| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ARHGAP35-ESR1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ARHGAP35-ESR1 | FusionPDB ID: 6088 | FusionGDB2.0 ID: 6088 | Hgene | Tgene | Gene symbol | ARHGAP35 | ESR1 | Gene ID | 2909 | 2099 |
| Gene name | Rho GTPase activating protein 35 | estrogen receptor 1 | |
| Synonyms | GRF-1|GRLF1|P190-A|P190A|p190ARhoGAP|p190RhoGAP | ER|ESR|ESRA|ESTRR|Era|NR3A1 | |
| Cytomap | 19q13.32 | 6q25.1-q25.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | rho GTPase-activating protein 35glucocorticoid receptor DNA-binding factor 1glucocorticoid receptor repression factor 1rho GAP p190A | estrogen receptorE2 receptor alphaER-alphaestradiol receptorestrogen nuclear receptor alphaestrogen receptor alpha E1-E2-1-2estrogen receptor alpha E1-N2-E2-1-2nuclear receptor subfamily 3 group A member 1oestrogen receptor alpha | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | Q9NRY4 | P03372 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000404338, ENST00000598548, | ENST00000406599, ENST00000427531, ENST00000482101, ENST00000206249, ENST00000338799, ENST00000440973, ENST00000443427, ENST00000456483, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 19 X 11 X 9=1881 | 10 X 10 X 6=600 |
| # samples | 19 | 12 | |
| ** MAII score | log2(19/1881*10)=-3.30742852519225 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/600*10)=-2.32192809488736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ARHGAP35 [Title/Abstract] AND ESR1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ARHGAP35(47440665)-ESR1(152201789), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ARHGAP35-ESR1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ARHGAP35-ESR1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ARHGAP35-ESR1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ARHGAP35-ESR1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ESR1 | GO:0006366 | transcription by RNA polymerase II | 15831516 |
| Tgene | ESR1 | GO:0010629 | negative regulation of gene expression | 21695196 |
| Tgene | ESR1 | GO:0030520 | intracellular estrogen receptor signaling pathway | 9841876 |
| Tgene | ESR1 | GO:0032355 | response to estradiol | 15304487 |
| Tgene | ESR1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling | 7651415|16043358 |
| Tgene | ESR1 | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 10816575 |
| Tgene | ESR1 | GO:0043627 | response to estrogen | 11581164 |
| Tgene | ESR1 | GO:0045893 | positive regulation of transcription, DNA-templated | 9841876|20074560 |
| Tgene | ESR1 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly | 9841876 |
| Tgene | ESR1 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 11544182|12047722|15345745|15831516|18563714 |
| Tgene | ESR1 | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 9328340|10681512 |
| Tgene | ESR1 | GO:0071392 | cellular response to estradiol stimulus | 15831516 |
 Fusion gene breakpoints across ARHGAP35 (5'-gene) Fusion gene breakpoints across ARHGAP35 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
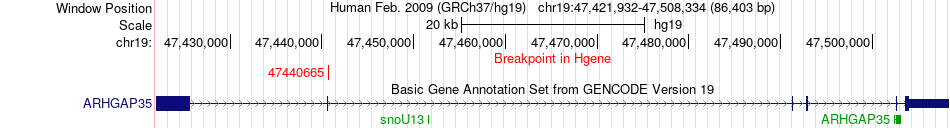 |
 Fusion gene breakpoints across ESR1 (3'-gene) Fusion gene breakpoints across ESR1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
