| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:NFATC2-EWSR1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: NFATC2-EWSR1 | FusionPDB ID: 58791 | FusionGDB2.0 ID: 58791 | Hgene | Tgene | Gene symbol | NFATC2 | EWSR1 | Gene ID | 4773 | 2130 |
| Gene name | nuclear factor of activated T cells 2 | EWS RNA binding protein 1 | |
| Synonyms | NFAT1|NFATP | EWS|EWS-FLI1|bK984G1.4 | |
| Cytomap | 20q13.2 | 22q12.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nuclear factor of activated T-cells, cytoplasmic 2NF-ATc2NFAT pre-existing subunitNFAT transcription complex, preexisting componentT cell transcription factor NFAT1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2nuclear fact | RNA-binding protein EWSEWS RNA-binding protein variant 6Ewing sarcoma breakpoint region 1Ewings sarcoma EWS-Fli1 (type 1) oncogene | |
| Modification date | 20200329 | 20200329 | |
| UniProtAcc | Q8NCF5 | Q01844 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000371564, ENST00000396009, ENST00000414705, ENST00000609507, ENST00000609943, ENST00000610033, | ENST00000331029, ENST00000332035, ENST00000332050, ENST00000333395, ENST00000397938, ENST00000406548, ENST00000414183, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 9 X 9 X 5=405 | 17 X 58 X 10=9860 |
| # samples | 8 | 48 | |
| ** MAII score | log2(8/405*10)=-2.33985000288462 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(48/9860*10)=-4.36048133565676 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: NFATC2 [Title/Abstract] AND EWSR1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | EWSR1(29684775)-NFATC2(50133495), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | EWSR1-NFATC2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. EWSR1-NFATC2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NFATC2-EWSR1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. NFATC2-EWSR1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. NFATC2-EWSR1 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. NFATC2-EWSR1 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. NFATC2-EWSR1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. NFATC2-EWSR1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | NFATC2 | GO:0016477 | cell migration | 21871017 |
| Hgene | NFATC2 | GO:0045893 | positive regulation of transcription, DNA-templated | 15790681 |
| Hgene | NFATC2 | GO:1905064 | negative regulation of vascular smooth muscle cell differentiation | 23853098 |
 Fusion gene breakpoints across NFATC2 (5'-gene) Fusion gene breakpoints across NFATC2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across EWSR1 (3'-gene) Fusion gene breakpoints across EWSR1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + |
| ChimerKB3 | . | . | NFATC2 | chr20 | 50139619 | - | EWSR1 | chr22 | 29683123 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000396009 | NFATC2 | chr20 | 50090516 | - | ENST00000332050 | EWSR1 | chr22 | 29674018 | + | 3817 | 1928 | 211 | 3453 | 1080 |
| ENST00000396009 | NFATC2 | chr20 | 50090516 | - | ENST00000397938 | EWSR1 | chr22 | 29674018 | + | 4037 | 1928 | 211 | 3672 | 1153 |
| ENST00000396009 | NFATC2 | chr20 | 50090516 | - | ENST00000406548 | EWSR1 | chr22 | 29674018 | + | 3845 | 1928 | 211 | 3669 | 1152 |
| ENST00000396009 | NFATC2 | chr20 | 50090516 | - | ENST00000331029 | EWSR1 | chr22 | 29674018 | + | 3923 | 1928 | 211 | 3558 | 1115 |
| ENST00000396009 | NFATC2 | chr20 | 50090516 | - | ENST00000414183 | EWSR1 | chr22 | 29674018 | + | 3852 | 1928 | 211 | 3669 | 1152 |
| ENST00000396009 | NFATC2 | chr20 | 50090516 | - | ENST00000333395 | EWSR1 | chr22 | 29674018 | + | 2948 | 1928 | 211 | 2766 | 851 |
| ENST00000396009 | NFATC2 | chr20 | 50090516 | - | ENST00000332035 | EWSR1 | chr22 | 29674018 | + | 3680 | 1928 | 211 | 3504 | 1097 |
| ENST00000609943 | NFATC2 | chr20 | 50090516 | - | ENST00000332050 | EWSR1 | chr22 | 29674018 | + | 3739 | 1850 | 202 | 3375 | 1057 |
| ENST00000609943 | NFATC2 | chr20 | 50090516 | - | ENST00000397938 | EWSR1 | chr22 | 29674018 | + | 3959 | 1850 | 202 | 3594 | 1130 |
| ENST00000609943 | NFATC2 | chr20 | 50090516 | - | ENST00000406548 | EWSR1 | chr22 | 29674018 | + | 3767 | 1850 | 202 | 3591 | 1129 |
| ENST00000609943 | NFATC2 | chr20 | 50090516 | - | ENST00000331029 | EWSR1 | chr22 | 29674018 | + | 3845 | 1850 | 202 | 3480 | 1092 |
| ENST00000609943 | NFATC2 | chr20 | 50090516 | - | ENST00000414183 | EWSR1 | chr22 | 29674018 | + | 3774 | 1850 | 202 | 3591 | 1129 |
| ENST00000609943 | NFATC2 | chr20 | 50090516 | - | ENST00000333395 | EWSR1 | chr22 | 29674018 | + | 2870 | 1850 | 202 | 2688 | 828 |
| ENST00000609943 | NFATC2 | chr20 | 50090516 | - | ENST00000332035 | EWSR1 | chr22 | 29674018 | + | 3602 | 1850 | 202 | 3426 | 1074 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >58791_58791_1_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000396009_EWSR1_chr22_29674018_ENST00000331029_length(amino acids)=1115AA_BP=573 MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFG EPDRVGPQKFLSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSAS FISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGA SPQRSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQ GERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGR KNTRVRLVFRVHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAA TAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQ SSYGQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMS RDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLK VSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGN QNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGP -------------------------------------------------------------- >58791_58791_2_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000396009_EWSR1_chr22_29674018_ENST00000332035_length(amino acids)=1097AA_BP=573 MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFG EPDRVGPQKFLSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSAS FISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGA SPQRSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQ GERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGR KNTRVRLVFRVHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAA TAPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYG QESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGSAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLN DSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLP PREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPE GFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGRRGGRGGP -------------------------------------------------------------- >58791_58791_3_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000396009_EWSR1_chr22_29674018_ENST00000332050_length(amino acids)=1080AA_BP=573 MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFG EPDRVGPQKFLSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSAS FISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGA SPQRSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQ GERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGR KNTRVRLVFRVHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAA TAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQ SSYGQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQRPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGV VKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGG PGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRG GPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGRRGGRGGPGKMDKGEHRQERRDRPY -------------------------------------------------------------- >58791_58791_4_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000396009_EWSR1_chr22_29674018_ENST00000333395_length(amino acids)=851AA_BP=573 MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFG EPDRVGPQKFLSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSAS FISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGA SPQRSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQ GERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGR KNTRVRLVFRVHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAA TAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQ SSYGQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMS -------------------------------------------------------------- >58791_58791_5_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000396009_EWSR1_chr22_29674018_ENST00000397938_length(amino acids)=1153AA_BP=573 MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFG EPDRVGPQKFLSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSAS FISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGA SPQRSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQ GERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGR KNTRVRLVFRVHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAA TAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQ SSYGQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMS RGGRGGGRGGMGSAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLD KETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGD RGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGG -------------------------------------------------------------- >58791_58791_6_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000396009_EWSR1_chr22_29674018_ENST00000406548_length(amino acids)=1152AA_BP=573 MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFG EPDRVGPQKFLSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSAS FISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGA SPQRSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQ GERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGR KNTRVRLVFRVHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAA TAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQ SSYGQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMS RGGRGGGRGGMGAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDK ETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDR GGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGP -------------------------------------------------------------- >58791_58791_7_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000396009_EWSR1_chr22_29674018_ENST00000414183_length(amino acids)=1152AA_BP=573 MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFG EPDRVGPQKFLSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSAS FISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGA SPQRSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQ GERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGR KNTRVRLVFRVHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAA TAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQ SSYGQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMS RGGRGGGRGGMGAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDK ETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDR GGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGP -------------------------------------------------------------- >58791_58791_8_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000609943_EWSR1_chr22_29674018_ENST00000331029_length(amino acids)=1092AA_BP=550 MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLS PRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGGPDD LCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGS PAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILLVPPTWPKPLV PAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSL QTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQSSTG GYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGS YSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRDPDEDSDNSAIYVQGLNDSVTL DDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGR GMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPP PFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGRRGGRGGPGKMDK -------------------------------------------------------------- >58791_58791_9_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000609943_EWSR1_chr22_29674018_ENST00000332035_length(amino