00000513543 | ENST00000391954 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000144347 | 0.99985564 |
| ENST00000513543 | ENST00000367218 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000144347 | 0.99985564 |
| ENST00000513543 | ENST00000367219 | NFASC | chr1 | 204957934 | + | ATP2B4 | chr1 | 203708674 | + | 0.000144347 | 0.99985564 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >58738_58738_1_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000338515_ATP2B4_chr1_203708674_ENST00000341360_length(amino acids)=1165AA_BP=1137 MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDPNPQHRLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQ ATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVI AINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPV YVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQ ASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFN -------------------------------------------------------------- >58738_58738_2_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000338586_ATP2B4_chr1_203708674_ENST00000367218_length(amino acids)=1144AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDPNPQHRLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQ ATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVI AINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPV YVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQ ASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFN -------------------------------------------------------------- >58738_58738_3_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000338586_ATP2B4_chr1_203708674_ENST00000367219_length(amino acids)=1144AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDPNPQHRLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQ ATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVI AINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPV YVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQ ASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFN -------------------------------------------------------------- >58738_58738_4_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000338586_ATP2B4_chr1_203708674_ENST00000391954_length(amino acids)=1144AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDPNPQHRLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQ ATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVI AINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPV YVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQ ASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFN -------------------------------------------------------------- >58738_58738_5_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000339876_ATP2B4_chr1_203708674_ENST00000367218_length(amino acids)=1037AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDPSIQNELTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQ ATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVI AINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPV YVPYEIRVQAENDFGKGPEPESVIGYSGEDLPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVENFSPNQT -------------------------------------------------------------- >58738_58738_6_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000339876_ATP2B4_chr1_203708674_ENST00000367219_length(amino acids)=1037AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDPSIQNELTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQ ATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVI AINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPV YVPYEIRVQAENDFGKGPEPESVIGYSGEDLPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVENFSPNQT -------------------------------------------------------------- >58738_58738_7_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000339876_ATP2B4_chr1_203708674_ENST00000391954_length(amino acids)=1037AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDPSIQNELTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQ ATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVI AINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPV YVPYEIRVQAENDFGKGPEPESVIGYSGEDLPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVENFSPNQT -------------------------------------------------------------- >58738_58738_8_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000360049_ATP2B4_chr1_203708674_ENST00000367218_length(amino acids)=1140AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNS RFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLP SPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRNHPDMYSARGVAER TPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVR VKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAF VSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAEN QVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDL AKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINE VGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPY EIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQASFP GDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKV -------------------------------------------------------------- >58738_58738_9_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000360049_ATP2B4_chr1_203708674_ENST00000367219_length(amino acids)=1140AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNS RFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLP SPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRNHPDMYSARGVAER TPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVR VKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAF VSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAEN QVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDL AKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINE VGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPY EIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQASFP GDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKV -------------------------------------------------------------- >58738_58738_10_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000360049_ATP2B4_chr1_203708674_ENST00000391954_length(amino acids)=1140AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNS RFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLP SPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRNHPDMYSARGVAER TPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVR VKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAF VSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAEN QVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDL AKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINE VGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPY EIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQASFP GDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKV -------------------------------------------------------------- >58738_58738_11_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000367169_ATP2B4_chr1_203708674_ENST00000341360_length(amino