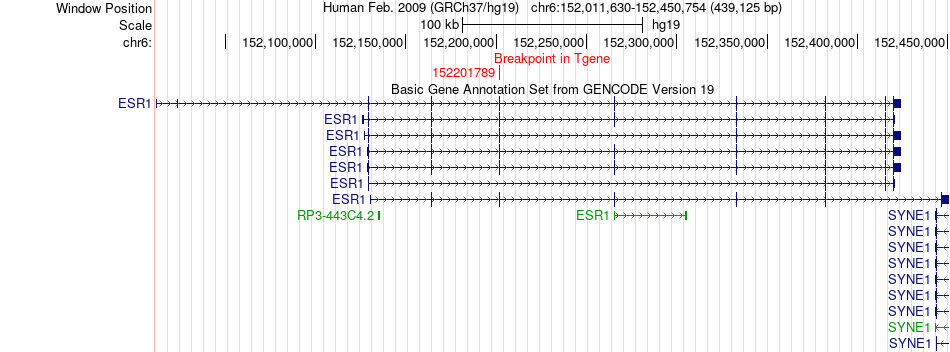 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A8-A06O | ARHGAP35 | chr19 | 47440665 | + | ESR1 | chr6 | 152201789 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000404338 | ARHGAP35 | chr19 | 47440665 | + | ENST00000440973 | ESR1 | chr6 | 152201789 | + | 9279 | 3826 | 0 | 4970 | 1656 |
| ENST00000404338 | ARHGAP35 | chr19 | 47440665 | + | ENST00000338799 | ESR1 | chr6 | 152201789 | + | 5058 | 3826 | 0 | 4970 | 1656 |
| ENST00000404338 | ARHGAP35 | chr19 | 47440665 | + | ENST00000456483 | ESR1 | chr6 | 152201789 | + | 8943 | 3826 | 0 | 4634 | 1544 |
| ENST00000404338 | ARHGAP35 | chr19 | 47440665 | + | ENST00000443427 | ESR1 | chr6 | 152201789 | + | 9279 | 3826 | 0 | 4970 | 1656 |
| ENST00000404338 | ARHGAP35 | chr19 | 47440665 | + | ENST00000206249 | ESR1 | chr6 | 152201789 | + | 9276 | 3826 | 0 | 4970 | 1656 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000404338 | ENST00000440973 | ARHGAP35 | chr19 | 47440665 | + | ESR1 | chr6 | 152201789 | + | 0.000241956 | 0.99975806 |
| ENST00000404338 | ENST00000338799 | ARHGAP35 | chr19 | 47440665 | + | ESR1 | chr6 | 152201789 | + | 0.000835508 | 0.99916446 |
| ENST00000404338 | ENST00000456483 | ARHGAP35 | chr19 | 47440665 | + | ESR1 | chr6 | 152201789 | + | 0.000138792 | 0.99986124 |
| ENST00000404338 | ENST00000443427 | ARHGAP35 | chr19 | 47440665 | + | ESR1 | chr6 | 152201789 | + | 0.000241956 | 0.99975806 |
| ENST00000404338 | ENST00000206249 | ARHGAP35 | chr19 | 47440665 | + | ESR1 | chr6 | 152201789 | + | 0.000242933 | 0.9997571 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >6088_6088_1_ARHGAP35-ESR1_ARHGAP35_chr19_47440665_ENST00000404338_ESR1_chr6_152201789_ENST00000206249_length(amino acids)=1656AA_BP=1275 MMMARKQDVRIPTYNISVVGLSGTEKEKGQCGIGKSCLCNRFVRPSADEFHLDHTSVLSTSDFGGRVVNNDHFLYWGEVSRSLEDCVECK MHIVEQTEFIDDQTFQPHRSTALQPYIKRAAATKLASAEKLMYFCTDQLGLEQDFEQKQMPDGKLLVDGFLLGIDVSRGMNRNFDDQLKF VSNLYNQLAKTKKPIVVVLTKCDEGVERYIRDAHTFALSKKNLQVVETSARSNVNVDLAFSTLVQLIDKSRGKTKIIPYFEALKQQSQQI ATAKDKYEWLVSRIVKNHNENWLSVSRKMQASPEYQDYVYLEGTQKAKKLFLQHIHRLKHEHIERRRKLYLAALPLAFEALIPNLDEIDH LSCIKAKKLLETKPEFLKWFVVLEETPWDATSHIDNMENERIPFDLMDTVPAEQLYEAHLEKLRNERKRVEMRRAFKENLETSPFITPGK PWEEARSFIMNEDFYQWLEESVYMDIYGKHQKQIIDKAKEEFQELLLEYSELFYELELDAKPSKEKMGVIQDVLGEEQRFKALQKLQAER DALILKHIHFVYHPTKETCPSCPACVDAKIEHLISSRFIRPSDRNQKNSLSDPNIDRINLVILGKDGLARELANEIRALCTNDDKYVIDG KMYELSLRPIEGNVRLPVNSFQTPTFQPHGCLCLYNSKESLSYVVESIEKSRESTLGRRDNHLVHLPLTLILVNKRGDTSGETLHSLIQQ GQQIASKLQCVFLDPASAGIGYGRNINEKQISQVLKGLLDSKRNLNLVSSTASIKDLADVDLRIVMCLMCGDPFSADDILFPVLQSQTCK SSHCGSNNSVLLELPIGLHKKRIELSVLSYHSSFSIRKSRLVHGYIVFYSAKRKASLAMLRAFLCEVQDIIPIQLVALTDGAVDVLDNDL SREQLTEGEEIAQEIDGRFTSIPCSQPQHKLEIFHPFFKDVVEKKNIIEATHMYDNAAEACSTTEEVFNSPRAGSPLCNSNLQDSEEDIE PSYSLFREDTSLPSLSKDHSKLSMELEGNDGLSFIMSNFESKLNNKVPPPVKPKPPVHFEITKGDLSYLDQGHRDGQRKSVSSSPWLPQD GFDPSDYAEPMDAVVKPRNEEENIYSVPHDSTQGKIITIRNINKAQSNGSGNGSDSEMDTSSLERGRKVSIVSKPVLYRTRCTRLGRFAS YRTSFSVGSDDELGPIRKKEEDQASQGYKGDNAVIPYETDEDPRRRNILRSLRRNTKKPKPKPRPSITKATWESNYFGVPLTTVVTPEKP IPIFIERCIEYIEATGHNDYMCPATNQCTIDKNRRKSCQACRLRKCYEVGMMKGGIRKDRRGGRMLKHKRQRDDGEGRGEVGSAGDMRAA NLWPSPLMIKRSKKNSLALSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINWAKRVPGFVDLTLHDQVHLL ECAWLEILMIGLVWRSMEHPGKLLFAPNLLLDRNQGKCVEGMVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFLSSTLKSL EEKDHIHRVLDKITDTLIHLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLLLEMLDAHRLHAPTSRGGAS -------------------------------------------------------------- >6088_6088_2_ARHGAP35-ESR1_ARHGAP35_chr19_47440665_ENST00000404338_ESR1_chr6_152201789_ENST00000338799_length(amino acids)=1656AA_BP=1275 MMMARKQDVRIPTYNISVVGLSGTEKEKGQCGIGKSCLCNRFVRPSADEFHLDHTSVLSTSDFGGRVVNNDHFLYWGEVSRSLEDCVECK MHIVEQTEFIDDQTFQPHRSTALQPYIKRAAATKLASAEKLMYFCTDQLGLEQDFEQKQMPDGKLLVDGFLLGIDVSRGMNRNFDDQLKF VSNLYNQLAKTKKPIVVVLTKCDEGVERYIRDAHTFALSKKNLQVVETSARSNVNVDLAFSTLVQLIDKSRGKTKIIPYFEALKQQSQQI ATAKDKYEWLVSRIVKNHNENWLSVSRKMQASPEYQDYVYLEGTQKAKKLFLQHIHRLKHEHIERRRKLYLAALPLAFEALIPNLDEIDH LSCIKAKKLLETKPEFLKWFVVLEETPWDATSHIDNMENERIPFDLMDTVPAEQLYEAHLEKLRNERKRVEMRRAFKENLETSPFITPGK PWEEARSFIMNEDFYQWLEESVYMDIYGKHQKQIIDKAKEEFQELLLEYSELFYELELDAKPSKEKMGVIQDVLGEEQRFKALQKLQAER DALILKHIHFVYHPTKETCPSCPACVDAKIEHLISSRFIRPSDRNQKNSLSDPNIDRINLVILGKDGLARELANEIRALCTNDDKYVIDG KMYELSLRPIEGNVRLPVNSFQTPTFQPHGCLCLYNSKESLSYVVESIEKSRESTLGRRDNHLVHLPLTLILVNKRGDTSGETLHSLIQQ GQQIASKLQCVFLDPASAGIGYGRNINEKQISQVLKGLLDSKRNLNLVSSTASIKDLADVDLRIVMCLMCGDPFSADDILFPVLQSQTCK SSHCGSNNSVLLELPIGLHKKRIELSVLSYHSSFSIRKSRLVHGYIVFYSAKRKASLAMLRAFLCEVQDIIPIQLVALTDGAVDVLDNDL SREQLTEGEEIAQEIDGRFTSIPCSQPQHKLEIFHPFFKDVVEKKNIIEATHMYDNAAEACSTTEEVFNSPRAGSPLCNSNLQDSEEDIE PSYSLFREDTSLPSLSKDHSKLSMELEGNDGLSFIMSNFESKLNNKVPPPVKPKPPVHFEITKGDLSYLDQGHRDGQRKSVSSSPWLPQD GFDPSDYAEPMDAVVKPRNEEENIYSVPHDSTQGKIITIRNINKAQSNGSGNGSDSEMDTSSLERGRKVSIVSKPVLYRTRCTRLGRFAS YRTSFSVGSDDELGPIRKKEEDQASQGYKGDNAVIPYETDEDPRRRNILRSLRRNTKKPKPKPRPSITKATWESNYFGVPLTTVVTPEKP IPIFIERCIEYIEATGHNDYMCPATNQCTIDKNRRKSCQACRLRKCYEVGMMKGGIRKDRRGGRMLKHKRQRDDGEGRGEVGSAGDMRAA NLWPSPLMIKRSKKNSLALSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINWAKRVPGFVDLTLHDQVHLL ECAWLEILMIGLVWRSMEHPGKLLFAPNLLLDRNQGKCVEGMVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFLSSTLKSL EEKDHIHRVLDKITDTLIHLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLLLEMLDAHRLHAPTSRGGAS -------------------------------------------------------------- >6088_6088_3_ARHGAP35-ESR1_ARHGAP35_chr19_47440665_ENST00000404338_ESR1_chr6_152201789_ENST00000440973_length(amino acids)=1656AA_BP=1275 MMMARKQDVRIPTYNISVVGLSGTEKEKGQCGIGKSCLCNRFVRPSADEFHLDHTSVLSTSDFGGRVVNNDHFLYWGEVSRSLEDCVECK MHIVEQTEFIDDQTFQPHRSTALQPYIKRAAATKLASAEKLMYFCTDQLGLEQDFEQKQMPDGKLLVDGFLLGIDVSRGMNRNFDDQLKF VSNLYNQLAKTKKPIVVVLTKCDEGVERYIRDAHTFALSKKNLQVVETSARSNVNVDLAFSTLVQLIDKSRGKTKIIPYFEALKQQSQQI ATAKDKYEWLVSRIVKNHNENWLSVSRKMQASPEYQDYVYLEGTQKAKKLFLQHIHRLKHEHIERRRKLYLAALPLAFEALIPNLDEIDH LSCIKAKKLLETKPEFLKWFVVLEETPWDATSHIDNMENERIPFDLMDTVPAEQLYEAHLEKLRNERKRVEMRRAFKENLETSPFITPGK PWEEARSFIMNEDFYQWLEESVYMDIYGKHQKQIIDKAKEEFQELLLEYSELFYELELDAKPSKEKMGVIQDVLGEEQRFKALQKLQAER DALILKHIHFVYHPTKETCPSCPACVDAKIEHLISSRFIRPSDRNQKNSLSDPNIDRINLVILGKDGLARELANEIRALCTNDDKYVIDG KMYELSLRPIEGNVRLPVNSFQTPTFQPHGCLCLYNSKESLSYVVESIEKSRESTLGRRDNHLVHLPLTLILVNKRGDTSGETLHSLIQQ GQQIASKLQCVFLDPASAGIGYGRNINEKQISQVLKGLLDSKRNLNLVSSTASIKDLADVDLRIVMCLMCGDPFSADDILFPVLQSQTCK