| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:PBRM1-TMEM110-MUSTN1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: PBRM1-TMEM110-MUSTN1 | FusionPDB ID: 63012 | FusionGDB2.0 ID: 63012 | Hgene | Tgene | Gene symbol | PBRM1 | TMEM110-MUSTN1 | Gene ID | 55193 | 100526772 |
| Gene name | polybromo 1 | STIMATE-MUSTN1 readthrough | |
| Synonyms | BAF180|PB1 | TMEM110-MUSTN1 | |
| Cytomap | 3p21.1 | 3p21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein polybromo-1BRG1-associated factor 180polybromo-1D | TMEM110-MUSTN1 fusion proteinTMEM110-MUSTN1 read-through transcriptTMEM110-MUSTN1 readthrough | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000394830, ENST00000296302, ENST00000337303, ENST00000356770, ENST00000409057, ENST00000409114, ENST00000409767, ENST00000410007, | ENST00000504329, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 15 X 15 X 10=2250 | 1 X 1 X 1=1 |
| # samples | 19 | 1 | |
| ** MAII score | log2(19/2250*10)=-3.56585367777345 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(1/1*10)=3.32192809488736 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: PBRM1 [Title/Abstract] AND TMEM110-MUSTN1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | PBRM1(52584437)-TMEM110-MUSTN1(52889460), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | PBRM1-TMEM110-MUSTN1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. PBRM1-TMEM110-MUSTN1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. PBRM1-TMEM110-MUSTN1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. PBRM1-TMEM110-MUSTN1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. PBRM1-TMEM110-MUSTN1 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. PBRM1-TMEM110-MUSTN1 seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. PBRM1-TMEM110-MUSTN1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. PBRM1-TMEM110-MUSTN1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. PBRM1-TMEM110-MUSTN1 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across PBRM1 (5'-gene) Fusion gene breakpoints across PBRM1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
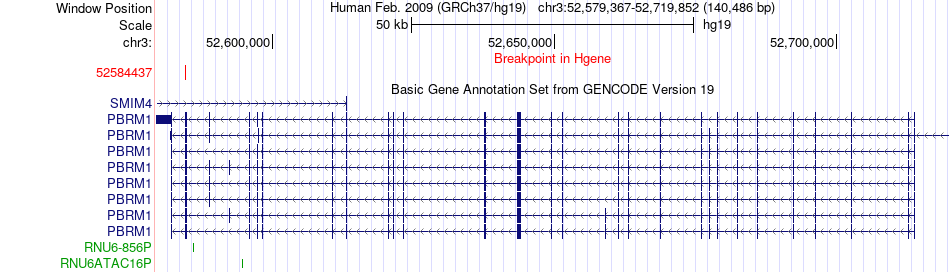 |
 Fusion gene breakpoints across TMEM110-MUSTN1 (3'-gene) Fusion gene breakpoints across TMEM110-MUSTN1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
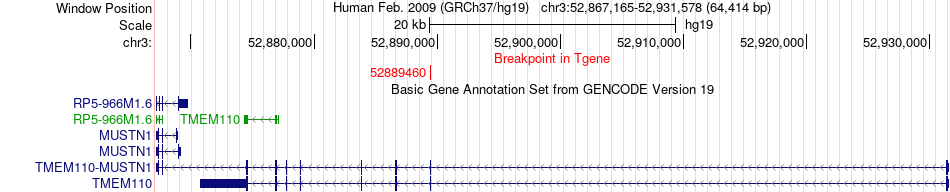 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-55-8085-01A | PBRM1 | chr3 | 52584437 | - | TMEM110-MUSTN1 | chr3 | 52889460 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000356770 | PBRM1 | chr3 | 52584437 | - | ENST00000504329 | TMEM110-MUSTN1 | chr3 | 52889460 | - | 5772 | 4639 | 3 | 5597 | 1864 |
| ENST00000296302 | PBRM1 | chr3 | 52584437 | - | ENST00000504329 | TMEM110-MUSTN1 | chr3 | 52889460 | - | 6032 | 4899 | 2 | 5857 | 1951 |
| ENST00000337303 | PBRM1 | chr3 | 52584437 | - | ENST00000504329 | TMEM110-MUSTN1 | chr3 | 52889460 | - | 5712 | 4579 | 3 | 5537 | 1844 |
| ENST00000410007 | PBRM1 | chr3 | 52584437 | - | ENST00000504329 | TMEM110-MUSTN1 | chr3 | 52889460 | - | 5793 | 4660 | 3 | 5618 | 1871 |