acids)=1074AA_BP=550 MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLS PRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGGPDD LCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGS PAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILLVPPTWPKPLV PAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSL QTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTSYSSTQPTSYDQSSYSQQN TYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGR GRGGFDRGGMSRGGRGGGRGGMGSAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKR TGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGG PMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMR -------------------------------------------------------------- >58791_58791_10_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000609943_EWSR1_chr22_29674018_ENST00000332050_length(amino acids)=1057AA_BP=550 MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLS PRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGGPDD LCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGS PAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILLVPPTWPKPLV PAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSL QTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQSSTG GYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGS YSQAPSQYSQQSSSYGQQRPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKP KGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPP RGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFR -------------------------------------------------------------- >58791_58791_11_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000609943_EWSR1_chr22_29674018_ENST00000333395_length(amino acids)=828AA_BP=550 MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLS PRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGGPDD LCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGS PAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILLVPPTWPKPLV PAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSL QTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQSSTG GYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGS YSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGLQSESLVYTSI -------------------------------------------------------------- >58791_58791_12_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000609943_EWSR1_chr22_29674018_ENST00000397938_length(amino acids)=1130AA_BP=550 MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLS PRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGGPDD LCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGS PAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILLVPPTWPKPLV PAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSL QTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQSSTG GYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGS YSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGSAGERGGFNKP GGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAA VEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQ HRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMD -------------------------------------------------------------- >58791_58791_13_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000609943_EWSR1_chr22_29674018_ENST00000406548_length(amino acids)=1129AA_BP=550 MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLS PRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGGPDD LCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGS PAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILLVPPTWPKPLV PAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSL QTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQSSTG GYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGS YSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGAGERGGFNKPG GPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAV EWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQH RAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDR -------------------------------------------------------------- >58791_58791_14_NFATC2-EWSR1_NFATC2_chr20_50090516_ENST00000609943_EWSR1_chr22_29674018_ENST00000414183_length(amino acids)=1129AA_BP=550 MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLS PRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGGPDD LCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGS PAGYPPVAGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILLVPPTWPKPLV PAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSL QTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQSSTG GYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGS YSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGAGERGGFNKPG GPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAV EWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQH RAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDR -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
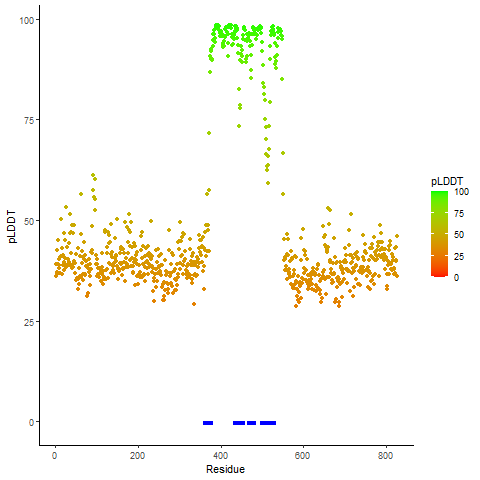
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:29684775/chr22:50133495) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| NFATC2 | EWSR1 |
| FUNCTION: In T-helper 2 (Th2) cells, regulates the magnitude of NFAT-driven transcription of a specific subset of cytokine genes, including IL3, IL4, IL5 and IL13, but not IL2. Recruits PRMT1 to the IL4 promoter; this leads to enhancement of histone H4 'Arg-3'-methylation and facilitates subsequent histone acetylation at the IL4 locus, thus promotes robust cytokine expression (By similarity). Down-regulates formation of poly-SUMO chains by UBE2I/UBC9 (By similarity). {ECO:0000250}. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
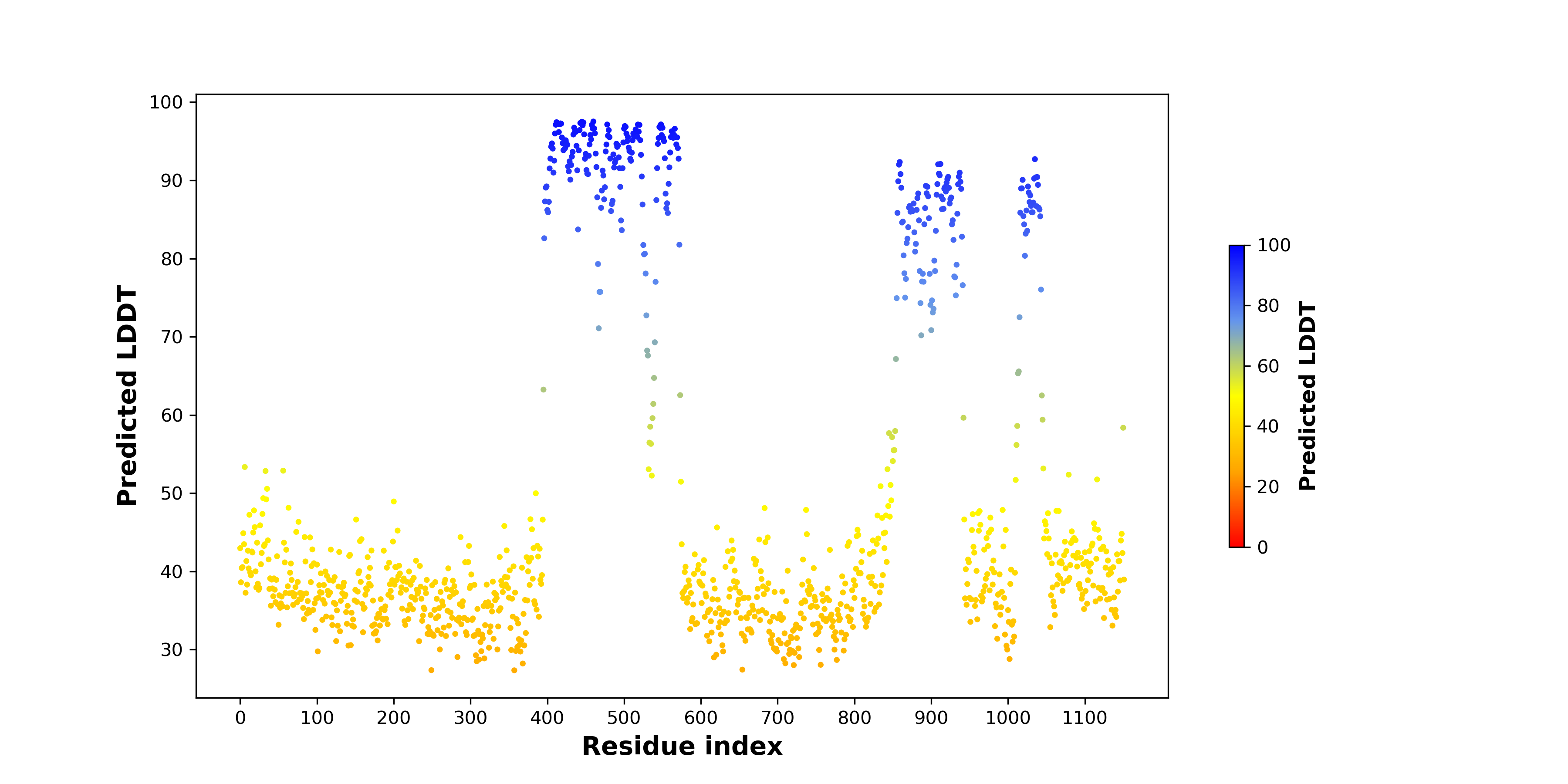