SSHCGSNNSVLLELPIGLHKKRIELSVLSYHSSFSIRKSRLVHGYIVFYSAKRKASLAMLRAFLCEVQDIIPIQLVALTDGAVDVLDNDL SREQLTEGEEIAQEIDGRFTSIPCSQPQHKLEIFHPFFKDVVEKKNIIEATHMYDNAAEACSTTEEVFNSPRAGSPLCNSNLQDSEEDIE PSYSLFREDTSLPSLSKDHSKLSMELEGNDGLSFIMSNFESKLNNKVPPPVKPKPPVHFEITKGDLSYLDQGHRDGQRKSVSSSPWLPQD GFDPSDYAEPMDAVVKPRNEEENIYSVPHDSTQGKIITIRNINKAQSNGSGNGSDSEMDTSSLERGRKVSIVSKPVLYRTRCTRLGRFAS YRTSFSVGSDDELGPIRKKEEDQASQGYKGDNAVIPYETDEDPRRRNILRSLRRNTKKPKPKPRPSITKATWESNYFGVPLTTVVTPEKP IPIFIERCIEYIEATGHNDYMCPATNQCTIDKNRRKSCQACRLRKCYEVGMMKGGIRKDRRGGRMLKHKRQRDDGEGRGEVGSAGDMRAA NLWPSPLMIKRSKKNSLALSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINWAKRVPGFVDLTLHDQVHLL ECAWLEILMIGLVWRSMEHPGKLLFAPNLLLDRNQGKCVEGMVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFLSSTLKSL EEKDHIHRVLDKITDTLIHLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLLLEMLDAHRLHAPTSRGGAS -------------------------------------------------------------- >6088_6088_4_ARHGAP35-ESR1_ARHGAP35_chr19_47440665_ENST00000404338_ESR1_chr6_152201789_ENST00000443427_length(amino acids)=1656AA_BP=1275 MMMARKQDVRIPTYNISVVGLSGTEKEKGQCGIGKSCLCNRFVRPSADEFHLDHTSVLSTSDFGGRVVNNDHFLYWGEVSRSLEDCVECK MHIVEQTEFIDDQTFQPHRSTALQPYIKRAAATKLASAEKLMYFCTDQLGLEQDFEQKQMPDGKLLVDGFLLGIDVSRGMNRNFDDQLKF VSNLYNQLAKTKKPIVVVLTKCDEGVERYIRDAHTFALSKKNLQVVETSARSNVNVDLAFSTLVQLIDKSRGKTKIIPYFEALKQQSQQI ATAKDKYEWLVSRIVKNHNENWLSVSRKMQASPEYQDYVYLEGTQKAKKLFLQHIHRLKHEHIERRRKLYLAALPLAFEALIPNLDEIDH LSCIKAKKLLETKPEFLKWFVVLEETPWDATSHIDNMENERIPFDLMDTVPAEQLYEAHLEKLRNERKRVEMRRAFKENLETSPFITPGK PWEEARSFIMNEDFYQWLEESVYMDIYGKHQKQIIDKAKEEFQELLLEYSELFYELELDAKPSKEKMGVIQDVLGEEQRFKALQKLQAER DALILKHIHFVYHPTKETCPSCPACVDAKIEHLISSRFIRPSDRNQKNSLSDPNIDRINLVILGKDGLARELANEIRALCTNDDKYVIDG KMYELSLRPIEGNVRLPVNSFQTPTFQPHGCLCLYNSKESLSYVVESIEKSRESTLGRRDNHLVHLPLTLILVNKRGDTSGETLHSLIQQ GQQIASKLQCVFLDPASAGIGYGRNINEKQISQVLKGLLDSKRNLNLVSSTASIKDLADVDLRIVMCLMCGDPFSADDILFPVLQSQTCK SSHCGSNNSVLLELPIGLHKKRIELSVLSYHSSFSIRKSRLVHGYIVFYSAKRKASLAMLRAFLCEVQDIIPIQLVALTDGAVDVLDNDL SREQLTEGEEIAQEIDGRFTSIPCSQPQHKLEIFHPFFKDVVEKKNIIEATHMYDNAAEACSTTEEVFNSPRAGSPLCNSNLQDSEEDIE PSYSLFREDTSLPSLSKDHSKLSMELEGNDGLSFIMSNFESKLNNKVPPPVKPKPPVHFEITKGDLSYLDQGHRDGQRKSVSSSPWLPQD GFDPSDYAEPMDAVVKPRNEEENIYSVPHDSTQGKIITIRNINKAQSNGSGNGSDSEMDTSSLERGRKVSIVSKPVLYRTRCTRLGRFAS YRTSFSVGSDDELGPIRKKEEDQASQGYKGDNAVIPYETDEDPRRRNILRSLRRNTKKPKPKPRPSITKATWESNYFGVPLTTVVTPEKP IPIFIERCIEYIEATGHNDYMCPATNQCTIDKNRRKSCQACRLRKCYEVGMMKGGIRKDRRGGRMLKHKRQRDDGEGRGEVGSAGDMRAA NLWPSPLMIKRSKKNSLALSLTADQMVSALLDAEPPILYSEYDPTRPFSEASMMGLLTNLADRELVHMINWAKRVPGFVDLTLHDQVHLL ECAWLEILMIGLVWRSMEHPGKLLFAPNLLLDRNQGKCVEGMVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFLSSTLKSL EEKDHIHRVLDKITDTLIHLMAKAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLLLEMLDAHRLHAPTSRGGAS -------------------------------------------------------------- >6088_6088_5_ARHGAP35-ESR1_ARHGAP35_chr19_47440665_ENST00000404338_ESR1_chr6_152201789_ENST00000456483_length(amino acids)=1544AA_BP=1275 MMMARKQDVRIPTYNISVVGLSGTEKEKGQCGIGKSCLCNRFVRPSADEFHLDHTSVLSTSDFGGRVVNNDHFLYWGEVSRSLEDCVECK MHIVEQTEFIDDQTFQPHRSTALQPYIKRAAATKLASAEKLMYFCTDQLGLEQDFEQKQMPDGKLLVDGFLLGIDVSRGMNRNFDDQLKF VSNLYNQLAKTKKPIVVVLTKCDEGVERYIRDAHTFALSKKNLQVVETSARSNVNVDLAFSTLVQLIDKSRGKTKIIPYFEALKQQSQQI