| ENST00000409057 | PBRM1 | chr3 | 52584437 | - | ENST00000504329 | TMEM110-MUSTN1 | chr3 | 52889460 | - | 5868 | 4735 | 3 | 5693 | 1896 |
| ENST00000409114 | PBRM1 | chr3 | 52584437 | - | ENST00000504329 | TMEM110-MUSTN1 | chr3 | 52889460 | - | 5922 | 4789 | 3 | 5747 | 1914 |
| ENST00000409767 | PBRM1 | chr3 | 52584437 | - | ENST00000504329 | TMEM110-MUSTN1 | chr3 | 52889460 | - | 5757 | 4624 | 3 | 5582 | 1859 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000356770 | ENST00000504329 | PBRM1 | chr3 | 52584437 | - | TMEM110-MUSTN1 | chr3 | 52889460 | - | 0.001051668 | 0.9989484 |
| ENST00000296302 | ENST00000504329 | PBRM1 | chr3 | 52584437 | - | TMEM110-MUSTN1 | chr3 | 52889460 | - | 0.001180482 | 0.99881953 |
| ENST00000337303 | ENST00000504329 | PBRM1 | chr3 | 52584437 | - | TMEM110-MUSTN1 | chr3 | 52889460 | - | 0.000524635 | 0.9994754 |
| ENST00000410007 | ENST00000504329 | PBRM1 | chr3 | 52584437 | - | TMEM110-MUSTN1 | chr3 | 52889460 | - | 0.000987623 | 0.99901235 |
| ENST00000409057 | ENST00000504329 | PBRM1 | chr3 | 52584437 | - | TMEM110-MUSTN1 | chr3 | 52889460 | - | 0.001097651 | 0.9989023 |
| ENST00000409114 | ENST00000504329 | PBRM1 | chr3 | 52584437 | - | TMEM110-MUSTN1 | chr3 | 52889460 | - | 0.000486713 | 0.99951327 |
| ENST00000409767 | ENST00000504329 | PBRM1 | chr3 | 52584437 | - | TMEM110-MUSTN1 | chr3 | 52889460 | - | 0.000554543 | 0.9994455 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >63012_63012_1_PBRM1-TMEM110-MUSTN1_PBRM1_chr3_52584437_ENST00000296302_TMEM110-MUSTN1_chr3_52889460_ENST00000504329_length(amino acids)=1951AA_BP=1630 MGSKRRRATSPSSSVSGDFDDGHHSVSTPGPSRKRRRLSNLPTVDPIAVCHELYNTIRDYKDEQGRLLCELFIRAPKRRNQPDYYEVVSQ PIDLMKIQQKLKMEEYDDVNLLTADFQLLFNNAKSYYKPDSPEYKAACKLWDLYLRTRNEFVQKGEADDEDDDEDGQDNQGTVTEGSSPA YLKEILEQLLEAIVVATNPSGRLISELFQKLPSKVQYPDYYAIIKEPIDLKTIAQRIQNGSYKSIHAMAKDIDLLAKNAKTYNEPGSQVF KDANSIKKIFYMKKAEIEHHEMAKSSLRMRTPSNLAAARLTGPSHSKGSLGEERNPTSKYYRNKRAVQGGRLSAITMALQYGSESEEDAA LAAARYEEGESEAESITSFMDVSNPFYQLYDTVRSCRNNQGQLIAEPFYHLPSKKKYPDYYQQIKMPISLQQIRTKLKNQEYETLDHLEC DLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGR RLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPD DDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQD IDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAV DPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHN DVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGE KWLYGCWFYRPNETFHLATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPI SSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHG LVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESR YNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLA SELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEERAAKVAEQQE RERAAQQQQPSASPRAGTPVGALMGVVPPPTPMGMLNQQLTPVAGMMGGYPPGLPPLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGVPGL PGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAE SNSISKWDQTLAVKRFREPKHERRPWRIWFLDTSKQAIGMLFIHFANVYLADLTEEDPCSLYLINFLLDATVGMLLIYVGVRAVSVLVEW QQWESLRFGEYGDPLQCGAWVGQCALYIVIMIFEKSVVFIVLLILQWKKVALLNPIENPDLKLAIVMLIVPFFVNALMFWVVDNFLMRKG