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (669) >>>669.pdbFusion protein BP residue: 550 CIF file (669) >>>669.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLK PYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLSPRIEITPSHE LIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPA SSGSSASFISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIM SPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQ RSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPP KMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILL VPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHH RAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILK LRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSLQTASNPIECC YTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPA YGQQPAATAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVP GSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSY GQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMG VYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGL | 828 |
| 3D view using mol* of 669 (AA BP:550) | ||||||||||
 | ||||||||||
| PDB file (677) >>>677.pdbFusion protein BP residue: 573 CIF file (677) >>>677.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPN AHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKF LSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAA SPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGG PDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSS PGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGSPAGYPPV AGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYP AVEFLGPCEQGERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPL EWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTK VLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFR VHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTA TVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQS STGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSY DQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGSYSQAPSQ YSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRG RGGFDRGGMSRGGRGGGRGGMGLQSESLVYTSILKKYPYSVLSRQHNEKW | 851 |
| 3D view using mol* of 677 (AA BP:573) | ||||||||||
 | ||||||||||
| PDB file (782) >>>782.pdbFusion protein BP residue: 550 CIF file (782) >>>782.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLK PYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLSPRIEITPSHE LIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPA SSGSSASFISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIM SPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQ RSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPP KMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILL VPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHH RAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILK LRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSLQTASNPIECC YTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPA YGQQPAATAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVP GSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSY GQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQRPMDEGPDLDLG PPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIH IYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKP PMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPP RGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPE GFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGF RGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGRRGGRGGPGKMDKGEHRQ | 1057 |
| 3D view using mol* of 782 (AA BP:550) | ||||||||||
 | ||||||||||
| PDB file (789) >>>789.pdbFusion protein BP residue: 550 CIF file (789) >>>789.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLK PYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLSPRIEITPSHE LIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPA SSGSSASFISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIM SPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQ RSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPP KMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILL VPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHH RAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILK LRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSLQTASNPIECC YTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPA YGQQPAATAPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSYGQQSSY GQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMGVYGQES GGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGSAGERGG FNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDLADFFK QCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDG KDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGG PMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQ NFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRG GPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGR | 1074 |
| 3D view using mol* of 789 (AA BP:550) | ||||||||||
 | ||||||||||
| PDB file (793) >>>793.pdbFusion protein BP residue: 573 CIF file (793) >>>793.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPN AHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKF LSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAA SPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGG PDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSS PGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGSPAGYPPV AGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYP AVEFLGPCEQGERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPL EWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTK VLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFR VHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTA TVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQS STGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSY DQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGSYSQAPSQ YSQQSSSYGQQRPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDD LADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAA VEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGG PGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPN PGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRG GLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLM | 1080 |