acids)=1058AA_BP=1030 MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDPNPQHRLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQ ATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVI AINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPV YVPYEIRVQAENDFGKGPEPESVIGYSGEDLPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVENFSPNQT -------------------------------------------------------------- >58738_58738_12_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000367170_ATP2B4_chr1_203708674_ENST00000341360_length(amino acids)=1165AA_BP=1137 MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDPNPQHRLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQ ATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVI AINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPV YVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQ ASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFN -------------------------------------------------------------- >58738_58738_13_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000367171_ATP2B4_chr1_203708674_ENST00000341360_length(amino acids)=1104AA_BP=1076 MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLSIETTVTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLGRP DRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINEVGSSHPSLPSE RYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPYEIRVQAENDFG KGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQASFPGDRLRGVVSRL FPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVACRCMAAAPGVKGPRSKW -------------------------------------------------------------- >58738_58738_14_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000367172_ATP2B4_chr1_203708674_ENST00000341360_length(amino acids)=1165AA_BP=1137 MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDPNPQHRLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSF HWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCN PPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTA SSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEP KNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRML SPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTR IYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQ ATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVI AINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPV YVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQ ASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFN -------------------------------------------------------------- >58738_58738_15_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000401399_ATP2B4_chr1_203708674_ENST00000367218_length(amino acids)=996AA_BP= LIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLEVTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGA IEIPMDPSIQNELTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEG EYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVML QDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKF ENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNP NREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLD GGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPL YIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDAN NSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNM EITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPYEIRVQAENDFGKGPEPESVIGYSGEDLPSAPRRFRVR QPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVENFSPNQTKFTVQRTDPVSRYRFTLSARTQVGSGEAVTEESPAPPNEDQ -------------------------------------------------------------- >58738_58738_16_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000401399_ATP2B4_chr1_203708674_ENST00000367219_length(amino acids)=996AA_BP= LIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLEVTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGA IEIPMDPSIQNELTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEG EYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVML QDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKF ENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNP NREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLD GGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPL YIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDAN NSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNM EITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPYEIRVQAENDFGKGPEPESVIGYSGEDLPSAPRRFRVR QPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVENFSPNQTKFTVQRTDPVSRYRFTLSARTQVGSGEAVTEESPAPPNEDQ -------------------------------------------------------------- >58738_58738_17_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000401399_ATP2B4_chr1_203708674_ENST00000391954_length(amino acids)=996AA_BP= LIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLEVTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGA IEIPMDPSIQNELTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEG EYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVML QDMQTDYSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKF ENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNP NREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLD GGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPL YIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLADQATPTNRLAALPKGRPDRPRDLELTDLAERSVRLTWIPGDAN NSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNM EITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPYEIRVQAENDFGKGPEPESVIGYSGEDLPSAPRRFRVR QPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVENFSPNQTKFTVQRTDPVSRYRFTLSARTQVGSGEAVTEESPAPPNEDQ -------------------------------------------------------------- >58738_58738_18_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000404076_ATP2B4_chr1_203708674_ENST00000367218_length(amino acids)=1082AA_BP= MIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLEVTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGA IEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFA RNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTD YSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKA LRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAG DTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHV YENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRM KKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVW HDHSKYPGSVNSAVLRLSPYVNYQFRVIAINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIV KWRRRETREAWNNVTVWGSRYVVGQTPVYVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQ GQLREYRAYYWRESSLLKNLWVSQKRQQASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNL ETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVENFSPNQTKFTVQRTDPVSRYRFTLSARTQVGSGEAVTEESPAPPNEDQSGQS -------------------------------------------------------------- >58738_58738_19_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000404076_ATP2B4_chr1_203708674_ENST00000367219_length(amino