ATAKDKYEWLVSRIVKNHNENWLSVSRKMQASPEYQDYVYLEGTQKAKKLFLQHIHRLKHEHIERRRKLYLAALPLAFEALIPNLDEIDH LSCIKAKKLLETKPEFLKWFVVLEETPWDATSHIDNMENERIPFDLMDTVPAEQLYEAHLEKLRNERKRVEMRRAFKENLETSPFITPGK PWEEARSFIMNEDFYQWLEESVYMDIYGKHQKQIIDKAKEEFQELLLEYSELFYELELDAKPSKEKMGVIQDVLGEEQRFKALQKLQAER DALILKHIHFVYHPTKETCPSCPACVDAKIEHLISSRFIRPSDRNQKNSLSDPNIDRINLVILGKDGLARELANEIRALCTNDDKYVIDG KMYELSLRPIEGNVRLPVNSFQTPTFQPHGCLCLYNSKESLSYVVESIEKSRESTLGRRDNHLVHLPLTLILVNKRGDTSGETLHSLIQQ GQQIASKLQCVFLDPASAGIGYGRNINEKQISQVLKGLLDSKRNLNLVSSTASIKDLADVDLRIVMCLMCGDPFSADDILFPVLQSQTCK SSHCGSNNSVLLELPIGLHKKRIELSVLSYHSSFSIRKSRLVHGYIVFYSAKRKASLAMLRAFLCEVQDIIPIQLVALTDGAVDVLDNDL SREQLTEGEEIAQEIDGRFTSIPCSQPQHKLEIFHPFFKDVVEKKNIIEATHMYDNAAEACSTTEEVFNSPRAGSPLCNSNLQDSEEDIE PSYSLFREDTSLPSLSKDHSKLSMELEGNDGLSFIMSNFESKLNNKVPPPVKPKPPVHFEITKGDLSYLDQGHRDGQRKSVSSSPWLPQD GFDPSDYAEPMDAVVKPRNEEENIYSVPHDSTQGKIITIRNINKAQSNGSGNGSDSEMDTSSLERGRKVSIVSKPVLYRTRCTRLGRFAS YRTSFSVGSDDELGPIRKKEEDQASQGYKGDNAVIPYETDEDPRRRNILRSLRRNTKKPKPKPRPSITKATWESNYFGVPLTTVVTPEKP IPIFIERCIEYIEATGHNDYMCPATNQCTIDKNRRKSCQACRLRKCYEVGMMKGGFVDLTLHDQVHLLECAWLEILMIGLVWRSMEHPGK LLFAPNLLLDRNQGKCVEGMVEIFDMLLATSSRFRMMNLQGEEFVCLKSIILLNSGVYTFLSSTLKSLEEKDHIHRVLDKITDTLIHLMA KAGLTLQQQHQRLAQLLLILSHIRHMSNKGMEHLYSMKCKNVVPLYDLLLEMLDAHRLHAPTSRGGASVEETDQSHLATAGSTSSHSLQK -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:47440665/chr6:152201789) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ARHGAP35 | ESR1 |
| FUNCTION: Rho GTPase-activating protein (GAP) (PubMed:19673492, PubMed:28894085). Binds several acidic phospholipids which inhibits the Rho GAP activity to promote the Rac GAP activity (PubMed:19673492). This binding is inhibited by phosphorylation by PRKCA (PubMed:19673492). Involved in cell differentiation as well as cell adhesion and migration, plays an important role in retinal tissue morphogenesis, neural tube fusion, midline fusion of the cerebral hemispheres and mammary gland branching morphogenesis (By similarity). Transduces signals from p21-ras to the nucleus, acting via the ras GTPase-activating protein (GAP) (By similarity). Transduces SRC-dependent signals from cell-surface adhesion molecules, such as laminin, to promote neurite outgrowth. Regulates axon outgrowth, guidance and fasciculation (By similarity). Modulates Rho GTPase-dependent F-actin polymerization, organization and assembly, is involved in polarized cell migration and in the positive regulation of ciliogenesis and cilia elongation (By similarity). During mammary gland development, is required in both the epithelial and stromal compartments for ductal outgrowth (By similarity). Represses transcription of the glucocorticoid receptor by binding to the cis-acting regulatory sequence 5'-GAGAAAAGAAACTGGAGAAACTC-3'; this function is however unclear and would need additional experimental evidences (PubMed:1894621). {ECO:0000250|UniProtKB:P81128, ECO:0000250|UniProtKB:Q91YM2, ECO:0000269|PubMed:1894621, ECO:0000269|PubMed:19673492, ECO:0000269|PubMed:28894085}. | FUNCTION: Nuclear hormone receptor. The steroid hormones and their receptors are involved in the regulation of eukaryotic gene expression and affect cellular proliferation and differentiation in target tissues. Ligand-dependent