| 3D view using mol* of 793 (AA BP:573) | ||||||||||
 | ||||||||||
| PDB file (798) >>>798.pdbFusion protein BP residue: 550 CIF file (798) >>>798.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLK PYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLSPRIEITPSHE LIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPA SSGSSASFISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIM SPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQ RSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPP KMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILL VPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHH RAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILK LRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSLQTASNPIECC YTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPA YGQQPAATAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVP GSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSY GQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMG VYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRDPDEDSDNSAIY VQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDAT VSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGR GMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGN VQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRG RGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGR | 1092 |
| 3D view using mol* of 798 (AA BP:550) | ||||||||||
 | ||||||||||
| PDB file (802) >>>802.pdbFusion protein BP residue: 573 CIF file (802) >>>802.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPN AHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKF LSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAA SPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGG PDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSS PGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGSPAGYPPV AGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYP AVEFLGPCEQGERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPL EWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTK VLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFR VHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTA TVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTSYSSTQPTSYDQSSYS QQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSS SYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRGRGGFDR GGMSRGGRGGGRGGMGSAGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSD NSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDKETGKP KGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLP PREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNP SGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPP GGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGG | 1097 |
| 3D view using mol* of 802 (AA BP:573) | ||||||||||
 | ||||||||||
| PDB file (807) >>>807.pdbFusion protein BP residue: 573 CIF file (807) >>>807.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPN AHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKF LSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAA SPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGG PDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSS PGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGSPAGYPPV AGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYP AVEFLGPCEQGERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPL EWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTK VLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFR VHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTA TVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQS STGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSY DQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGSYSQAPSQ YSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRG RGGFDRGGMSRDPDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNK RTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLK VSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRG GDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECN QCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGG RGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLMEQMGGRRGGRGGPGK | 1115 |
| 3D view using mol* of 807 (AA BP:573) | ||||||||||
 | ||||||||||
| PDB file (818) >>>818.pdbFusion protein BP residue: 550 CIF file (818) >>>818.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLK PYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLSPRIEITPSHE LIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPA SSGSSASFISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIM SPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQ RSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPP KMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILL VPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHH RAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILK LRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSLQTASNPIECC YTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPA YGQQPAATAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVP GSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSY GQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMG VYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGA GERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDDL ADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAAV EWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGGP GGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPNP GCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRGG LMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLME | 1129 |
| 3D view using mol* of 818 (AA BP:550) | ||||||||||
 | ||||||||||