acids)=1082AA_BP= MIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLEVTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGA IEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFA RNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTD YSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKA LRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAG DTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHV YENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRM KKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVW HDHSKYPGSVNSAVLRLSPYVNYQFRVIAINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIV KWRRRETREAWNNVTVWGSRYVVGQTPVYVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQ GQLREYRAYYWRESSLLKNLWVSQKRQQASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNL ETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVENFSPNQTKFTVQRTDPVSRYRFTLSARTQVGSGEAVTEESPAPPNEDQSGQS -------------------------------------------------------------- >58738_58738_20_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000404076_ATP2B4_chr1_203708674_ENST00000391954_length(amino acids)=1082AA_BP= MIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLEVTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGA IEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFA RNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTD YSCNARFHFTHTIQQKNPFTLKVLTTRGVAERTPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKA LRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAG DTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHV YENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRM KKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVW HDHSKYPGSVNSAVLRLSPYVNYQFRVIAINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIV KWRRRETREAWNNVTVWGSRYVVGQTPVYVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQ GQLREYRAYYWRESSLLKNLWVSQKRQQASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNL ETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVENFSPNQTKFTVQRTDPVSRYRFTLSARTQVGSGEAVTEESPAPPNEDQSGQS -------------------------------------------------------------- >58738_58738_21_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000404907_ATP2B4_chr1_203708674_ENST00000367218_length(amino acids)=1063AA_BP= LSAVQEAQLKRLEVTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAK GNPAPSFHWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGA PLTLQCNPPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRN HPDMYSARGVAERTPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLAS NKMGSIRHTISVRVKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCN TSNEHGYLLANAFVSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIY TCVATNILGKAENQVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGS YTCVASTELDQDLAKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSP YVNYQFRVIAINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGS RYVVGQTPVYVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKN LWVSQKRQQASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIG -------------------------------------------------------------- >58738_58738_22_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000404907_ATP2B4_chr1_203708674_ENST00000367219_length(amino acids)=1063AA_BP= LSAVQEAQLKRLEVTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAK GNPAPSFHWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGA PLTLQCNPPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRN HPDMYSARGVAERTPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLAS NKMGSIRHTISVRVKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCN TSNEHGYLLANAFVSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIY TCVATNILGKAENQVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGS YTCVASTELDQDLAKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSP YVNYQFRVIAINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGS RYVVGQTPVYVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKN LWVSQKRQQASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIG -------------------------------------------------------------- >58738_58738_23_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000404907_ATP2B4_chr1_203708674_ENST00000391954_length(amino acids)=1063AA_BP= LSAVQEAQLKRLEVTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAK GNPAPSFHWTRNSRFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGA PLTLQCNPPPGLPSPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRN HPDMYSARGVAERTPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLAS NKMGSIRHTISVRVKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCN TSNEHGYLLANAFVSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIY TCVATNILGKAENQVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGS YTCVASTELDQDLAKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSP YVNYQFRVIAINEVGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGS RYVVGQTPVYVPYEIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKN LWVSQKRQQASFPGDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIG -------------------------------------------------------------- >58738_58738_24_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000513543_ATP2B4_chr1_203708674_ENST00000367218_length(amino acids)=1044AA_BP= MGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNSRFFNIA KDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLPSPVIFW MSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRNHPDMYSARGVAERTPSFMY PQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPY WLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDV PPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEV KDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLT VLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINEVGSSHP SLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPYEIRVQA ENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQASFPGDRLRG VVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVE -------------------------------------------------------------- >58738_58738_25_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000513543_ATP2B4_chr1_203708674_ENST00000367219_length(amino acids)=1044AA_BP= MGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNSRFFNIA KDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLPSPVIFW MSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRNHPDMYSARGVAERTPSFMY PQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPY WLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDV PPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEV KDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLT VLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINEVGSSHP SLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPYEIRVQA ENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQASFPGDRLRG VVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVE -------------------------------------------------------------- >58738_58738_26_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000513543_ATP2B4_chr1_203708674_ENST00000391954_length(amino acids)=1044AA_BP= MGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNSRFFNIA KDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLPSPVIFW MSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRNHPDMYSARGVAERTPSFMY PQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVRVKAAPY WLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAFVSVLDV PPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAENQVRLEV KDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDLAKAYLT VLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINEVGSSHP SLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPYEIRVQA ENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQASFPGDRLRG VVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKVGKQIVE -------------------------------------------------------------- >58738_58738_27_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000539706_ATP2B4_chr1_203708674_ENST00000367218_length(amino acids)=1140AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNS RFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLP SPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRNHPDMYSARGVAER TPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVR VKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAF VSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAEN QVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDL AKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINE VGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPY EIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQASFP GDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKV -------------------------------------------------------------- >58738_58738_28_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000539706_ATP2B4_chr1_203708674_ENST00000367219_length(amino acids)=1140AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNS RFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLP SPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRNHPDMYSARGVAER TPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVR VKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAF VSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAEN QVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDL AKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINE VGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPY EIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQASFP GDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKV -------------------------------------------------------------- >58738_58738_29_NFASC-ATP2B4_NFASC_chr1_204957934_ENST00000539706_ATP2B4_chr1_203708674_ENST00000391954_length(amino acids)=1140AA_BP= MVSALMRRLAARGAAGDAGEVAAPAADSSDSAQGRSPSPWRLIDLCAIWDAGVYLPSAAWNRASSGVARKRLNEAEKLSAVQEAQLKRLE VTRPRVLGSREQGQVPRMARQPPPPWVHAAFLLCLLSLGGAIEIPMDLTQPPTITKQSAKDHIVDPRDNILIECEAKGNPAPSFHWTRNS RFFNIAKDPRVSMRRRSGTLVIDFRSGGRPEEYEGEYQCFARNKFGTALSNRIRLQVSKSPLWPKENLDPVVVQEGAPLTLQCNPPPGLP SPVIFWMSSSMEPITQDKRVSQGHNGDLYFSNVMLQDMQTDYSCNARFHFTHTIQQKNPFTLKVLTNHPYNDSSLRNHPDMYSARGVAER TPSFMYPQGTASSQMVLRGMDLLLECIASGVPTPDIAWYKKGGDLPSDKAKFENFNKALRITNVSEEDSGEYFCLASNKMGSIRHTISVR VKAAPYWLDEPKNLILAPGEDGRLVCRANGNPKPTVQWMVNGEPLQSAPPNPNREVAGDTIIFRDTQISSRAVYQCNTSNEHGYLLANAF VSVLDVPPRMLSPRNQLIRVILYNRTRLDCPFFGSPIPTLRWFKNGQGSNLDGGNYHVYENGSLEIKMIRKEDQGIYTCVATNILGKAEN QVRLEVKDPTRIYRMPEDQVARRGTTVQLECRVKHDPSLKLTVSWLKDDEPLYIGNRMKKEDDSLTIFGVAERDQGSYTCVASTELDQDL AKAYLTVLGRPDRPRDLELTDLAERSVRLTWIPGDANNSPITDYVVQFEEDQFQPGVWHDHSKYPGSVNSAVLRLSPYVNYQFRVIAINE VGSSHPSLPSERYRTSGAPPESNPGDVKGEGTRKNNMEITWTPMNATSAFGPNLRYIVKWRRRETREAWNNVTVWGSRYVVGQTPVYVPY EIRVQAENDFGKGPEPESVIGYSGEDYPRAAPTEVKVRVMNSTAISLQWNRVYSDTVQGQLREYRAYYWRESSLLKNLWVSQKRQQASFP GDRLRGVVSRLFPYSNYKLEMVVVNGRGDGPRSETKEFTTPEGVPSAPRRFRVRQPNLETINLEWDHPEHPNGIMIGYTLKYVAFNGTKV -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:204797910/chr1:203667285) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NFASC | ATP2B4 |
| FUNCTION: Cell adhesion, ankyrin-binding protein which may be involved in neurite extension, axonal guidance, synaptogenesis, myelination and neuron-glial cell interactions. {ECO:0000250}. | FUNCTION: Calcium/calmodulin-regulated and magnesium-dependent enzyme that catalyzes the hydrolysis of ATP coupled with the transport of calcium out of the cell (PubMed:8530416). By regulating sperm cell calcium homeostasis, may play a role in sperm motility (By similarity). {ECO:0000250|UniProtKB:Q6Q477, ECO:0000269|PubMed:8530416}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 143_230 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 244_332 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 337_424 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 41_137 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 429_517 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 521_603 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 630_725 | 922.3333333333334 | 1241.0 | Domain | Fibronectin type-III 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 730_823 | 922.3333333333334 | 1241.0 | Domain | Fibronectin type-III 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 143_230 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 244_332 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 337_424 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 41_137 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 429_517 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 521_603 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 630_725 | 1025.3333333333333 | 1170.0 | Domain | Fibronectin type-III 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 730_823 | 1025.3333333333333 | 1170.0 | Domain | Fibronectin type-III 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 828_930 | 1025.3333333333333 | 1170.0 | Domain | Fibronectin type-III 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 143_230 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 244_332 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 337_424 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 41_137 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 429_517 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 521_603 | 922.3333333333334 | 1241.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 630_725 | 922.3333333333334 | 1241.0 | Domain | Fibronectin type-III 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 730_823 | 922.3333333333334 | 1241.0 | Domain | Fibronectin type-III 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 143_230 | 1008.3333333333334 | 1158.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 244_332 | 1008.3333333333334 | 1158.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 337_424 | 1008.3333333333334 | 1158.