nuclear transactivation involves either direct homodimer binding to a palindromic estrogen response element (ERE) sequence or association with other DNA-binding transcription factors, such as AP-1/c-Jun, c-Fos, ATF-2, Sp1 and Sp3, to mediate ERE-independent signaling. Ligand binding induces a conformational change allowing subsequent or combinatorial association with multiprotein coactivator complexes through LXXLL motifs of their respective components. Mutual transrepression occurs between the estrogen receptor (ER) and NF-kappa-B in a cell-type specific manner. Decreases NF-kappa-B DNA-binding activity and inhibits NF-kappa-B-mediated transcription from the IL6 promoter and displace RELA/p65 and associated coregulators from the promoter. Recruited to the NF-kappa-B response element of the CCL2 and IL8 promoters and can displace CREBBP. Present with NF-kappa-B components RELA/p65 and NFKB1/p50 on ERE sequences. Can also act synergistically with NF-kappa-B to activate transcription involving respective recruitment adjacent response elements; the function involves CREBBP. Can activate the transcriptional activity of TFF1. Also mediates membrane-initiated estrogen signaling involving various kinase cascades. Isoform 3 is involved in activation of NOS3 and endothelial nitric oxide production. Isoforms lacking one or several functional domains are thought to modulate transcriptional activity by competitive ligand or DNA binding and/or heterodimerization with the full-length receptor. Essential for MTA1-mediated transcriptional regulation of BRCA1 and BCAS3. Isoform 3 can bind to ERE and inhibit isoform 1. {ECO:0000269|PubMed:10681512, ECO:0000269|PubMed:10816575, ECO:0000269|PubMed:11477071, ECO:0000269|PubMed:11682626, ECO:0000269|PubMed:14764652, ECO:0000269|PubMed:15078875, ECO:0000269|PubMed:15891768, ECO:0000269|PubMed:16043358, ECO:0000269|PubMed:16617102, ECO:0000269|PubMed:16684779, ECO:0000269|PubMed:17922032, ECO:0000269|PubMed:17932106, ECO:0000269|PubMed:18247370, ECO:0000269|PubMed:19350539, ECO:0000269|PubMed:20074560, ECO:0000269|PubMed:20705611, ECO:0000269|PubMed:21330404, ECO:0000269|PubMed:22083956, ECO:0000269|PubMed:7651415, ECO:0000269|PubMed:9328340}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 270_327 | 1275.3333333333333 | 1500.0 | Domain | Note=FF 1 |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 368_422 | 1275.3333333333333 | 1500.0 | Domain | Note=FF 2 |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 429_483 | 1275.3333333333333 | 1500.0 | Domain | Note=FF 3 |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 485_550 | 1275.3333333333333 | 1500.0 | Domain | Note=FF 4 |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 592_767 | 1275.3333333333333 | 1500.0 | Domain | pG1 pseudoGTPase |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 783_947 | 1275.3333333333333 | 1500.0 | Domain | pG2 pseudoGTPase |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 201_203 | 1275.3333333333333 | 1500.0 | Nucleotide binding | GTP |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 229_231 | 1275.3333333333333 | 1500.0 | Nucleotide binding | GTP |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 