| PDB file (820) >>>820.pdbFusion protein BP residue: 550 CIF file (820) >>>820.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MQREAAFRLGHCHPLRIMGSVDQEEPNAHKVASPPSGPAYPDDVLDYGLK PYSPLASLSGEPPGRFGEPDRVGPQKFLSAAKPAGASGLSPRIEITPSHE LIQAVGPLRMRDAGLLVEQPPLAGVAASPRFTLPVPGFEGYREPLCLSPA SSGSSASFISDTFSPYTSPCVSPNNGGPDDLCPQFQNIPAHYSPRTSPIM SPRTSLAEDSCLGRHSPVPRPASRSSSPGAKRRHSCAEALVALPPGASPQ RSRSPSPQPSSHVAPQDHGSPAGYPPVAGSAVIMDALNSLATDSPCGIPP KMWKTSPDPSPVSAAPSKAGLPRHIYPAVEFLGPCEQGERRNSAPESILL VPPTWPKPLVPAIPICSIPVTASLPPLEWPLSSQSGSYELRIEVQPKPHH RAHYETEGSRGAVKAPTGGHPVVQLHGYMENKPLGLQIFIGTADERILKP HAFYQVHRITGKTVTTTSYEKIVGNTKVLEIPLEPKNNMRATIDCAGILK LRNADIELRKGETDIGRKNTRVRLVFRVHIPESSGRIVSLQTASNPIECC YTTPTAPQAYSQPVQGYGTGAYDTTTATVTTTQASYAAQSAYGTQPAYPA YGQQPAATAPTRPQDGNKPTETSQPQSSTGGYNQPSLGYGQSNYSYPQVP GSYPMQPVTAPPSYPPTSYSSTQPTSYDQSSYSQQNTYGQPSSYGQQSSY GQQSSYGQQPPTSYPPQTGSYSQAPSQYSQQSSSYGQQSSFRQDHPSSMG VYGQESGGFSGPGENRSMSGPDNRGRGRGGFDRGGMSRGGRGGGRGGMGS AGERGGFNKPGGPMDEGPDLDLGPPVDPDEDSDNSAIYVQGLNDSVTLDD LADFFKQCGVVKMNKRTGQPMIHIYLDKETGKPKGDATVSYEDPPTAKAA VEWFDGKDFQGSKLKVSLARKKPPMNSMRGGLPPREGRGMPPPLRGGPGG PGGPGGPMGRMGGRGGDRGGFPPRGPRGSRGNPSGGGNVQHRAGDWQCPN PGCGNQNFAWRTECNQCKAPKPEGFLPPPFPPPGGDRGRGGPGGMRGGRG GLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGGPGGPPGPLM | 1130 |
| 3D view using mol* of 820 (AA BP:550) | ||||||||||
 | ||||||||||
| PDB file (831) >>>831.pdbFusion protein BP residue: 573 CIF file (831) >>>831.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPN AHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKF LSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAA SPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGG PDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSS PGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGSPAGYPPV AGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYP AVEFLGPCEQGERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPL EWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTK VLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFR VHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTA TVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQS STGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSY DQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGSYSQAPSQ YSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRG RGGFDRGGMSRGGRGGGRGGMGAGERGGFNKPGGPMDEGPDLDLGPPVDP DEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLDK ETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNSM RGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPRG SRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLPP PFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRG MDRGGFGGGRRGGPGGPPGPLMEQMGGRRGGRGGPGKMDKGEHRQERRDR | 1152 |
| 3D view using mol* of 831 (AA BP:573) | ||||||||||
 | ||||||||||
| PDB file (833) >>>833.pdbFusion protein BP residue: 573 CIF file (833) >>>833.cif | NFATC2 | chr20 | 50090516 | - | EWSR1 | chr22 | 29674018 | + | MRAMNAPERQPQPDGGDAPGHEPGGSPQDELDFSILFDYEYLNPNEEEPN AHKVASPPSGPAYPDDVLDYGLKPYSPLASLSGEPPGRFGEPDRVGPQKF LSAAKPAGASGLSPRIEITPSHELIQAVGPLRMRDAGLLVEQPPLAGVAA SPRFTLPVPGFEGYREPLCLSPASSGSSASFISDTFSPYTSPCVSPNNGG PDDLCPQFQNIPAHYSPRTSPIMSPRTSLAEDSCLGRHSPVPRPASRSSS PGAKRRHSCAEALVALPPGASPQRSRSPSPQPSSHVAPQDHGSPAGYPPV AGSAVIMDALNSLATDSPCGIPPKMWKTSPDPSPVSAAPSKAGLPRHIYP AVEFLGPCEQGERRNSAPESILLVPPTWPKPLVPAIPICSIPVTASLPPL EWPLSSQSGSYELRIEVQPKPHHRAHYETEGSRGAVKAPTGGHPVVQLHG YMENKPLGLQIFIGTADERILKPHAFYQVHRITGKTVTTTSYEKIVGNTK VLEIPLEPKNNMRATIDCAGILKLRNADIELRKGETDIGRKNTRVRLVFR VHIPESSGRIVSLQTASNPIECCYTTPTAPQAYSQPVQGYGTGAYDTTTA TVTTTQASYAAQSAYGTQPAYPAYGQQPAATAPTRPQDGNKPTETSQPQS STGGYNQPSLGYGQSNYSYPQVPGSYPMQPVTAPPSYPPTSYSSTQPTSY DQSSYSQQNTYGQPSSYGQQSSYGQQSSYGQQPPTSYPPQTGSYSQAPSQ YSQQSSSYGQQSSFRQDHPSSMGVYGQESGGFSGPGENRSMSGPDNRGRG RGGFDRGGMSRGGRGGGRGGMGSAGERGGFNKPGGPMDEGPDLDLGPPVD PDEDSDNSAIYVQGLNDSVTLDDLADFFKQCGVVKMNKRTGQPMIHIYLD KETGKPKGDATVSYEDPPTAKAAVEWFDGKDFQGSKLKVSLARKKPPMNS MRGGLPPREGRGMPPPLRGGPGGPGGPGGPMGRMGGRGGDRGGFPPRGPR GSRGNPSGGGNVQHRAGDWQCPNPGCGNQNFAWRTECNQCKAPKPEGFLP PPFPPPGGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGR GMDRGGFGGGRRGGPGGPPGPLMEQMGGRRGGRGGPGKMDKGEHRQERRD | 1153 |
| 3D view using mol* of 833 (AA BP:573) | ||||||||||
 | ||||||||||
Top |
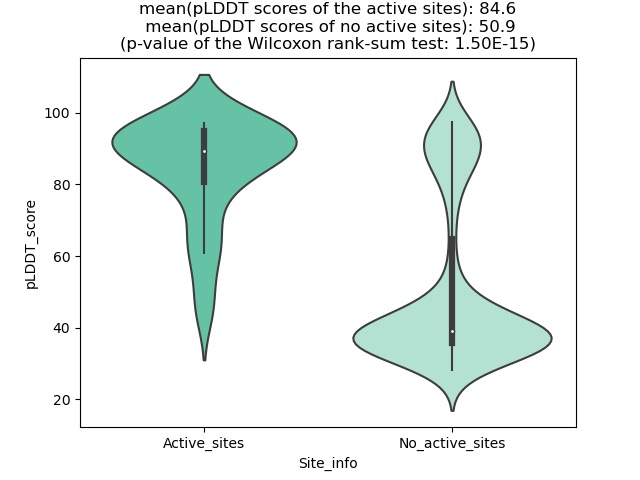
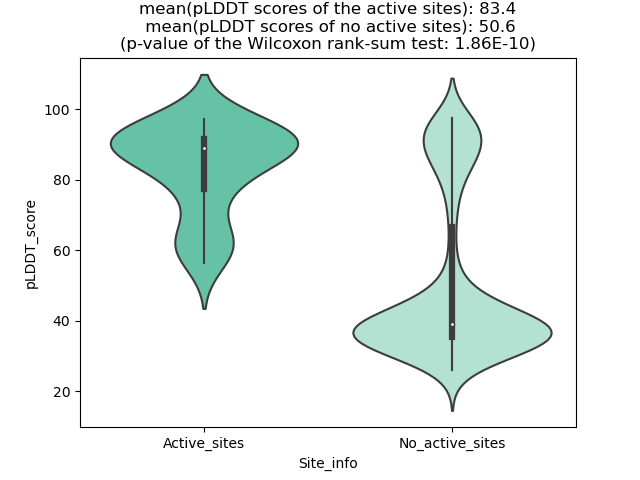
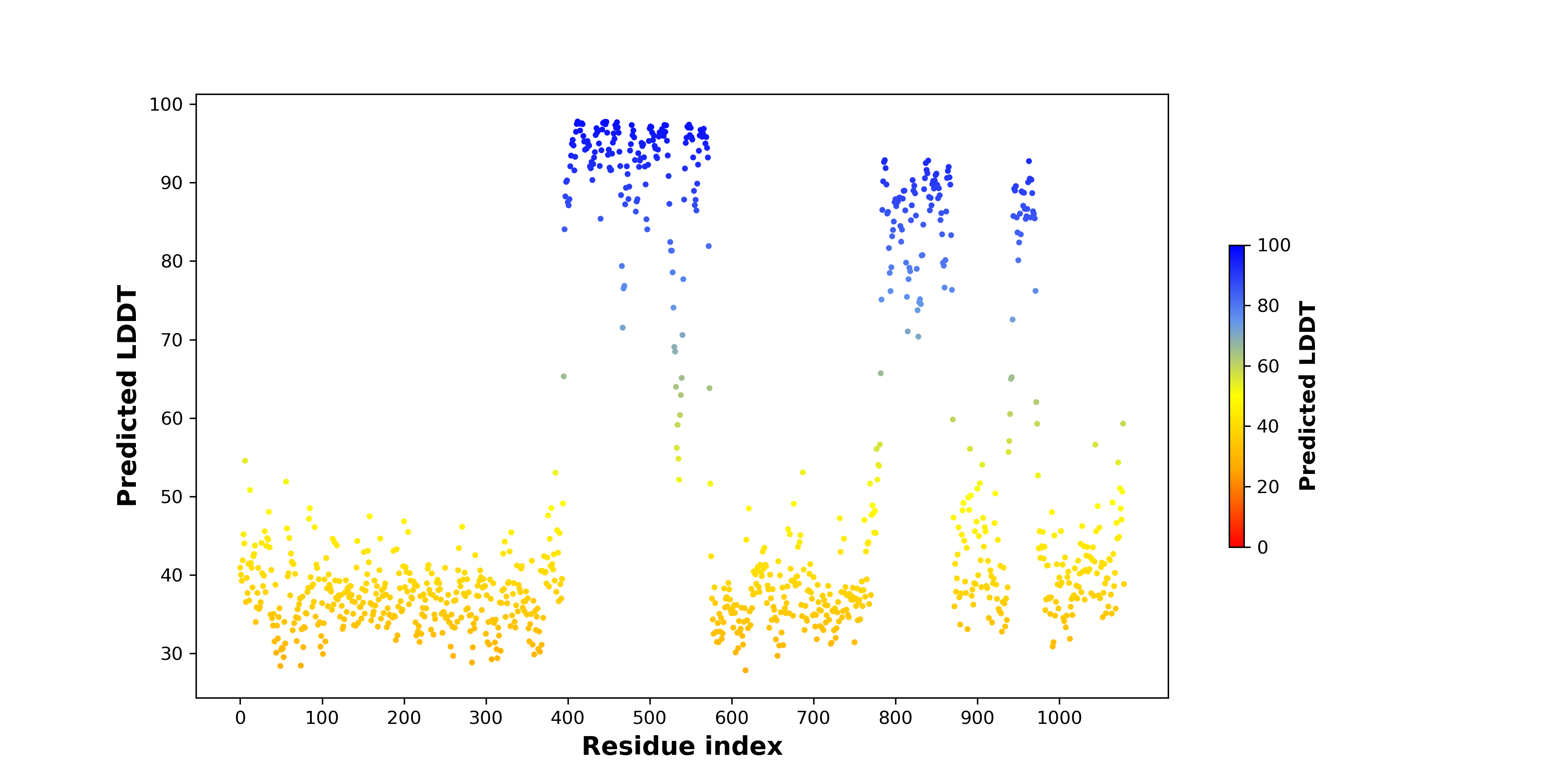
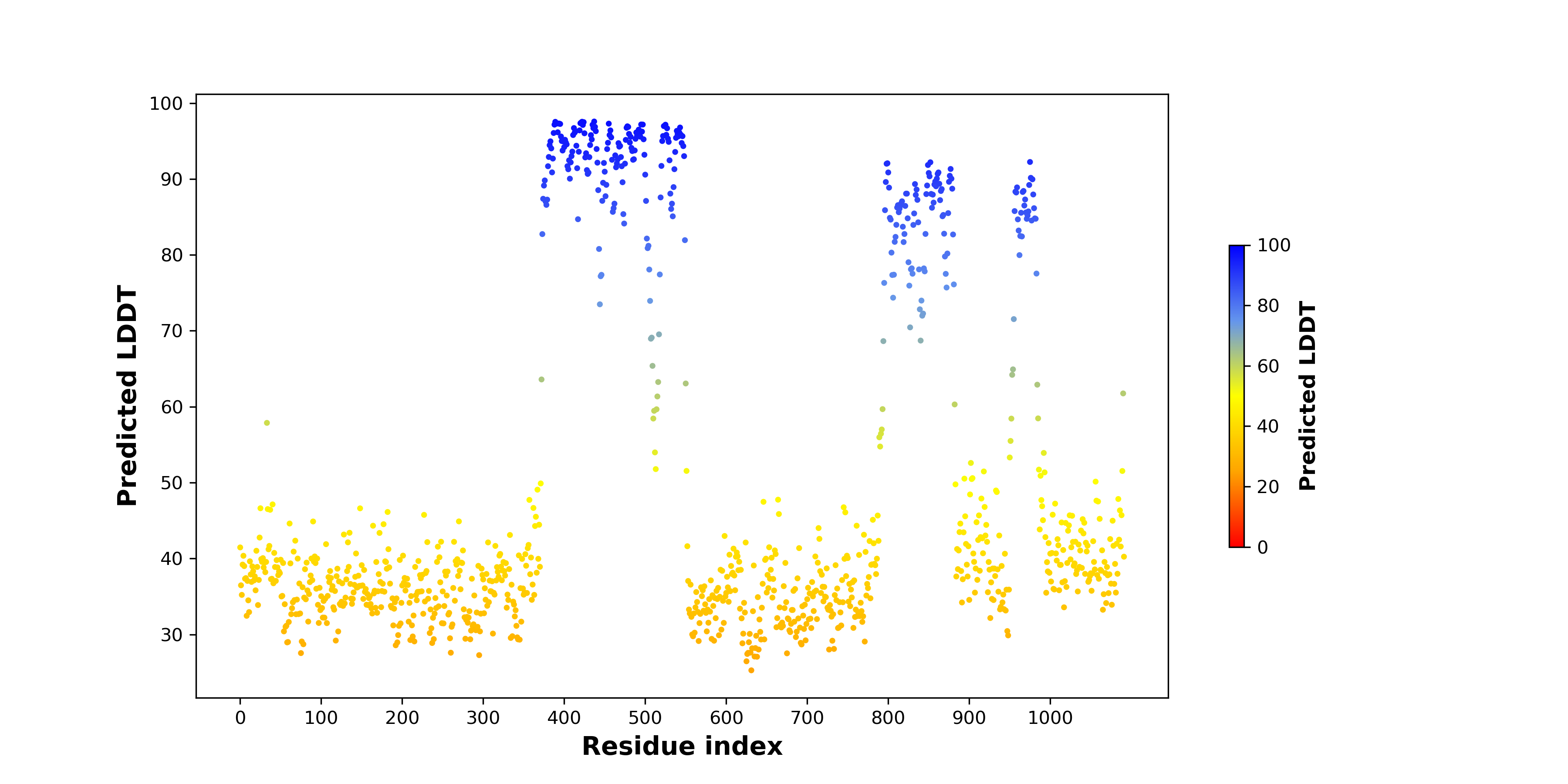
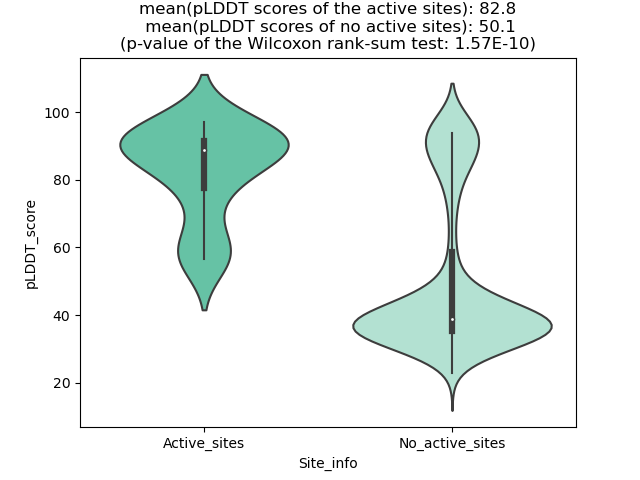
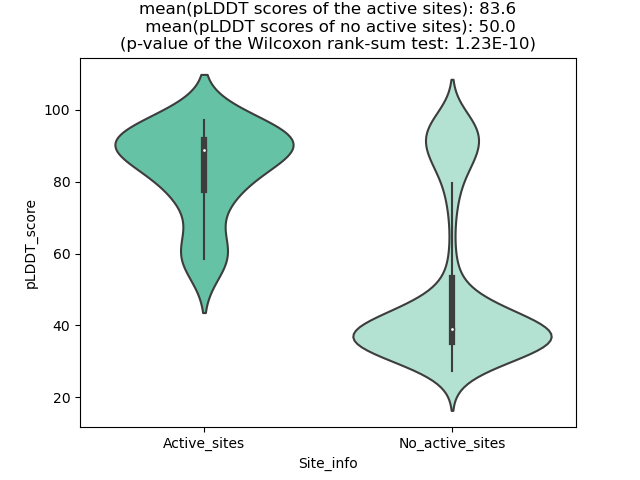
pLDDT score distribution |
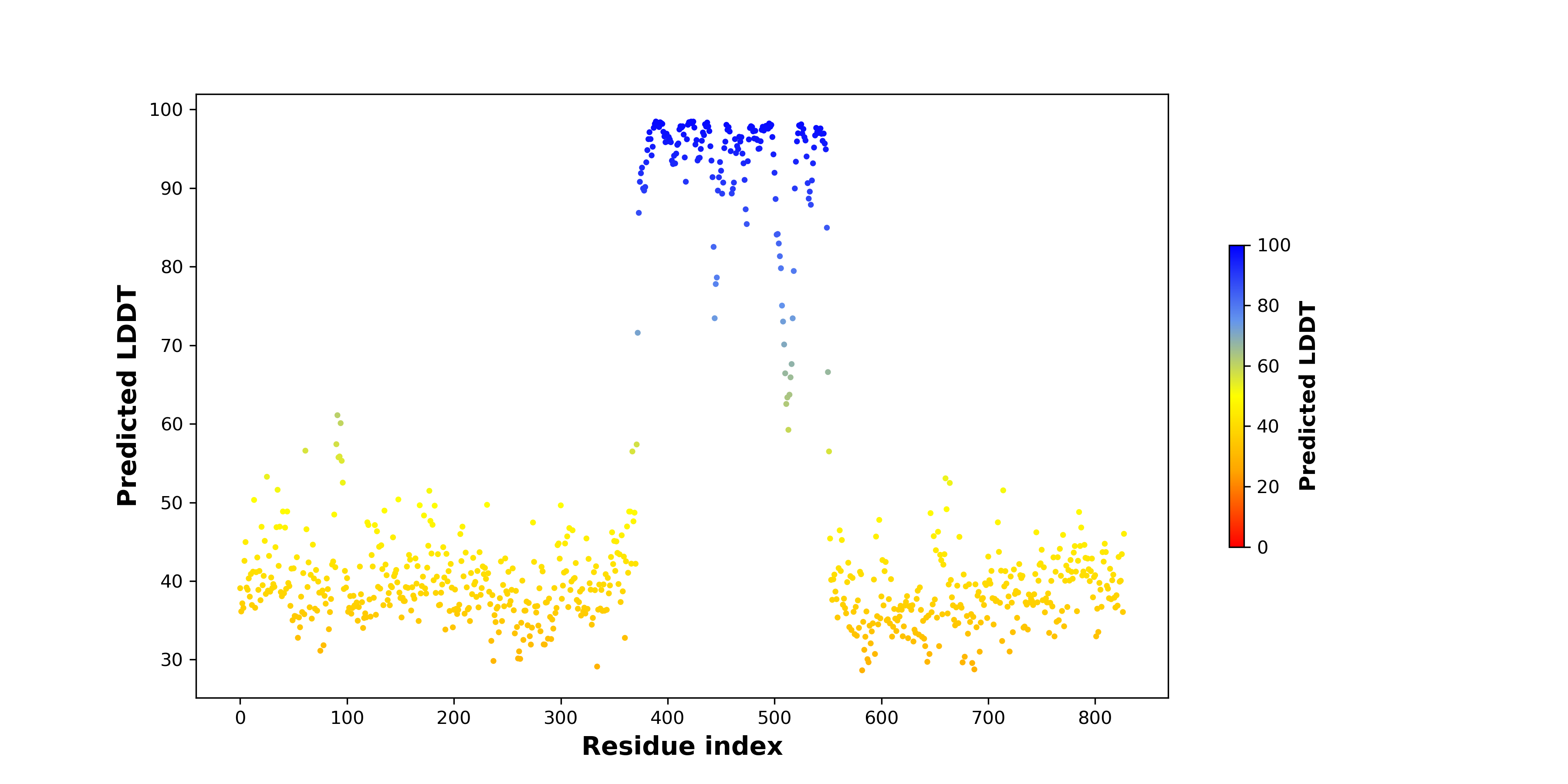
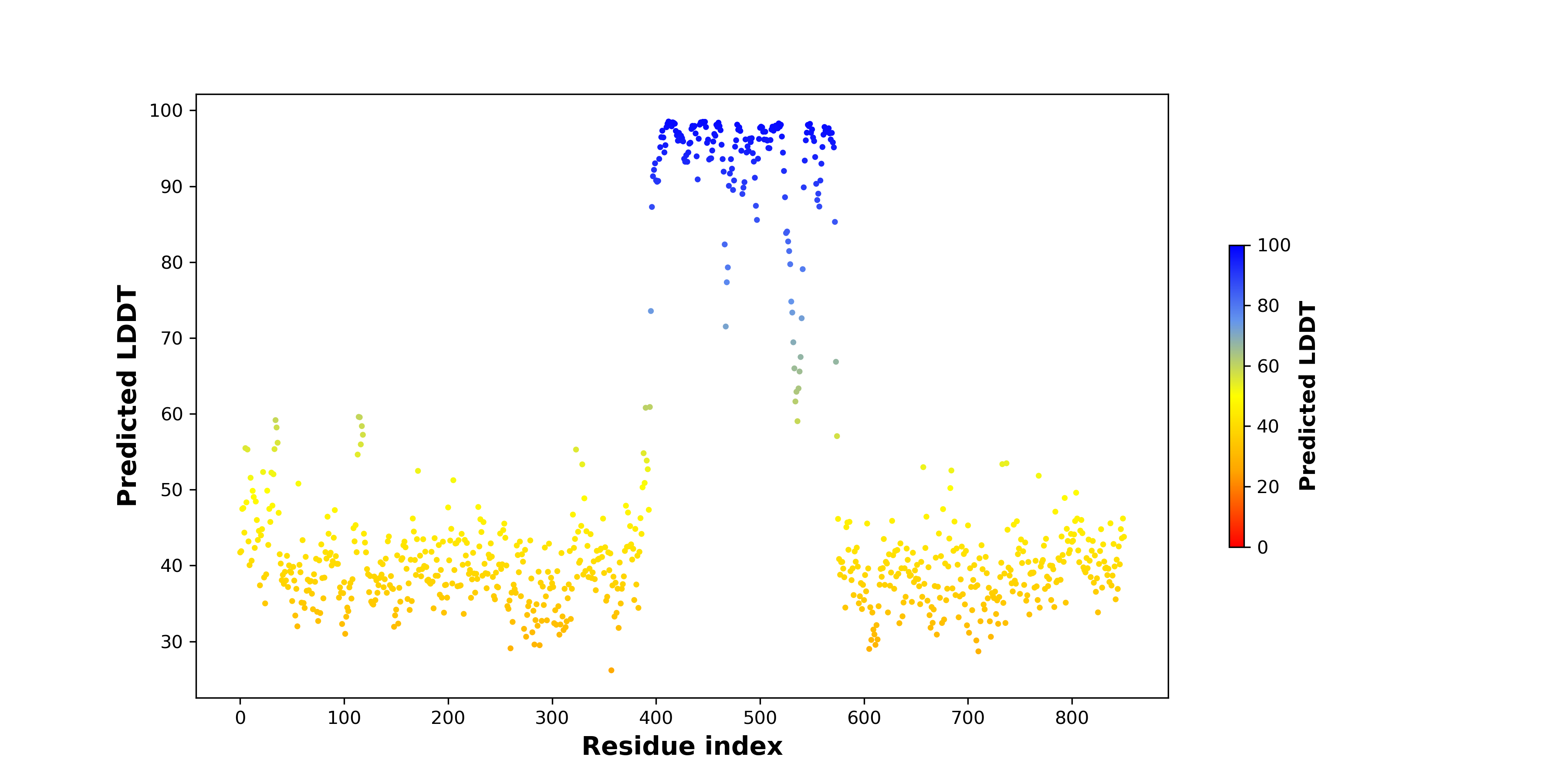
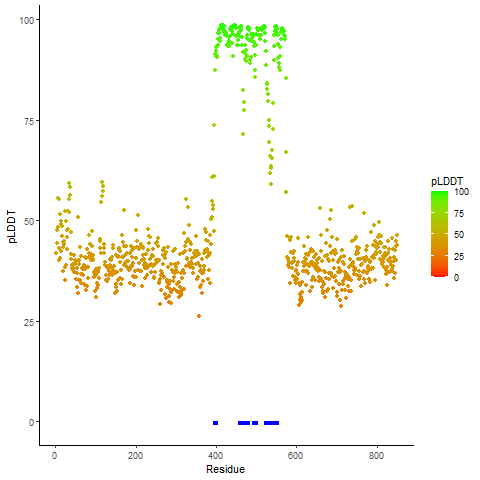
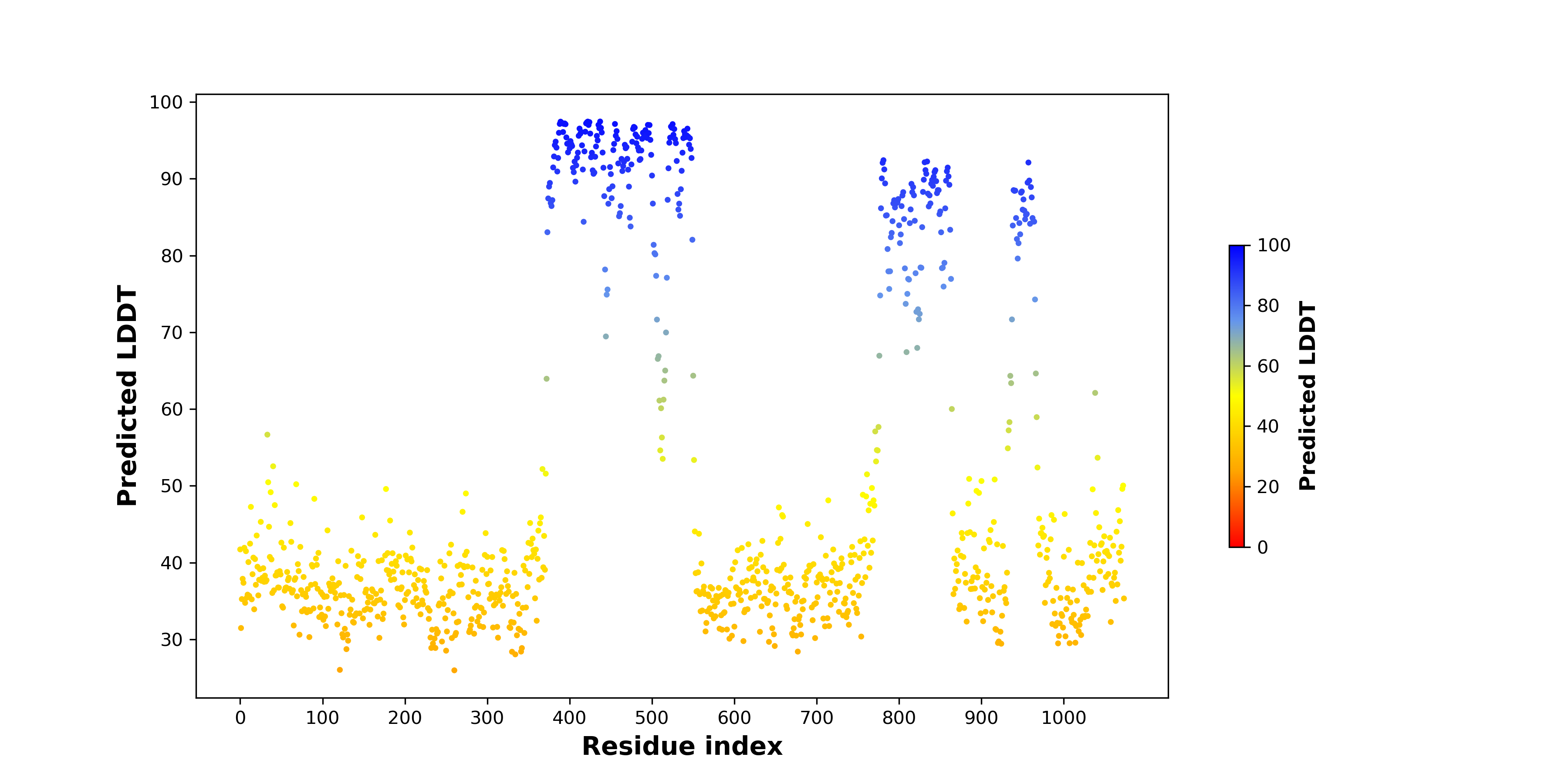
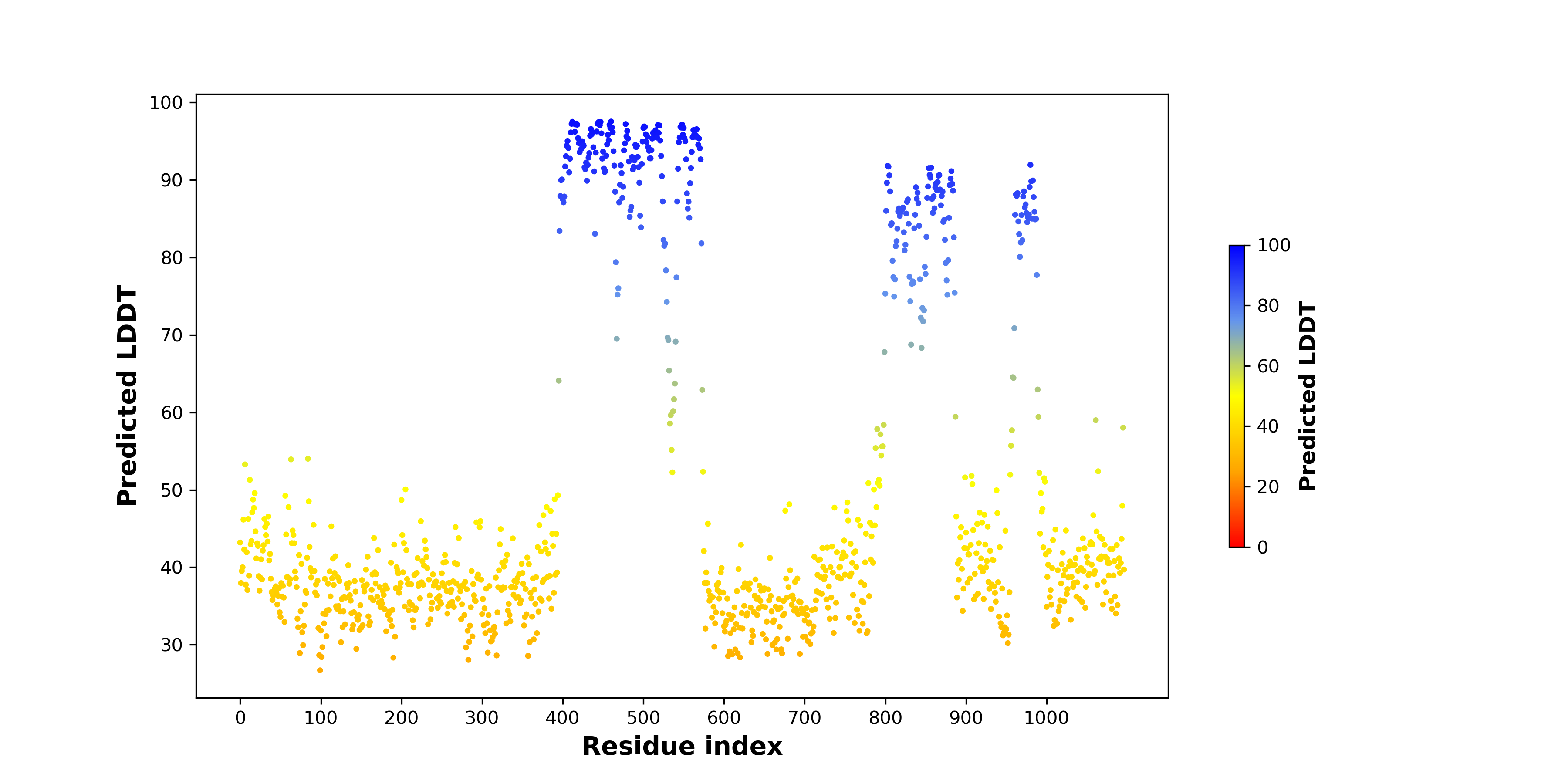
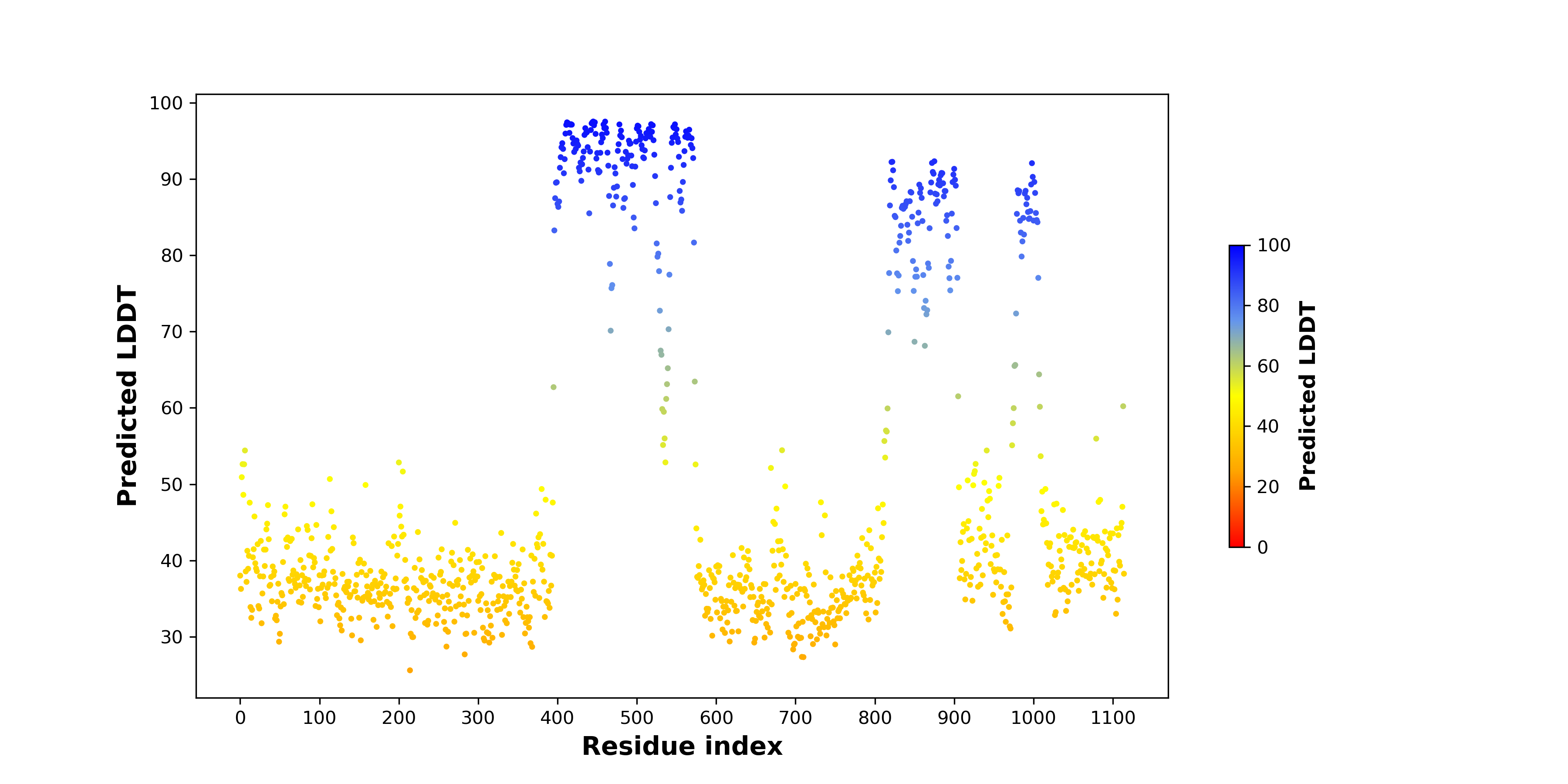
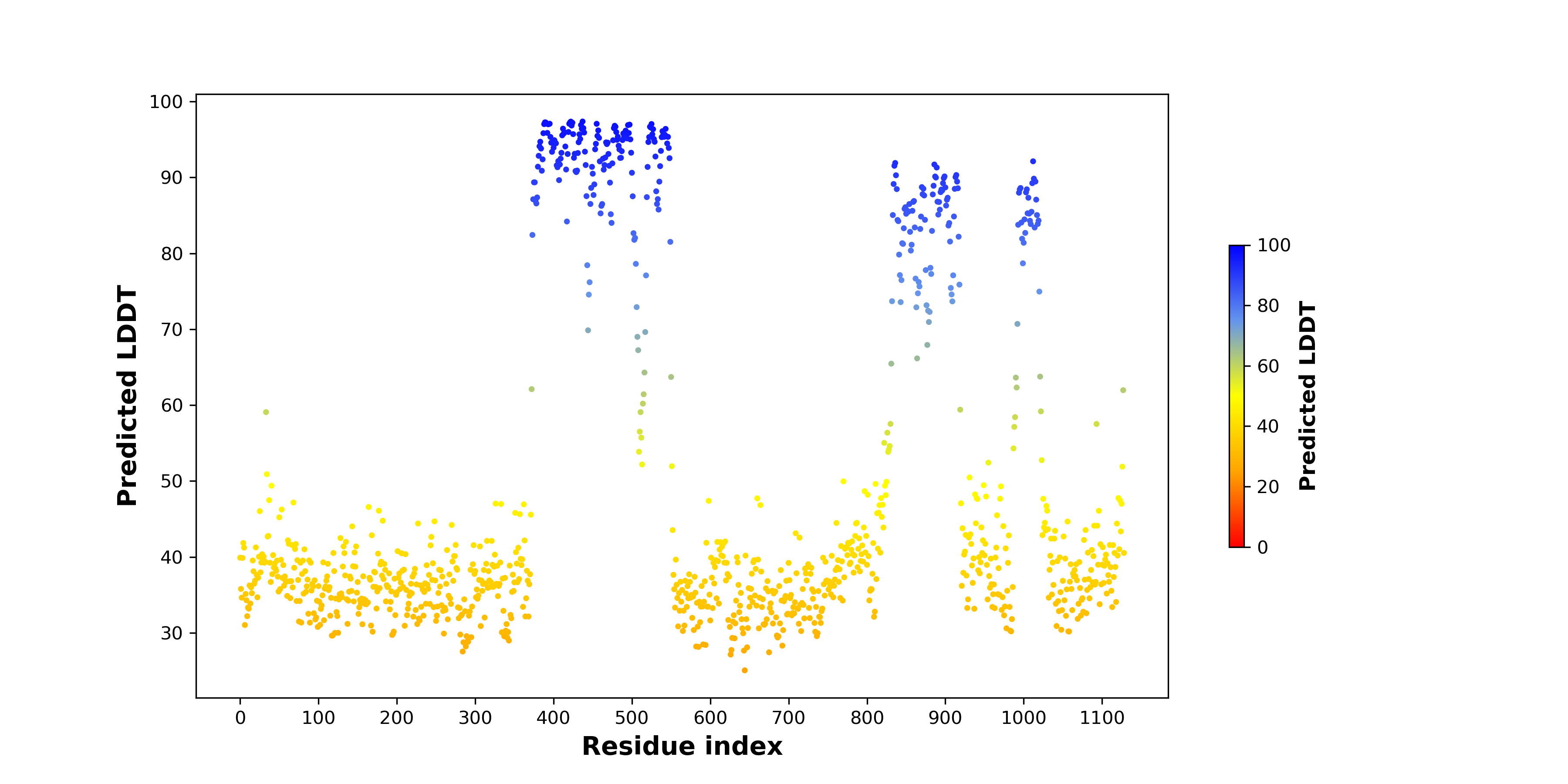
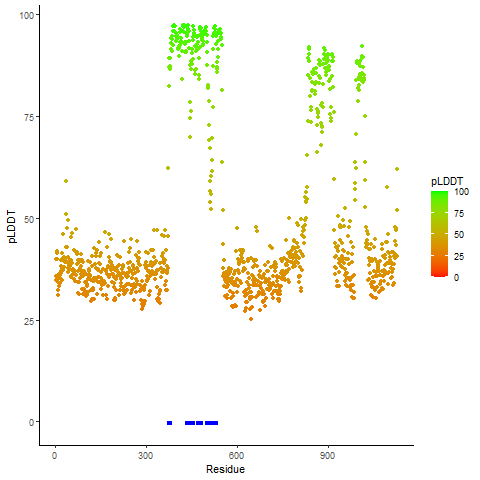
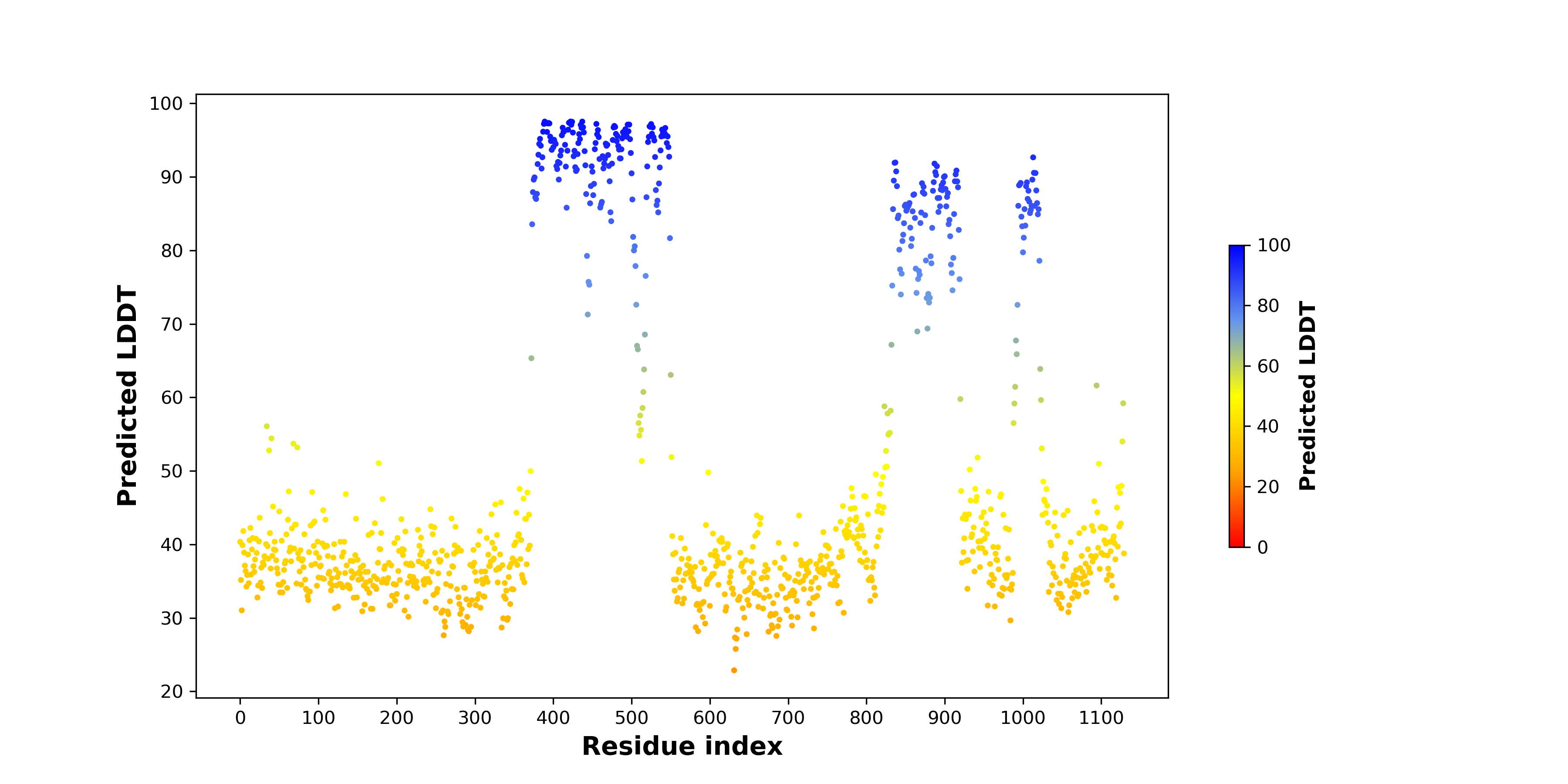
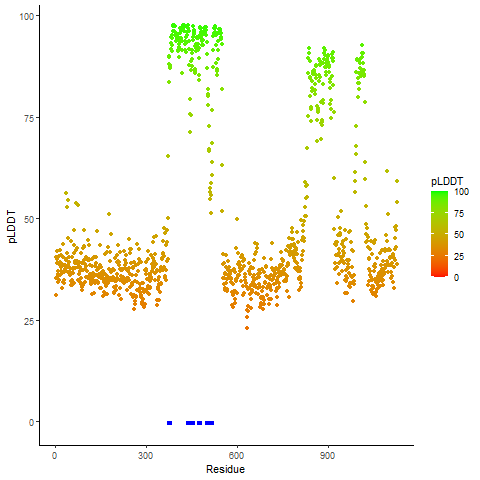
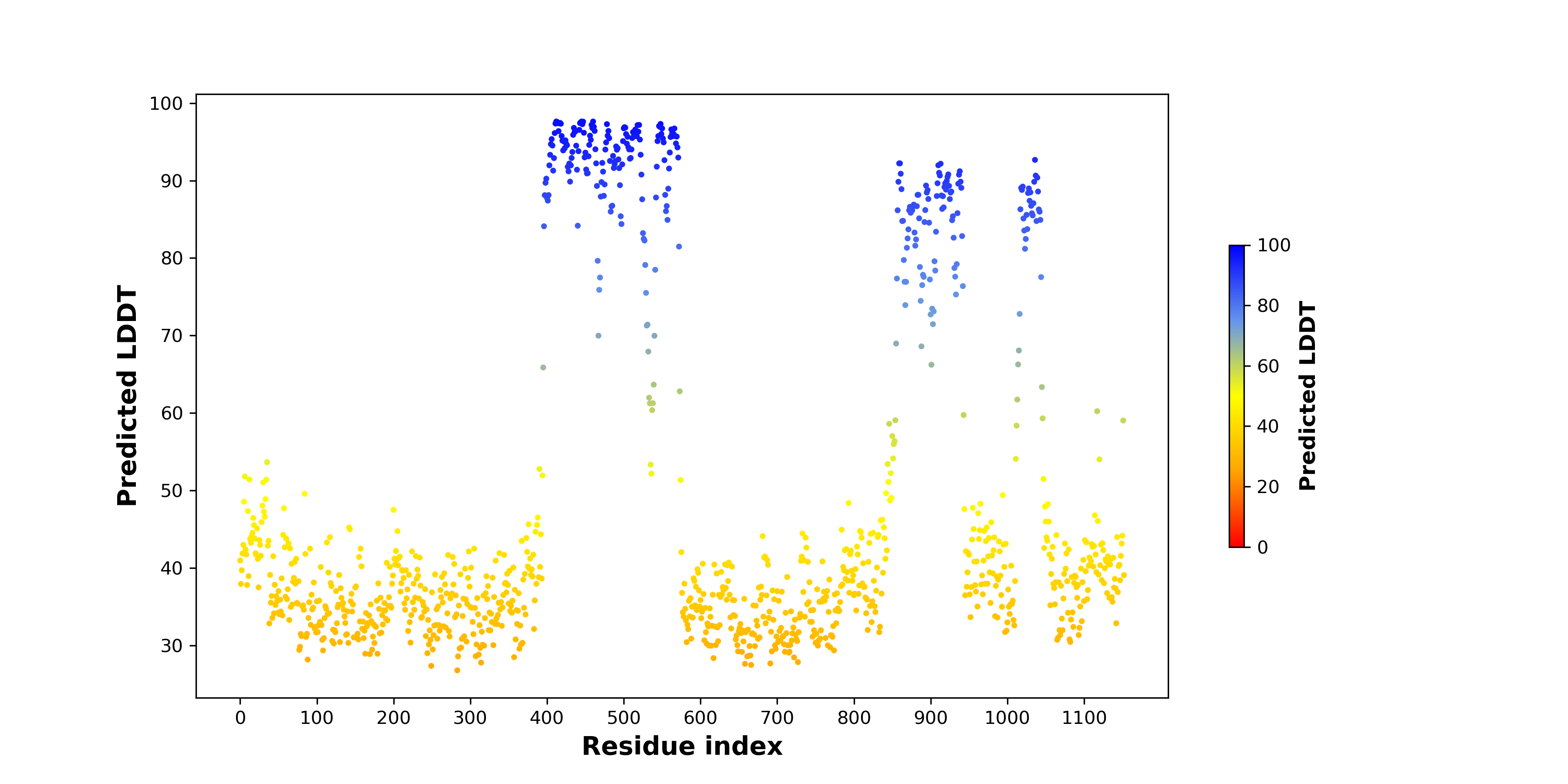
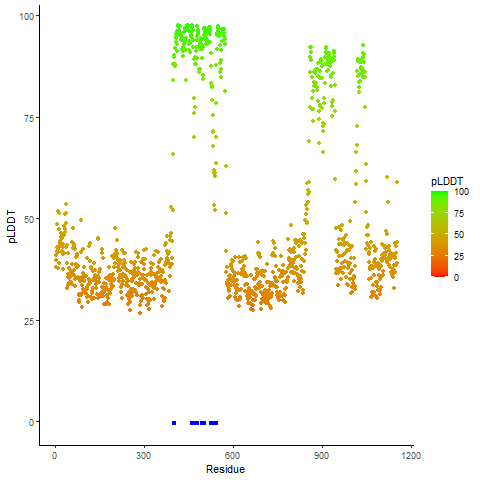
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
EWSR1_pLDDT.png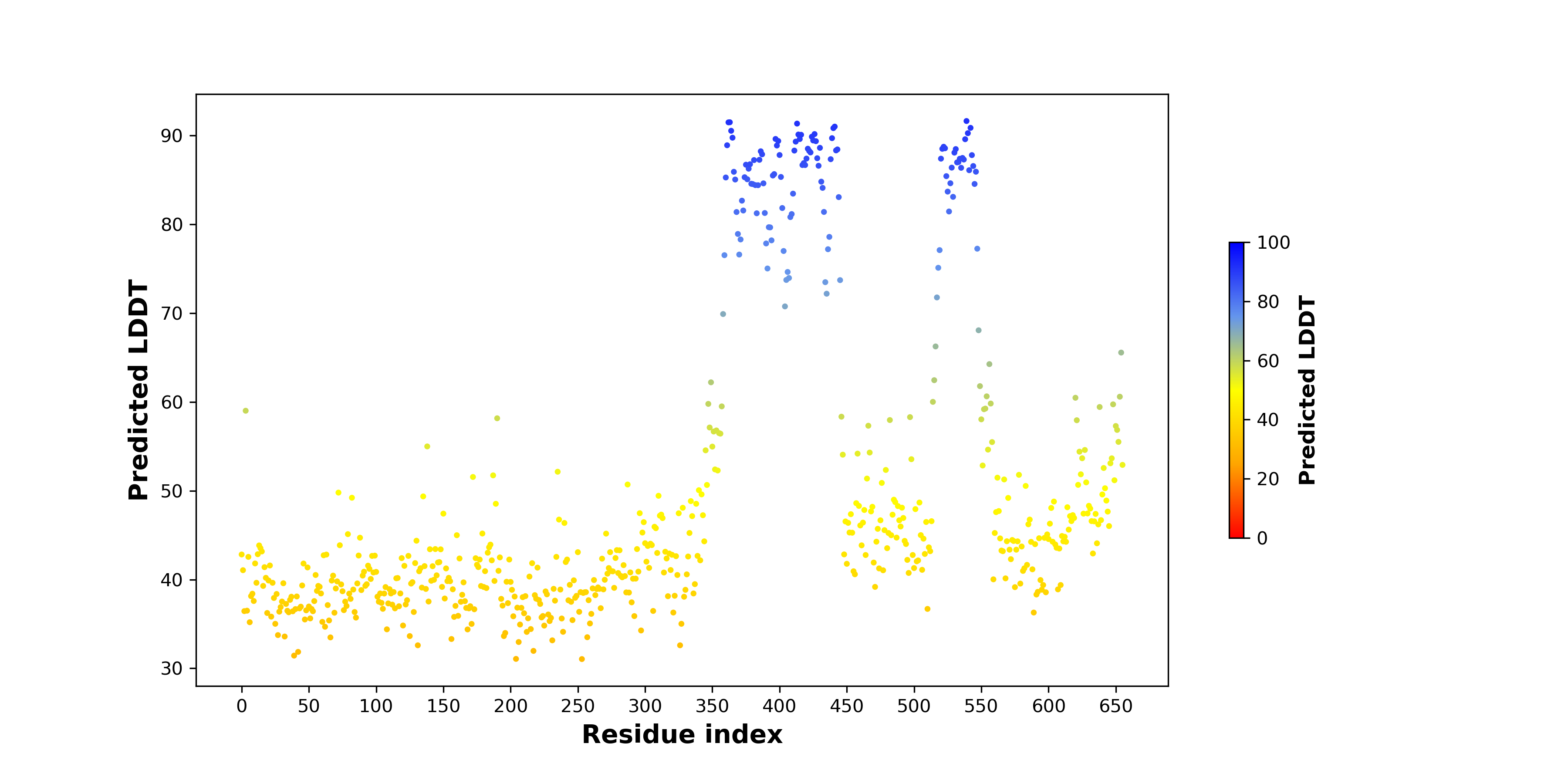 |
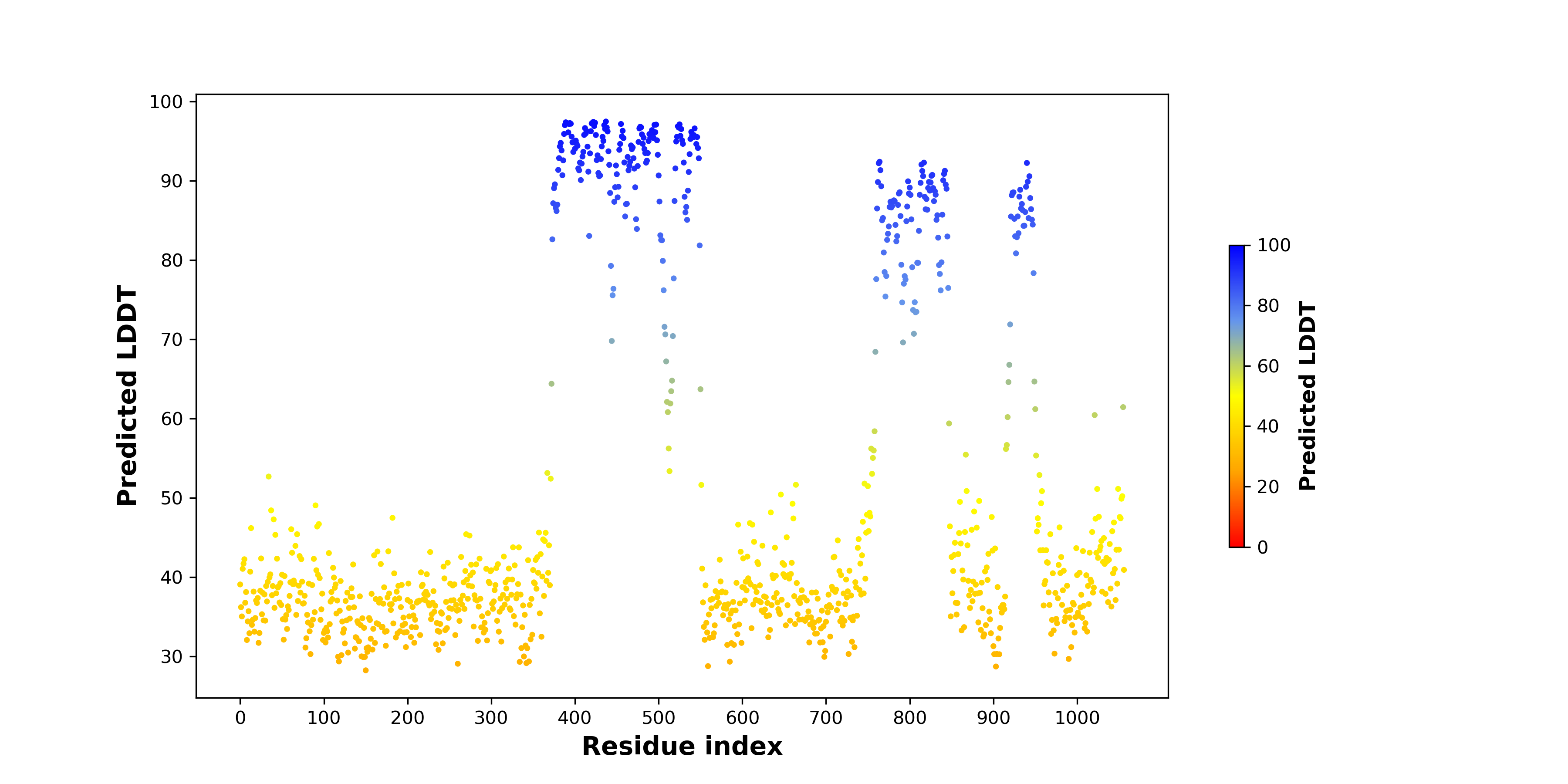
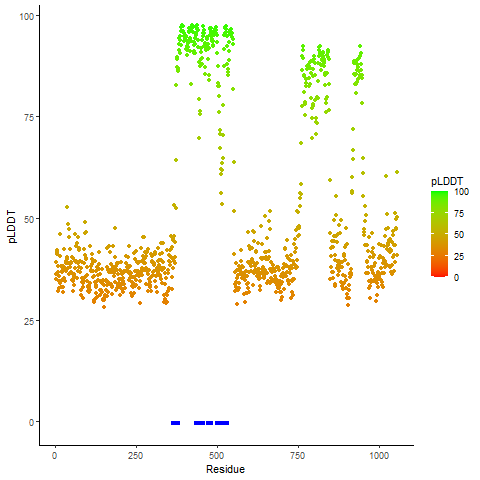
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
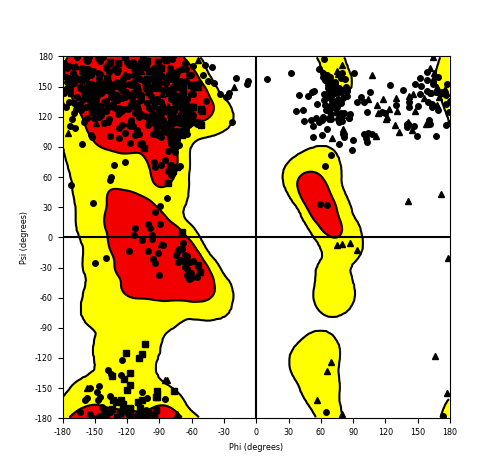
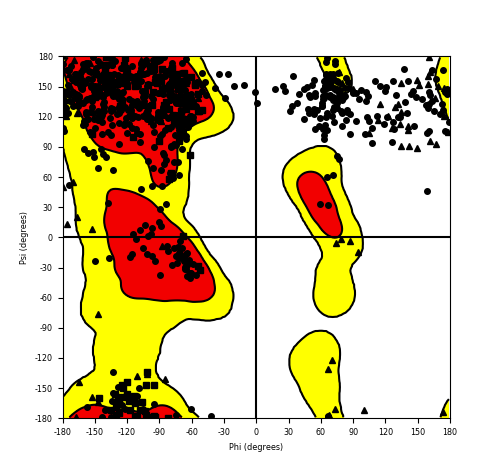
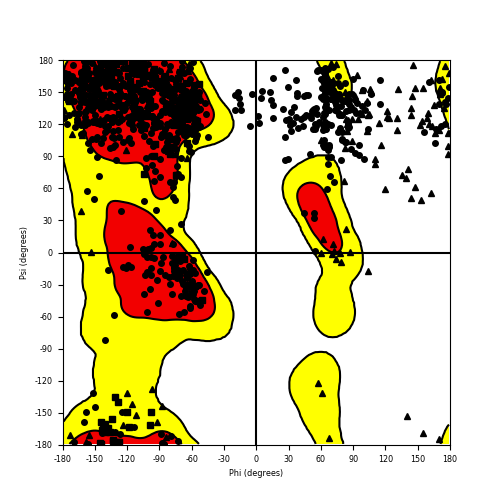
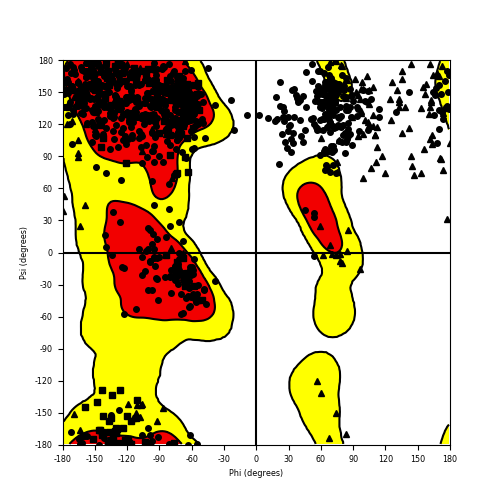
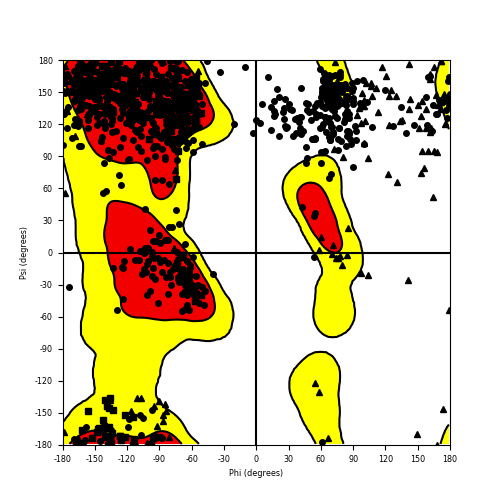
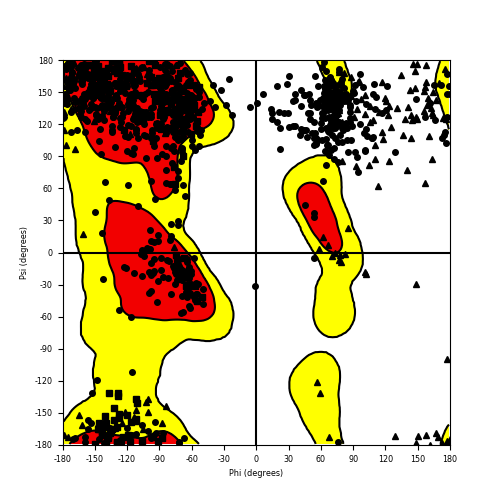
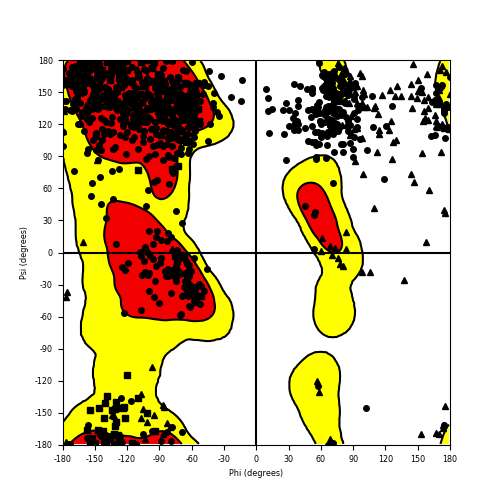
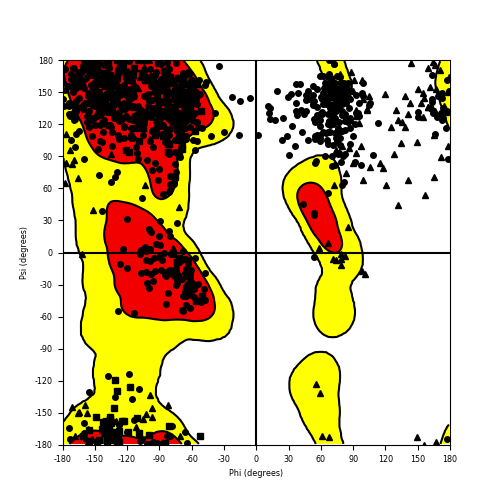
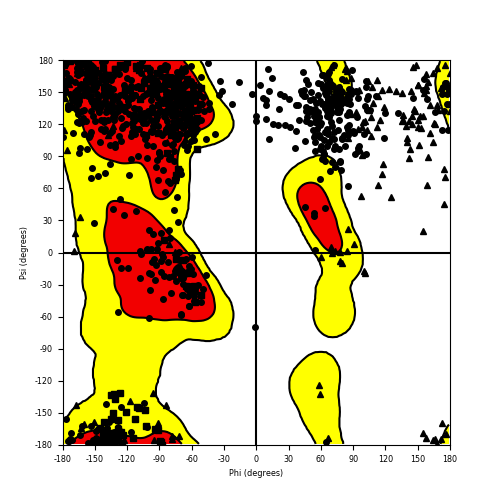
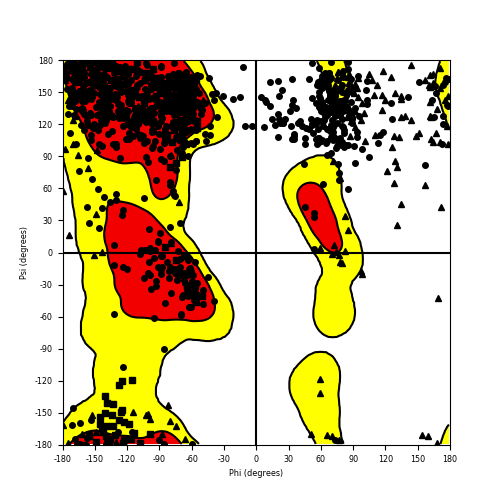
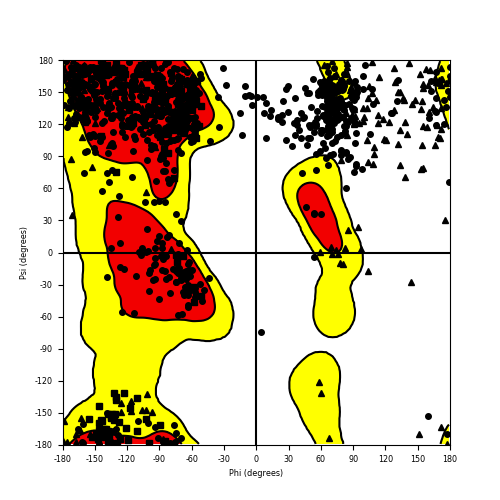
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
Top |
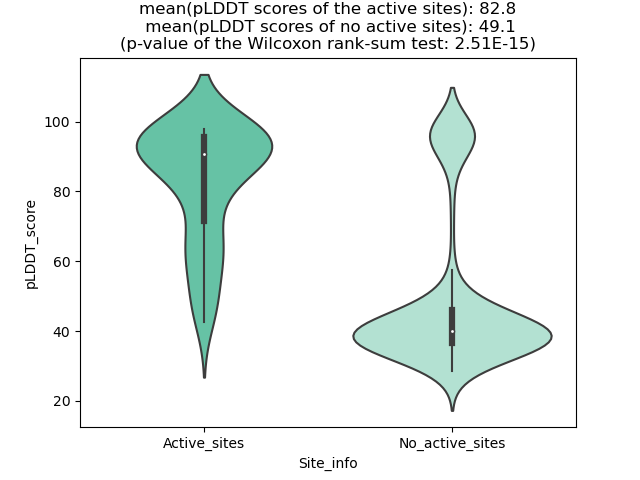
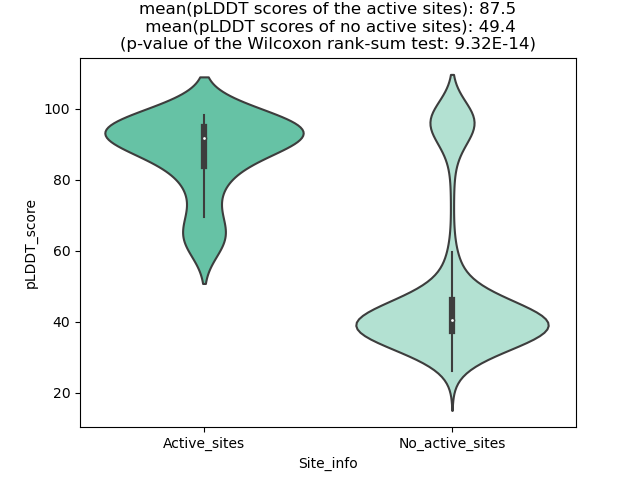
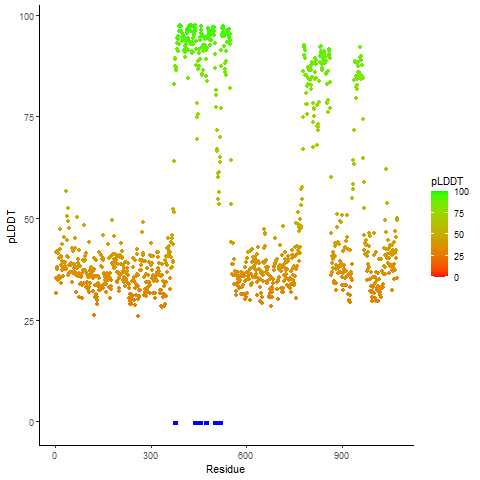
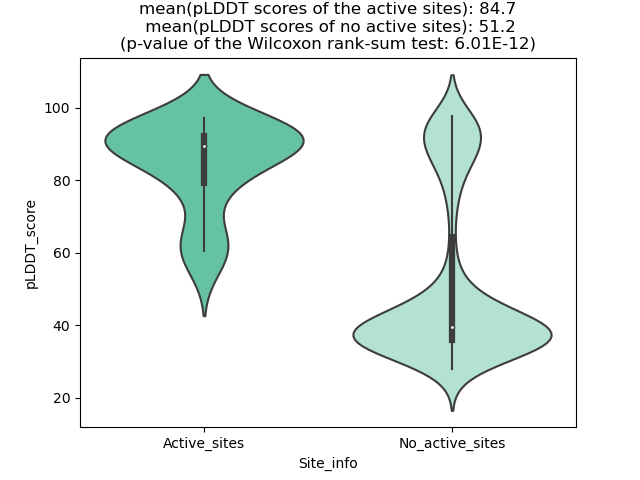
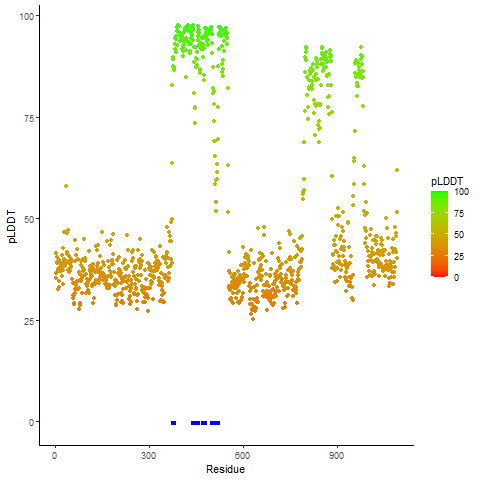
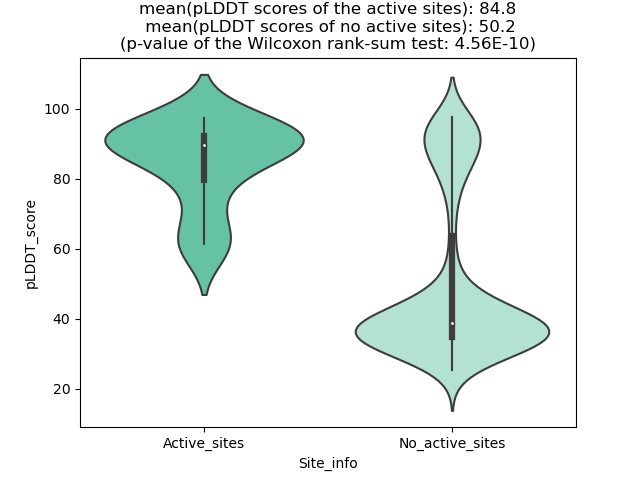
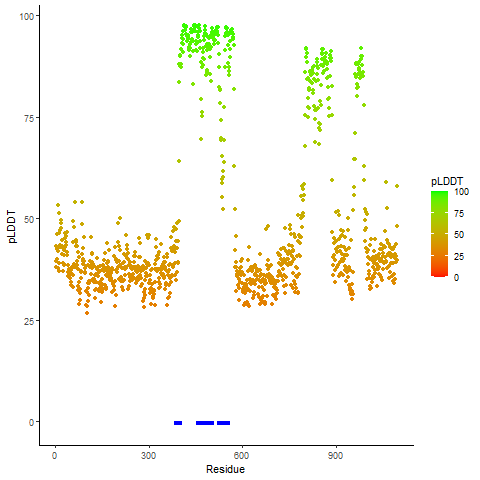
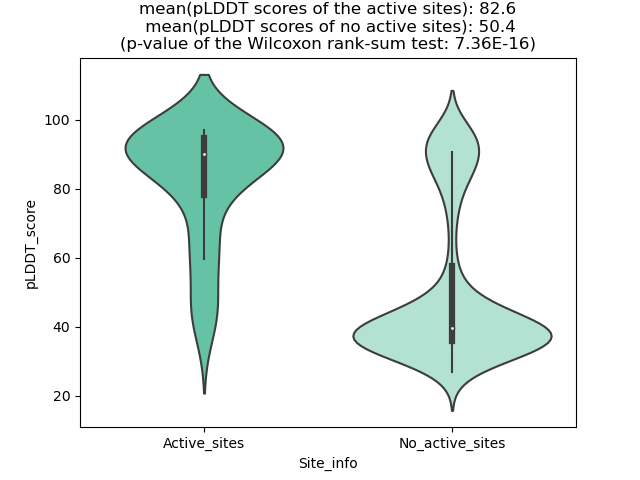
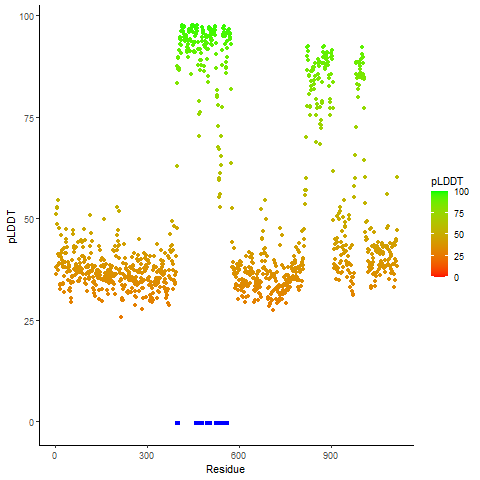
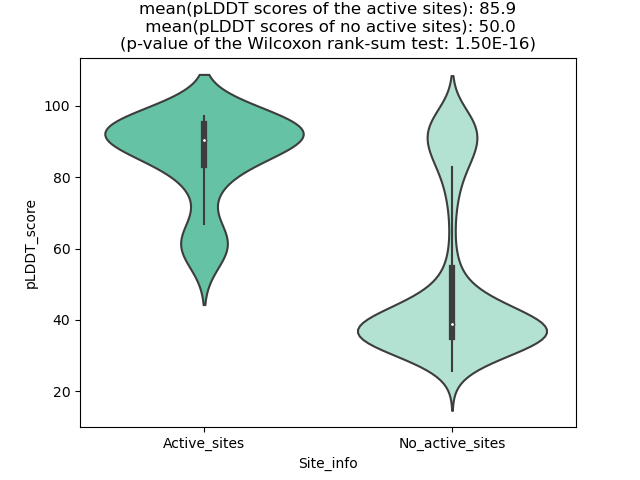
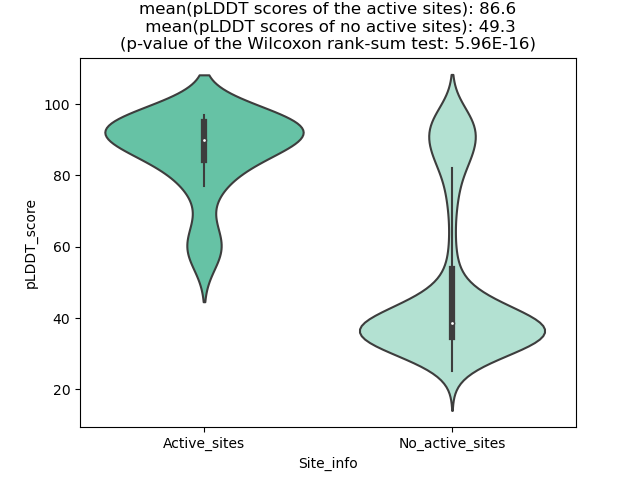
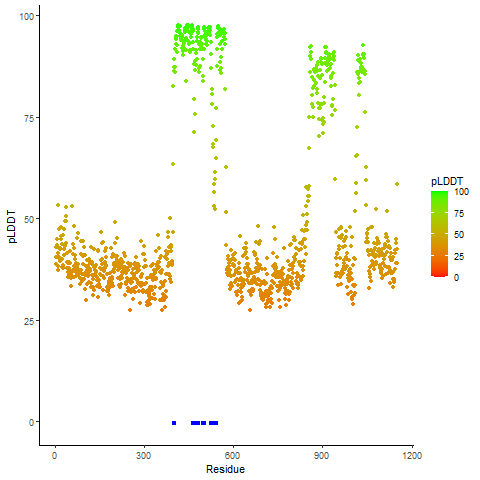