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 41_137 | 1008.3333333333334 | 1158.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 429_517 | 1008.3333333333334 | 1158.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 521_603 | 1008.3333333333334 | 1158.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 630_725 | 1008.3333333333334 | 1158.0 | Domain | Fibronectin type-III 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 730_823 | 1008.3333333333334 | 1158.0 | Domain | Fibronectin type-III 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 828_930 | 1008.3333333333334 | 1158.0 | Domain | Fibronectin type-III 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 143_230 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 244_332 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 337_424 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 41_137 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 429_517 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 521_603 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 630_725 | 1025.3333333333333 | 1175.0 | Domain | Fibronectin type-III 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 730_823 | 1025.3333333333333 | 1175.0 | Domain | Fibronectin type-III 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 828_930 | 1025.3333333333333 | 1175.0 | Domain | Fibronectin type-III 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 143_230 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 244_332 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 337_424 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 41_137 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 429_517 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 521_603 | 1025.3333333333333 | 1170.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 630_725 | 1025.3333333333333 | 1170.0 | Domain | Fibronectin type-III 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 730_823 | 1025.3333333333333 | 1170.0 | Domain | Fibronectin type-III 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 828_930 | 1025.3333333333333 | 1170.0 | Domain | Fibronectin type-III 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 143_230 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 244_332 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 337_424 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 41_137 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 429_517 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 521_603 | 1025.3333333333333 | 1175.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 630_725 | 1025.3333333333333 | 1175.0 | Domain | Fibronectin type-III 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 730_823 | 1025.3333333333333 | 1175.0 | Domain | Fibronectin type-III 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 828_930 | 1025.3333333333333 | 1175.0 | Domain | Fibronectin type-III 3 |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 1104_1113 | 1103.0 | 1206.0 | Region | Calmodulin-binding subdomain B |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 1031_1108 | 922.3333333333334 | 1241.0 | Compositional bias | Note=Thr-rich |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 1031_1108 | 1025.3333333333333 | 1170.0 | Compositional bias | Note=Thr-rich |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 1031_1108 | 922.3333333333334 | 1241.0 | Compositional bias | Note=Thr-rich |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 1031_1108 | 0 | 620.0 | Compositional bias | Note=Thr-rich |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 1031_1108 | 1008.3333333333334 | 1158.0 | Compositional bias | Note=Thr-rich |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 1031_1108 | 1025.3333333333333 | 1175.0 | Compositional bias | Note=Thr-rich |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 1031_1108 | 1025.3333333333333 | 1170.0 | Compositional bias | Note=Thr-rich |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 1031_1108 | 1025.3333333333333 | 1175.0 | Compositional bias | Note=Thr-rich |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 1114_1206 | 922.3333333333334 | 1241.0 | Domain | Fibronectin type-III 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 828_930 | 922.3333333333334 | 1241.0 | Domain | Fibronectin type-III 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 934_1030 | 922.3333333333334 | 1241.0 | Domain | Fibronectin type-III 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 1114_1206 | 1025.3333333333333 | 1170.0 | Domain | Fibronectin type-III 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 934_1030 | 1025.3333333333333 | 1170.0 | Domain | Fibronectin type-III 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 1114_1206 | 922.3333333333334 | 1241.0 | Domain | Fibronectin type-III 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 828_930 | 922.3333333333334 | 1241.0 | Domain | Fibronectin type-III 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 934_1030 | 922.3333333333334 | 1241.0 | Domain | Fibronectin type-III 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 1114_1206 | 0 | 620.0 | Domain | Fibronectin type-III 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 143_230 | 0 | 620.0 | Domain | Note=Ig-like C2-type 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 244_332 | 0 | 620.0 | Domain | Note=Ig-like C2-type 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 337_424 | 0 | 620.0 | Domain | Note=Ig-like C2-type 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 41_137 | 0 | 620.0 | Domain | Note=Ig-like C2-type 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 429_517 | 0 | 620.0 | Domain | Note=Ig-like C2-type 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 521_603 | 0 | 620.0 | Domain | Note=Ig-like C2-type 6 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 630_725 | 0 | 620.0 | Domain | Fibronectin type-III 1 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 730_823 | 0 | 620.0 | Domain | Fibronectin type-III 2 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 828_930 | 0 | 620.0 | Domain | Fibronectin type-III 3 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 934_1030 | 0 | 620.0 | Domain | Fibronectin type-III 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 1114_1206 | 1008.3333333333334 | 1158.0 | Domain | Fibronectin type-III 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 934_1030 | 1008.3333333333334 | 1158.0 | Domain | Fibronectin type-III 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 1114_1206 | 1025.3333333333333 | 1175.0 | Domain | Fibronectin type-III 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 934_1030 | 1025.3333333333333 | 1175.0 | Domain | Fibronectin type-III 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 1114_1206 | 1025.3333333333333 | 1170.0 | Domain | Fibronectin type-III 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 934_1030 | 1025.3333333333333 | 1170.0 | Domain | Fibronectin type-III 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 1114_1206 | 1025.3333333333333 | 1175.0 | Domain | Fibronectin type-III 5 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 934_1030 | 1025.3333333333333 | 1175.0 | Domain | Fibronectin type-III 4 |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 1239_1347 | 922.3333333333334 | 1241.0 | Topological domain | Cytoplasmic |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 25_1217 | 922.3333333333334 | 1241.0 | Topological domain | Extracellular |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 1239_1347 | 1025.3333333333333 | 1170.0 | Topological domain | Cytoplasmic |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 25_1217 | 1025.3333333333333 | 1170.0 | Topological domain | Extracellular |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 1239_1347 | 922.3333333333334 | 1241.0 | Topological domain | Cytoplasmic |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 25_1217 | 922.3333333333334 | 1241.0 | Topological domain | Extracellular |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 1239_1347 | 0 | 620.0 | Topological domain | Cytoplasmic |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 25_1217 | 0 | 620.0 | Topological domain | Extracellular |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 1239_1347 | 1008.3333333333334 | 1158.0 | Topological domain | Cytoplasmic |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 25_1217 | 1008.3333333333334 | 1158.0 | Topological domain | Extracellular |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 1239_1347 | 1025.3333333333333 | 1175.0 | Topological domain | Cytoplasmic |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 25_1217 | 1025.3333333333333 | 1175.0 | Topological domain | Extracellular |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 1239_1347 | 1025.3333333333333 | 1170.0 | Topological domain | Cytoplasmic |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 25_1217 | 1025.3333333333333 | 1170.0 | Topological domain | Extracellular |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 1239_1347 | 1025.3333333333333 | 1175.0 | Topological domain | Cytoplasmic |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 25_1217 | 1025.3333333333333 | 1175.0 | Topological domain | Extracellular |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000339876 | + | 23 | 30 | 1218_1238 | 922.3333333333334 | 1241.0 | Transmembrane | Helical |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000360049 | + | 24 | 27 | 1218_1238 | 1025.3333333333333 | 1170.0 | Transmembrane | Helical |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000401399 | + | 22 | 29 | 1218_1238 | 922.3333333333334 | 1241.0 | Transmembrane | Helical |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000403080 | + | 1 | 16 | 1218_1238 | 0 | 620.0 | Transmembrane | Helical |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404076 | + | 23 | 27 | 1218_1238 | 1008.3333333333334 | 1158.0 | Transmembrane | Helical |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000404907 | + | 22 | 26 | 1218_1238 | 1025.3333333333333 | 1175.0 | Transmembrane | Helical |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000513543 | + | 22 | 25 | 1218_1238 | 1025.3333333333333 | 1170.0 | Transmembrane | Helical |
| Hgene | NFASC | chr1:204957934 | chr1:203708674 | ENST00000539706 | + | 24 | 28 | 1218_1238 | 1025.3333333333333 | 1175.0 | Transmembrane | Helical |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 1086_1103 | 1162.3333333333333 | 1171.0 | Region | Calmodulin-binding subdomain A | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 1104_1113 | 1162.3333333333333 | 1171.0 | Region | Calmodulin-binding subdomain B | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 1086_1103 | 1103.0 | 1206.0 | Region | Calmodulin-binding subdomain A | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 1086_1103 | 1162.3333333333333 | 1171.0 | Region | Calmodulin-binding subdomain A | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 1104_1113 | 1162.3333333333333 | 1171.0 | Region | Calmodulin-binding subdomain B | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 1086_1103 | 1150.3333333333333 | 1159.0 | Region | Calmodulin-binding subdomain A | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 1104_1113 | 1150.3333333333333 | 1159.0 | Region | Calmodulin-binding subdomain B | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 1086_1103 | 1126.3333333333333 | 1135.0 | Region | Calmodulin-binding subdomain A | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 1104_1113 | 1126.3333333333333 | 1135.0 | Region | Calmodulin-binding subdomain B | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 1016_1025 | 1162.3333333333333 | 1171.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 1048_1241 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 114_150 | 1162.3333333333333 | 1171.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 172_356 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 1_92 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 377_409 | 1162.3333333333333 | 1171.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 428_840 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 861_870 | 1162.3333333333333 | 1171.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 892_911 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 935_952 | 1162.3333333333333 | 1171.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 975_993 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 1016_1025 | 1103.0 | 1206.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 1048_1241 | 1103.0 | 1206.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 114_150 | 1103.0 | 1206.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 172_356 | 1103.0 | 1206.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 1_92 | 1103.0 | 1206.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 377_409 | 1103.0 | 1206.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 428_840 | 1103.0 | 1206.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 861_870 | 1103.0 | 1206.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 892_911 | 1103.0 | 1206.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 935_952 | 1103.0 | 1206.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 975_993 | 1103.0 | 1206.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 1016_1025 | 1162.3333333333333 | 1171.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 1048_1241 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 114_150 | 1162.3333333333333 | 1171.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 172_356 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 1_92 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 377_409 | 1162.3333333333333 | 1171.