33_37 | 1275.3333333333333 | 1500.0 | Nucleotide binding | GTP |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 95_97 | 1275.3333333333333 | 1500.0 | Nucleotide binding | GTP |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 1213_1236 | 1275.3333333333333 | 1500.0 | Region | Required for phospholipid binding and regulation of the substrate preference |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 1_266 | 1275.3333333333333 | 1500.0 | Region | Has GTPase activity%2C required for proper localization |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000206249 | 1 | 8 | 311_547 | 214.33333333333334 | 596.0 | Domain | NR LBD | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000338799 | 2 | 9 | 311_547 | 214.33333333333334 | 596.0 | Domain | NR LBD | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000440973 | 3 | 10 | 311_547 | 214.33333333333334 | 596.0 | Domain | NR LBD | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000443427 | 2 | 9 | 311_547 | 214.33333333333334 | 596.0 | Domain | NR LBD | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000456483 | 2 | 8 | 311_547 | 214.33333333333334 | 484.0 | Domain | NR LBD | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000206249 | 1 | 8 | 251_310 | 214.33333333333334 | 596.0 | Region | Note=Hinge | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000206249 | 1 | 8 | 264_595 | 214.33333333333334 | 596.0 | Region | Note=Self-association | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000206249 | 1 | 8 | 311_595 | 214.33333333333334 | 596.0 | Region | Note=Transactivation AF-2 | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000338799 | 2 | 9 | 251_310 | 214.33333333333334 | 596.0 | Region | Note=Hinge | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000338799 | 2 | 9 | 264_595 | 214.33333333333334 | 596.0 | Region | Note=Self-association | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000338799 | 2 | 9 | 311_595 | 214.33333333333334 | 596.0 | Region | Note=Transactivation AF-2 | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000440973 | 3 | 10 | 251_310 | 214.33333333333334 | 596.0 | Region | Note=Hinge | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000440973 | 3 | 10 | 264_595 | 214.33333333333334 | 596.0 | Region | Note=Self-association | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000440973 | 3 | 10 | 311_595 | 214.33333333333334 | 596.0 | Region | Note=Transactivation AF-2 | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000443427 | 2 | 9 | 251_310 | 214.33333333333334 | 596.0 | Region | Note=Hinge | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000443427 | 2 | 9 | 264_595 | 214.33333333333334 | 596.0 | Region | Note=Self-association | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000443427 | 2 | 9 | 311_595 | 214.33333333333334 | 596.0 | Region | Note=Transactivation AF-2 | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000456483 | 2 | 8 | 251_310 | 214.33333333333334 | 484.0 | Region | Note=Hinge | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000456483 | 2 | 8 | 264_595 | 214.33333333333334 | 484.0 | Region | Note=Self-association | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000456483 | 2 | 8 | 311_595 | 214.33333333333334 | 484.0 | Region | Note=Transactivation AF-2 | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000206249 | 1 | 8 | 221_245 | 214.33333333333334 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000338799 | 2 | 9 | 221_245 | 214.33333333333334 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000440973 | 3 | 10 | 221_245 | 214.33333333333334 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000443427 | 2 | 9 | 221_245 | 214.33333333333334 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000456483 | 2 | 8 | 221_245 | 214.33333333333334 | 484.0 | Zinc finger | NR C4-type |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 1440_1490 | 1275.3333333333333 | 1500.0 | Compositional bias | Note=Pro-rich |
| Hgene | ARHGAP35 | chr19:47440665 | chr6:152201789 | ENST00000404338 | + | 2 | 6 | 1249_1436 | 1275.3333333333333 | 1500.0 | Domain | Rho-GAP |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000206249 | 1 | 8 | 185_250 | 214.33333333333334 | 596.0 | DNA binding | Nuclear receptor | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000338799 | 2 | 9 | 185_250 | 214.33333333333334 | 596.0 | DNA binding | Nuclear receptor | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000440973 | 3 | 10 | 185_250 | 214.33333333333334 | 596.0 | DNA binding | Nuclear receptor | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000443427 | 2 | 9 | 185_250 | 214.33333333333334 | 596.0 | DNA binding | Nuclear receptor | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000456483 | 2 | 8 | 185_250 | 214.33333333333334 | 484.0 | DNA binding | Nuclear receptor | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000206249 | 1 | 8 | 185_205 | 214.33333333333334 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000338799 | 2 | 9 | 185_205 | 214.33333333333334 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000440973 | 3 | 10 | 185_205 | 214.33333333333334 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000443427 | 2 | 9 | 185_205 | 214.33333333333334 | 596.0 | Zinc finger | NR C4-type | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000456483 | 2 | 8 | 185_205 | 214.33333333333334 | 484.0 | Zinc finger | NR C4-type |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ARHGAP35 | |
| ESR1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000206249 | 1 | 8 | 35_174 | 214.33333333333334 | 596.0 | DDX5%3B self-association | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000338799 | 2 | 9 | 35_174 | 214.33333333333334 | 596.0 | DDX5%3B self-association | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000440973 | 3 | 10 | 35_174 | 214.33333333333334 | 596.0 | DDX5%3B self-association | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000443427 | 2 | 9 | 35_174 | 214.33333333333334 | 596.0 | DDX5%3B self-association | |
| Tgene | ESR1 | chr19:47440665 | chr6:152201789 | ENST00000456483 | 2 | 8 | 35_174 | 214.33333333333334 | 484.0 | DDX5%3B self-association |
Top |
Related Drugs to ARHGAP35-ESR1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ARHGAP35-ESR1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