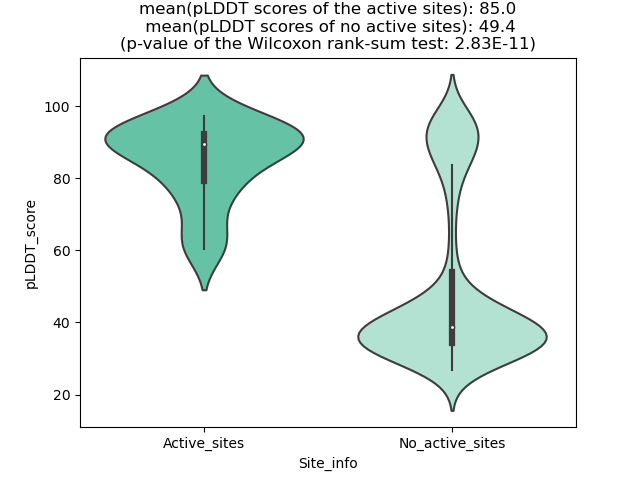
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 669 | 0.996 | 154 | 0.996 | 491.176 | 0.636 | 0.692 | 0.874 | 0.417 | 1.101 | 0.379 | 0.777 | Chain A: 362,363,365,366,368,374,375,376,377,378,4 34,435,437,440,442,444,450,451,452,453,454,455,469 ,471,473,474,475,476,477,480,501,506,509,510,512,5 13,514,515,516,519,527,528,529 |
| 677 | 0.982 | 137 | 0.965 | 351.575 | 0.586 | 0.671 | 0.895 | 0.364 | 1.16 | 0.314 | 0.596 | Chain A: 397,398,399,400,401,460,462,463,465,467,4 72,473,474,475,476,477,478,479,494,496,497,498,499 ,500,524,529,532,533,535,536,538,539,542,550,552 |
| 782 | 0.994 | 196 | 1.02 | 423.948 | 0.561 | 0.677 | 0.896 | 0.52 | 1.002 | 0.519 | 0.889 | Chain A: 365,366,368,374,375,376,377,378,434,435,4 37,439,440,442,444,449,450,451,452,453,454,455,456 ,471,473,474,475,476,477,480,501,506,509,510,512,5 15,516,519,527,528,529,531 |
| 789 | 1.024 | 134 | 1.017 | 298.067 | 0.483 | 0.734 | 1.048 | 0.806 | 1.116 | 0.723 | 0.508 | Chain A: 376,377,378,439,440,442,444,449,450,451,4 52,453,454,455,456,473,474,475,476,477,501,506,509 ,510,512,513,515,516,519 |
| 793 | 1.017 | 161 | 1.066 | 327.565 | 0.536 | 0.666 | 0.903 | 0.816 | 0.84 | 0.971 | 0.522 | Chain A: 397,398,399,400,401,460,462,463,465,467,4 72,473,474,475,476,477,478,494,496,497,498,499,524 ,529,532,533,535,536,538,539,542,550,552 |
| 798 | 1.013 | 118 | 0.937 | 254.506 | 0.482 | 0.718 | 1.038 | 0.37 | 1.33 | 0.278 | 0.653 | Chain A: 377,378,439,440,442,449,450,451,452,453,4 54,455,456,473,474,475,476,477,501,506,509,510,512 ,515,516,519 |
| 802 | 1.0092 | 186 | 1.0404 | 451.731 | 0.6009 | 0.6878 | 0.8983 | 0.5254 | 0.9579 | 0.5485 | 0.7996 | Chain A: 386,388,389,390,391,397,398,399,400,401,4 57,458,460,462,463,465,467,472,473,474,475,476,477 ,478,479,492,494,496,497,498,499,500,503,524,529,5 32,533,535,538,539,542,550,551,552,554 |
| 807 | 1.02 | 179 | 1.055 | 421.204 | 0.564 | 0.695 | 0.894 | 0.982 | 0.926 | 1.06 | 0.778 | Chain A: 396,397,398,399,400,401,457,458,460,462,4 63,465,472,473,474,475,476,477,478,479,494,496,497 ,498,499,500,503,524,529,532,533,535,536,538,539,5 42,550,551,552,554,560 |
| 818 | 0.972 | 163 | 1.006 | 407.827 | 0.644 | 0.633 | 0.789 | 0.44 | 0.965 | 0.456 | 0.81 | Chain A: 374,375,376,377,378,434,435,437,439,440,4 42,444,449,450,451,452,453,454,455,456,471,473,474 ,475,476,477,480,501,506,509,512,513,515,516,519,5 27,528,529 |
| 820 | 1.033 | 134 | 0.996 | 333.396 | 0.457 | 0.747 | 1.043 | 0.54 | 1.204 | 0.449 | 0.87 | Chain A: 377,378,439,440,442,444,449,450,451,452,4 53,454,455,456,473,474,475,476,477,501,506,509,510 ,512,513,515,516,519 |
| 831 | 0.9878 | 123 | 1.0013 | 288.12 | 0.5305 | 0.6798 | 0.9243 | 0.5865 | 1.0631 | 0.5517 | 0.7099 | Chain A: 400,401,462,463,465,467,470,472,473,474,4 75,476,477,478,479,496,497,498,499,500,524,529,532 ,535,536,538,539,542 |
| 833 | 0.9995 | 107 | 0.9375 | 231.868 | 0.5447 | 0.6973 | 0.9926 | 0.2829 | 1.2911 | 0.2191 | 0.5905 | Chain A: 399,400,401,462,463,465,467,470,472,473,4 74,475,476,477,478,479,496,497,498,499,500,524,529 ,532,533,535,538,539,542 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| EWSR1 | PRTFDC1, ZDHHC3, MSC, SLC22A24, MYO1F, KXD1, KHDRBS2, DNAJB3, FASN, NLE1, MNS1, PRUNE2, WWP2, NDUFB1, BNIP3L, NSUN4, KRR1, WWP1, RMND5B, SLC1A1, RASL11B, DFFA, WDR37, RPS15A, CPSF6, C11orf16, YY1AP1, RNF183, MTCP1, TULP2, RBPMS, KEL, MYOZ2, FAM131C, HMGA1, NPPB, HERPUD1, CD177, RPL31, VPS72, ACTL6A, RAD23A, MAGEA11, CFDP1, DMRTB1, CXADR, ZNF165, SSBP2, TPGS2, RAB37, CETP, NDUFV1, DYNLL2, NBPF3, CEACAM5, GPBP1L1, SERP2, GNPDA1, C19orf57, ELAVL3, ELAVL4, LILRA3, BAD, CCDC7, MRPS18B, CUEDC2, CNST, TSPAN3, CCDC91, TRIM37, NINL, NTNG2, CPSF7, PGLS, EPT1, MYL6, SMAD4, TMSB4Y, TRPV5, MVK, MAPK1IP1L, MDFI, MTMR9, PLSCR1, RALYL, PDHX, C10orf12, RHOXF2, MATK, SALL2, AGT, KCNMB1, SUV39H2, SMNDC1, ARHGDIA, PUF60, GSK3B, ILK, CD2BP2, BARD1, CREBBP, BTK, SF1, SNRPC, ELK1, PTK2B, CALM1, POU4F1, EZH2, IRF3, TONSL, RFX3, HLTF, SUPT4H1, ZNF184, HIST1H2BN, BLZF1, HDAC3, FXR2, HMGN4, POLR3A, ECD, ZBTB1, SCMH1, SUZ12, E2F8, TRIM5, ZNF383, DHX9, SMN1, PCM1, RAD21, NDRG1, CEBPA, ELAVL1, SIRT7, HNRNPA1, TSG101, TP53, POLR2A, YBX1, TDRD3, CUL3, CUL4A, CUL4B, CUL5, CUL2, CUL1, COPS5, COPS6, DCUN1D1, CAND1, NEDD8, KCND3, ATN1, ATXN3, ERCC5, HBP1, HSPA2, PCBP1, USP7, SRSF5, TMEM126A, SAP30BP, GORASP2, MBD3, MRPS9, HAX1, SFXN1, ITGA5, TCIRG1, RNF168, GEMIN5, HAS1, MTCH1, NDUFA5, MCAT, MRPL57, HDAC2, ESR1, FN1, VCAM1, ITGA4, CD81, PRMT1, SF3B4, EP300, PRRC2A, EWSR1, FUS, ITGB5, NONO, TRAF3, HNRNPUL1, EPAS1, CHERP, CDK12, ITCH, WBP4, rev, RPA3, RPA2, RPA1, HSPA5, RIOK2, TRAF1, TRAF2, SEC24D, TFG, SEC24A, SSBP3, PRR13, ATPAF2, PEF1, JUN, CUL7, OBSL1, CCDC8, RNF2, BMI1, EGFR, ABL1, SRPK1, ABCE1, PRMT8, RPS6KB2, ACAA2, ACAT1, EIF4H, ANXA2, HIP1R, PICALM, POR, NTRK1, SCARNA22, NPM1, KRT2, PRDX2, S100A9, YWHAZ, DDX17, PCLO, ANXA6, SEC24C, GIPC1, CSRP2, FHL1, HNRNPA3, CTTN, MARCKSL1, PARP1, SERPINB3, CCDC50, KIF2A, KRT6B, HIST1H1D, STK38, H2AFV, PACSIN2, U2AF1, RSL24D1, XRCC5, ESYT3, RPL29, SDF2L1, LRP1B, MSN, SEC23A, RPL7A, SNX18, PPIL1, RPS27A, IGHM, SUMO3, GTF2I, RUVBL1, KRT16, RBM8A, RPL8, SRSF9, ZC3HAV1L, GAPDH, ETV3, IGF2R, COL5A2, HNRNPD, ANP32B, WAC, TFAP2A, TTN, CBR3, ARGLU1, HNRNPAB, SRRT, ATP1B3, COPS7B, PRR12, ATF3, NOMO3, NOMO1, NOMO2, EYA3, C1orf198, MAZ, U2AF2, SSB, TRA2B, C1orf52, HMGB1, HMGB1P10, RPRD1A, HNRNPH3, EIF5A2, C1orf131, SEC13, MAPRE1, CSTF2T, SRSF3, LENG1, UPF1, HDGFRP2, RPS10P7, RPS10P11, RPS10, RPS10P13, RPS10P4, RPS10P22, DACH1, ANO1, NCOR1, MLX, SUMO1P3, SUMO1, WIZ, PFDN6, ARFIP2, ZHX3, EEF1B2, MBIP, BAG6, DENND2A, PRCC, SRSF1, EIF4ENIF1, SPTAN1, CDCA8, PLS3, API5P1, API5, PSMA4, DNTTIP1, AKAP8, NCOA3, SMTN, FBRS, SMARCE1, ERC1, WBP11, SPTBN1, NFRKB, OLA1, ZNF207, R3HDM1, TRIM33, SAFB, UBFD1, SRSF7, SRSF2, GATA6, VDAC1P1, VDAC1, IL16, GMNN, ILF2, MED4, QKI, VCL, MFAP1, SNAP29, PADI1, BCL9, BCL9L, PKM, GPATCH11, CASC3, PSMC6, CACYBP, RPL12P6