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 428_840 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 861_870 | 1162.3333333333333 | 1171.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 892_911 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 935_952 | 1162.3333333333333 | 1171.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 975_993 | 1162.3333333333333 | 1171.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 1016_1025 | 1150.3333333333333 | 1159.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 1048_1241 | 1150.3333333333333 | 1159.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 114_150 | 1150.3333333333333 | 1159.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 172_356 | 1150.3333333333333 | 1159.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 1_92 | 1150.3333333333333 | 1159.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 377_409 | 1150.3333333333333 | 1159.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 428_840 | 1150.3333333333333 | 1159.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 861_870 | 1150.3333333333333 | 1159.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 892_911 | 1150.3333333333333 | 1159.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 935_952 | 1150.3333333333333 | 1159.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 975_993 | 1150.3333333333333 | 1159.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 1016_1025 | 1126.3333333333333 | 1135.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 1048_1241 | 1126.3333333333333 | 1135.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 114_150 | 1126.3333333333333 | 1135.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 172_356 | 1126.3333333333333 | 1135.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 1_92 | 1126.3333333333333 | 1135.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 377_409 | 1126.3333333333333 | 1135.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 428_840 | 1126.3333333333333 | 1135.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 861_870 | 1126.3333333333333 | 1135.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 892_911 | 1126.3333333333333 | 1135.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 935_952 | 1126.3333333333333 | 1135.0 | Topological domain | Extracellular | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 975_993 | 1126.3333333333333 | 1135.0 | Topological domain | Cytoplasmic | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 1026_1047 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 151_171 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 357_376 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 410_427 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 841_860 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 871_891 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 912_934 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 93_113 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 953_974 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000341360 | 19 | 21 | 994_1015 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 1026_1047 | 1103.0 | 1206.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 151_171 | 1103.0 | 1206.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 357_376 | 1103.0 | 1206.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 410_427 | 1103.0 | 1206.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 841_860 | 1103.0 | 1206.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 871_891 | 1103.0 | 1206.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 912_934 | 1103.0 | 1206.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 93_113 | 1103.0 | 1206.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 953_974 | 1103.0 | 1206.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000357681 | 19 | 21 | 994_1015 | 1103.0 | 1206.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 1026_1047 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 151_171 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 357_376 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 410_427 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 841_860 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 871_891 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 912_934 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 93_113 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 953_974 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367218 | 20 | 22 | 994_1015 | 1162.3333333333333 | 1171.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 1026_1047 | 1150.3333333333333 | 1159.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 151_171 | 1150.3333333333333 | 1159.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 357_376 | 1150.3333333333333 | 1159.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 410_427 | 1150.3333333333333 | 1159.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 841_860 | 1150.3333333333333 | 1159.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 871_891 | 1150.3333333333333 | 1159.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 912_934 | 1150.3333333333333 | 1159.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 93_113 | 1150.3333333333333 | 1159.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 953_974 | 1150.3333333333333 | 1159.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000367219 | 19 | 21 | 994_1015 | 1150.3333333333333 | 1159.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 1026_1047 | 1126.3333333333333 | 1135.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 151_171 | 1126.3333333333333 | 1135.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 357_376 | 1126.3333333333333 | 1135.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 410_427 | 1126.3333333333333 | 1135.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 841_860 | 1126.3333333333333 | 1135.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 871_891 | 1126.3333333333333 | 1135.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 912_934 | 1126.3333333333333 | 1135.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 93_113 | 1126.3333333333333 | 1135.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 953_974 | 1126.3333333333333 | 1135.0 | Transmembrane | Helical | |
| Tgene | ATP2B4 | chr1:204957934 | chr1:203708674 | ENST00000391954 | 19 | 21 | 994_1015 | 1126.3333333333333 | 1135.0 | Transmembrane | Helical |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| NFASC | |
| ATP2B4 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to NFASC-ATP2B4 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to NFASC-ATP2B4 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