, RPL12P32, RPL12P14, RPL12, RPL12P2, RPL12P35, RPL12P19, TPI1, TPI1P1, CHAF1A, MIA3, CIC, SDCBP, CA2, FKBP3, ACE, NKX2-5, CSTF2, PFDN2, UBTF, FAM207A, LOC729774, BRD8, C12orf45, C1orf35, TCF20, SOD1, SPAG7, MED8, ETS2, ALDOC, FKBP4, INCENP, CEP85, CECR2, TFE3, SUPT16HP1, SUPT16H, MAPT, HTATSF1, RPS18P12, RPS18P5, KPNA2, TMX1, CKAP4, HSPE1, COPRS, PTGES3, LAMP2, ERLIN2, CTNNBL1, TOMM22, NRBF2, C9orf78, NCOA6, MED26, RANBP1, LOC389842, LOC727803, HMGB3, CANX, PUS7, RPSAP19, RPSA, RPSAP18, RPSAP58, RPSAP15, RPSAP8, RPSAP9, RPSAP12, RPSAP29, RPSAP61, ARHGAP17, USF1, PSIP1, SNRPEP2, SNRPE, NUDT5, PPM1G, OTUB1, AHCY, COPS3, NSMCE2, SAE1, PROSER1, GRWD1, CREB5, TAF9B, RBM33, EDF1, PGK1, FAM114A2, SRRM1, RAD23B, CIAPIN1, CIAPIN1P, LRRC59, PABPN1, KMT2A, RPRD1B, GPATCH8, CCDC43, DGCR14, PPP1R2, ERICH1, EIF5A, EIF5AL1, BAG3, PCNA, SOX7, PNISR, FAM168A, MED15, SRSF11, SIRT1, RSF1, MAML1, HPRT1, SPDL1, CRTC3, CEP55, CDV3, ALYREF, RNF40, STOML2, DGCR8, NUCKS1, UBN2, PSMD7, WNT10A, HMBS, KHDRBS1, VBP1, NCSTN, CDCA2, SFSWAP, ZRANB2, DDB1, RBBP6, ZEB1, SRSF6, LOC644422, EIF2S1, RFX5, RPS19, RPS19P3, TALDO1, CWC15, CDCA5, LOC645086, C11orf58, TXN, STX12, PHRF1, BSG, TAF4, SH3GL1, LIN37, HRNR, FAM192A, RRBP1, KIAA0907, GOLGB1, PAX9, P4HB, CHMP5, LDHB, CALR, SUMO2P1, SUMO2, PDIA6, AHSA1, EN2, CCDC124, RPLP0P6, RPLP0P2, RPLP0P3, RPLP0, NIPBL, PDLIM4, PRKCSH, C15orf39, HNRNPDL, PDIA4, NUP210, RPLP2P3, RPLP2, PRDX4, DAZAP1, UBE2T, PHAX, AMOT, MARCKS, LOC284685, SMARCC1, BCORL1, RFC4, GLRX3, ANP32E, HYOU1, NPM3, ATF7IP, SARNP, TRA2A, HDGF, STIP1, PELP1, KCTD12, GLO1, PCF11, CLIC1, DNAJC8, RNF114, SLC4A1AP, FAM50A, GTF2A1, PRPF40A, CDC37, PPIAP22, PPIA, SMARCC2, MEGF11, KIAA1143, DENR, LAMP1, MYBL2, PITX1, UBE2MP1, UBE2M, CHTF8, OTX1, NACA, FNBP4, GTF2F2, GLTSCR1, GTF2E1, PQBP1, EMD, RNF113A, GPALPP1, SNRPA, RRP15, RPS25, RPS25P8, GMEB2, LNPEP, DNAJB1, IGBP1P1, IGBP1, HINT1, ARID1A, PPIB, ANXA11, MATR3, Sgol2, PPARGC1A, MCM2, Ksr1, UBASH3B, SFPQ, CAPN13, HEY1, BRCA1, MTCH2, PPIE, TBX3, BMP4, CTNNB1, GSK3A, HNF1B, TCF7L2, TRIP4, YAF2, ZNF217, AAR2, PIH1D1, EFTUD2, TNIP2, CHD3, CHD4, HEXIM1, MEPCE, LARP7, RUNX1, AGR2, RECQL4, REST, CDK9, SMARCA4, DDIT3, FLI1, TP53BP1, MDC1, METTL3, METTL14, KIAA1429, RC3H1, RC3H2, ATG16L1, PHB, DISC1, NR2C2, UBQLN2, ZFYVE21, XRCC6, AGRN, USP19, HIST1H4A, APEX1, DDX5, SNRNP70, SNRPB, SNRPD1, SNRPD2, SNRPD3, RNU1-1, RBMX, HNRNPM, HNRNPA2B1, TAF15, DDX3X, TARDBP, CLINT1, HNRNPL, NUMA1, ZFR, SNRNP200, ZNF326, HNRNPK, SF3B1, TOE1, HSPA8, SNRPB2, DDX20, GOT2, ILF3, PRPF6, ZNF638, HNRNPF, HNRNPH1, HNRNPR, VCP, CAD, CCAR2, DDX23, GEMIN4, HSPA1A, PCMTD1, POTEF, PRMT5, RBM45, SAFB2, SF3B2, SF3B3, SNRPF, SRSF4, THRAP3, TIA1, TTC7A, ZCCHC8, ITFG1, ARAF, BRD7, SOX2, ARIH2, PLEKHA4, NGB, OPTN, ZC3H18, CELF1, MKI67, INS, Apc2, FBP1, N, ZNF768, SYNCRIP, KDM5C, DDX58, OGT, SPOP, UFL1, DDRGK1, WDR5, TPX2, MALL, SOX21, POU3F3, PTP4A3, TRIM8, RCHY1, nsp14, SOX5, |
| NFATC2 | NFATC2, EP300, CREBBP, MED31, VIM, PRKCZ, PIN1, MEF2D, IRF4, FOXP3, MAPK9, MAPK8, SUV39H1, TUBA1A, KPNB1, TAF9, HNRNPA1, TFAP4, IRF2BP2, JUN, LRRK2, IQGAP1, CSE1L, TNPO1, PPP2R1A, USP22, GNB2, HCVgp1, CREB1, IKZF1, RPTOR, FOS, CHEK1, SCAI, IKZF2, GSK3B, RUNX1, SATB1, WDR48, NFATC1, ZNF384, ATF7, ZNF131, MAFK, XRCC5, PRKDC, CSNK1D, JUNB, NFYC, PLK1, LIG3, XRCC1, HSPA1A, HLTF, BTAF1, YWHAB, CHAF1B, LEF1, IRF2BP1, JUND, TFAM, MAFG, SSBP1, RANBP9, CHAMP1, RPA1, WDR26, ZNF148, FOXK1, ZNF217, DEK, RPA3, POLG, CACYBP, RPA2, NFYB, PARP1, ETV6, XRCC6, KIF4A, RFC4, RFC3, EGR1, CAD, RFC5, CTBP1, RFC2, BCL11B, YWHAG, KIF2C, RNGTT, PRDM15, ALDH18A1, MTHFD2, YWHAE, MRPL39, TUFM, IFI16, TP53BP1, ERAL1, SSRP1, TRRAP, YWHAQ, HIRA, ERCC3, UBE2S, SUPT6H, ARID1A, HSPA9, POGZ, UHRF1, SUCLA2, HSPA5, DPF2, YWHAZ, GATAD2A, LRRC47, ZMYM2, DNAJA1, IDH3A, SDHA, CBX5, CABIN1, VAPA, CBX3, MRE11A, SMARCA5, EEF2, RB1, MCRS1, EMD, GTF2H2C, GTF2H2, GTF2H2C_2, SUPT5H, RCOR1, VAPB, MRPL17, UQCRC2, TCF12, BEND3, SMARCA4, KDM1A, SMARCD2, AIFM1, MCM2, H2AFY, HSPA8, POLDIP2, RAD50, SMARCC1, POLR1C, TIMM50, MTA2, HADHA, TLK2, TLK1, RBBP4, GATAD2B, RANBP2, CEBPE, ADNP, SMARCE1, CNP, CBX1, SMARCC2, ZNF24, CHD4, SMARCD1, MBD3, NUDC, ACTL6A, MTA1, IDH3B, CSTF1, RPL23, PPP1CA, HSPH1, PMPCB, GTF3C5, TCP1, GTF3C4, DCAF7, HDAC1, NKX2-1, KIAA1429, ESR1, EGFR, CIC, SOCS3, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| NFATC2 | 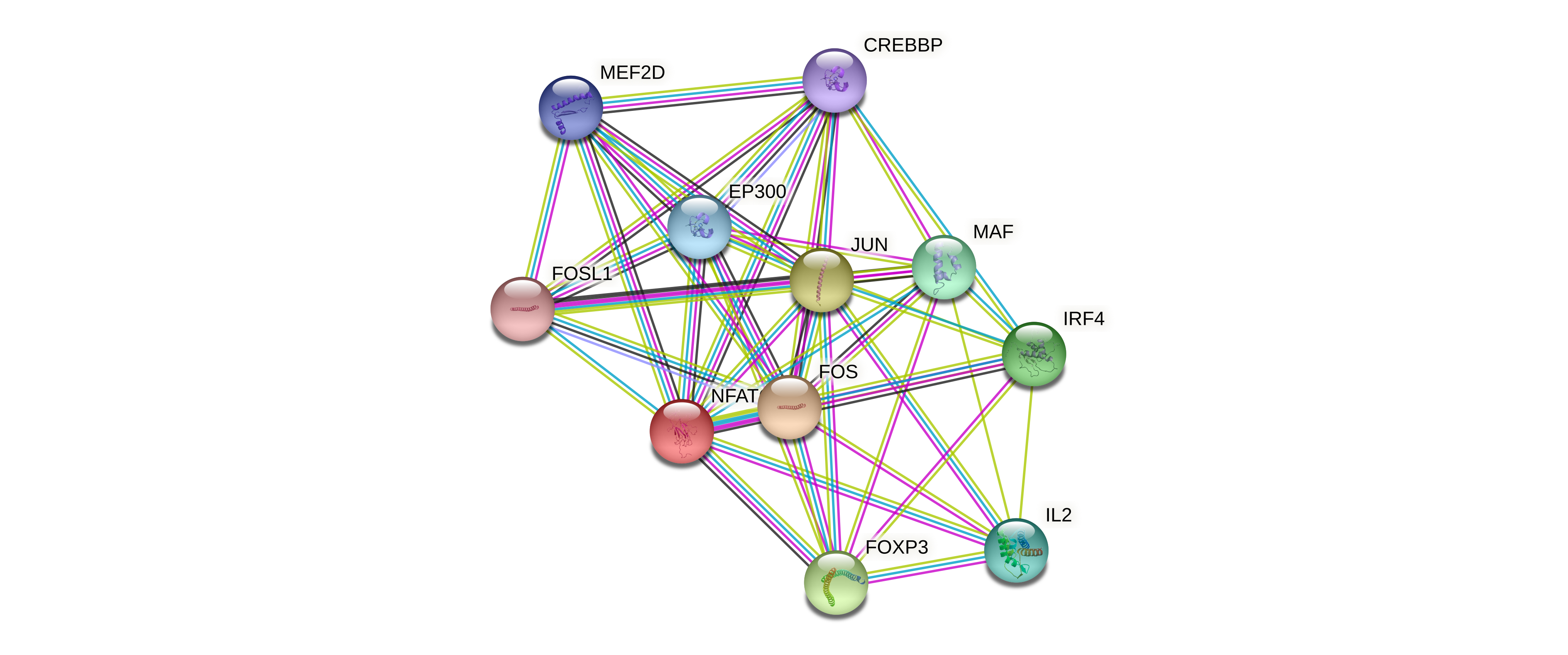 |
| EWSR1 | 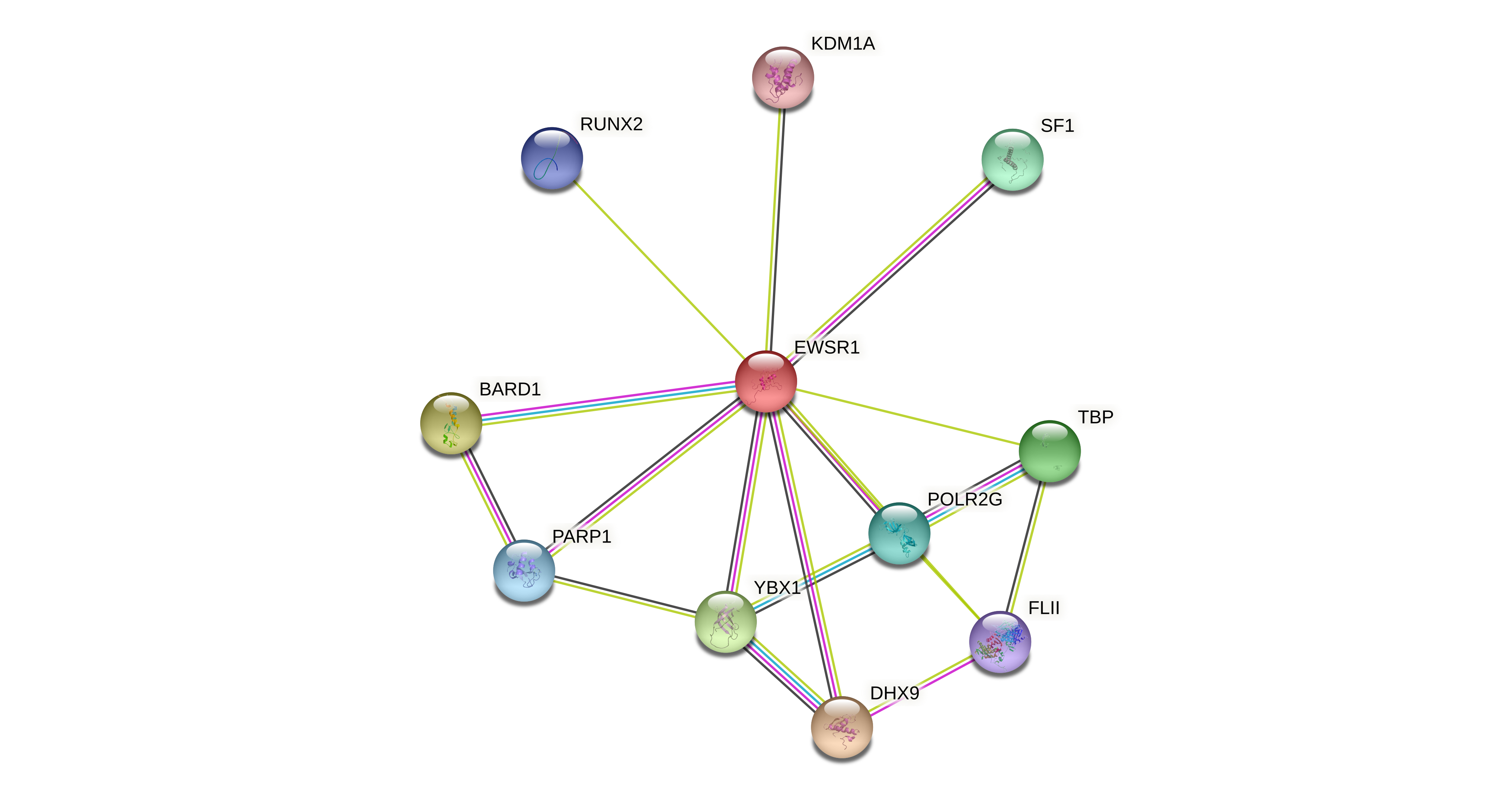 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to NFATC2-EWSR1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
| NFATC2 | EWSR1 | Combination Mtor And Vegf Inhibition | PubMed | 34021224 |
Top |
Related Diseases to NFATC2-EWSR1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| NFATC2 | EWSR1 | Ewing Family Of Tumors | PubMed | 34021224 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | NFATC2 | C0011860 | Diabetes Mellitus, Non-Insulin-Dependent | 1 | CTD_human |
| Hgene | NFATC2 | C0020542 | Pulmonary Hypertension | 1 | CTD_human |
| Hgene | NFATC2 | C0027765 | nervous system disorder | 1 | CTD_human |
| Tgene | EWSR1 | C0553580 | Ewings sarcoma | 3 | CTD_human;ORPHANET |
| Tgene | EWSR1 | C0002736 | Amyotrophic Lateral Sclerosis | 1 | GENOMICS_ENGLAND |
| Tgene | EWSR1 | C0033578 | Prostatic Neoplasms | 1 | CTD_human |
| Tgene | EWSR1 | C0206651 | Clear Cell Sarcoma of Soft Tissue | 1 | ORPHANET |
| Tgene | EWSR1 | C0206663 | Neuroectodermal Tumor, Primitive | 1 | CTD_human |
| Tgene | EWSR1 | C0279980 | Extra-osseous Ewing's sarcoma | 1 | ORPHANET |
| Tgene | EWSR1 | C0334584 | Spongioblastoma | 1 | CTD_human |
| Tgene | EWSR1 | C0334596 | Medulloepithelioma | 1 | CTD_human |
| Tgene | EWSR1 | C0376358 | Malignant neoplasm of prostate | 1 | CTD_human |
| Tgene | EWSR1 | C0700367 | Ependymoblastoma | 1 | CTD_human |
| Tgene | EWSR1 | C0751675 | Cerebral Primitive Neuroectodermal Tumor | 1 | CTD_human |
| Tgene | EWSR1 | C1275278 | Extraskeletal Myxoid Chondrosarcoma | 1 | ORPHANET |