KTKAKLEERGANQDSRNGSKVRYRRAASHEESESEILISADDEMEESDVEEDLRRLTPLKPVKKKKHRFGLPAGAQEAPIKKKRPPVKDE -------------------------------------------------------------- >63012_63012_2_PBRM1-TMEM110-MUSTN1_PBRM1_chr3_52584437_ENST00000337303_TMEM110-MUSTN1_chr3_52889460_ENST00000504329_length(amino acids)=1844AA_BP=1523 MGSKRRRATSPSSSVSGDFDDGHHSVSTPGPSRKRRRLSNLPTVDPIAVCHELYNTIRDYKDEQGRLLCELFIRAPKRRNQPDYYEVVSQ PIDLMKIQQKLKMEEYDDVNLLTADFQLLFNNAKSYYKPDSPEYKAACKLWDLYLRTRNEFVQKGEADDEDDDEDGQDNQGTVTEGSSPA YLKEILEQLLEAIVVATNPSGRLISELFQKLPSKVQYPDYYAIIKEPIDLKTIAQRIQNGSYKSIHAMAKDIDLLAKNAKTYNEPGSQVF KDANSIKKIFYMKKAEIEHHEMAKSSLRMRTPSNLAAARLTGPSHSKGSLGEERNPTSKYYRNKRAVQGGRLSAITMALQYGSESEEDAA LAAARYEEGESEAESITSFMDVSNPFYQLYDTVRSCRNNQGQLIAEPFYHLPSKKKYPDYYQQIKMPISLQQIRTKLKNQEYETLDHLEC DLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGR RLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPD DDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQD IDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAV DPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHN DVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGE KWLYGCWFYRPNETFHLATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPI SSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHG LVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESR YNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLA SELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGVMNQGVAPMV GTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAVKRFR EPKHERRPWRIWFLDTSKQAIGMLFIHFANVYLADLTEEDPCSLYLINFLLDATVGMLLIYVGVRAVSVLVEWQQWESLRFGEYGDPLQC GAWVGQCALYIVIMIFEKSVVFIVLLILQWKKVALLNPIENPDLKLAIVMLIVPFFVNALMFWVVDNFLMRKGKTKAKLEERGANQDSRN GSKVRYRRAASHEESESEILISADDEMEESDVEEDLRRLTPLKPVKKKKHRFGLPAGAQEAPIKKKRPPVKDEDLKGARGNLTKNQEIKS -------------------------------------------------------------- >63012_63012_3_PBRM1-TMEM110-MUSTN1_PBRM1_chr3_52584437_ENST00000356770_TMEM110-MUSTN1_chr3_52889460_ENST00000504329_length(amino acids)=1864AA_BP=1543 MGSKRRRATSPSSSVSGDFDDGHHSVSTPGPSRKRRRLSNLPTVDPIAVCHELYNTIRDYKDEQGRLLCELFIRAPKRRNQPDYYEVVSQ PIDLMKIQQKLKMEEYDDVNLLTADFQLLFNNAKSYYKPDSPEYKAACKLWDLYLRTRNEFVQKGEADDEDDDEDGQDNQGTVTEGSSPA YLKEILEQLLEAIVVATNPSGRLISELFQKLPSKVQYPDYYAIIKEPIDLKTIAQRIQNGSYKSIHAMAKDIDLLAKNAKTYNEPGSQVF KDANSIKKIFYMKKAEIEHHEMAKSSLRMSNKRAVQGGRLSAITMALQYGSESEEDAALAAARYEEGESEAESITSFMDVSNPFYQLYDT VRSCRNNQGQLIAEPFYHLPSKKKYPDYYQQIKMPISLQQIRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNSAIYKRVLKLQQVMQA KKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYKIILEPMDLKIIE HNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPKKSKYMTPMQQKL NEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNEPESLIYKDALVL HKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVENNRYRRLDLFQE HMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLKREEEKREAEKSE DSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHLATRKFLEKEVFKSDY YNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKN TDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEET EHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFR KPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKGSAKKEGSKRKIN MSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGMMGGYPPGLPPLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGV PGLPGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGL SAESNSISKWDQTLAVKRFREPKHERRPWRIWFLDTSKQAIGMLFIHFANVYLADLTEEDPCSLYLINFLLDATVGMLLIYVGVRAVSVL VEWQQWESLRFGEYGDPLQCGAWVGQCALYIVIMIFEKSVVFIVLLILQWKKVALLNPIENPDLKLAIVMLIVPFFVNALMFWVVDNFLM RKGKTKAKLEERGANQDSRNGSKVRYRRAASHEESESEILISADDEMEESDVEEDLRRLTPLKPVKKKKHRFGLPAGAQEAPIKKKRPPV -------------------------------------------------------------- >63012_63012_4_PBRM1-TMEM110-MUSTN1_PBRM1_chr3_52584437_ENST00000409057_TMEM110-MUSTN1_chr3_52889460_ENST00000504329_length(amino acids)=1896AA_BP=1575 MGSKRRRATSPSSSVSGDFDDGHHSVSTPGPSRKRRRLSNLPTVDPIAVCHELYNTIRDYKDEQGRLLCELFIRAPKRRNQPDYYEVVSQ PIDLMKIQQKLKMEEYDDVNLLTADFQLLFNNAKSYYKPDSPEYKAACKLWDLYLRTRNEFVQKGEADDEDDDEDGQDNQGTVTEGSSPA YLKEILEQLLEAIVVATNPSGRLISELFQKLPSKVQYPDYYAIIKEPIDLKTIAQRIQNGSYKSIHAMAKDIDLLAKNAKTYNEPGSQVF KDANSIKKIFYMKKAEIEHHEMAKSSLRMRTPSNLAAARLTGPSHSKGSLGEERNPTSKYYRNKRAVQGGRLSAITMALQYGSESEEDAA LAAARYEEGESEAESITSFMDVSNPFYQLYDTVRSCRNNQGQLIAEPFYHLPSKKKYPDYYQQIKMPISLQQIRTKLKNQEYETLDHLEC DLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGR RLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPD DDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQD IDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAV DPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHN DVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGE KWLYGCWFYRPNETFHLATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPI SSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHG LVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESR YNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLA SELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGMMGGYPPGLP PLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGVPGLPGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPV IQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAVKRFREPKHERRPWRIWFLDTSKQAIGMLFIHFANVYLADLTE EDPCSLYLINFLLDATVGMLLIYVGVRAVSVLVEWQQWESLRFGEYGDPLQCGAWVGQCALYIVIMIFEKSVVFIVLLILQWKKVALLNP IENPDLKLAIVMLIVPFFVNALMFWVVDNFLMRKGKTKAKLEERGANQDSRNGSKVRYRRAASHEESESEILISADDEMEESDVEEDLRR LTPLKPVKKKKHRFGLPAGAQEAPIKKKRPPVKDEDLKGARGNLTKNQEIKSKTYQVMRECEQAGSAAPSVFSRTRTGTETVFEKPKAGP -------------------------------------------------------------- >63012_63012_5_PBRM1-TMEM110-MUSTN1_PBRM1_chr3_52584437_ENST00000409114_TMEM110-MUSTN1_chr3_52889460_ENST00000504329_length(amino acids)=1914AA_BP=1593 MGSKRRRATSPSSSVSGDFDDGHHSVSTPGPSRKRRRLSNLPTVDPIAVCHELYNTIRDYKDEQGRLLCELFIRAPKRRNQPDYYEVVSQ PIDLMKIQQKLKMEEYDDVNLLTADFQLLFNNAKSYYKPDSPEYKAACKLWDLYLRTRNEFVQKGEADDEDDDEDGQDNQGTVTEGSSPA YLKEILEQLLEAIVVATNPSGRLISELFQKLPSKVQYPDYYAIIKEPIDLKTIAQRIQNGSYKSIHAMAKDIDLLAKNAKTYNEPGSQVF KDANSIKKIFYMKKAEIEHHEMAKSSLRMRTPSNLAAARLTGPSHSKGSLGEERNPTSKYYRNKRAVQGGRLSAITMALQYGSESEEDAA LAAARYEEGESEAESITSFMDVSNPFYQLYDTVRSCRNNQGQLIAEPFYHLPSKKKYPDYYQQIKMPISLQQIRTKLKNQEYETLDHLEC DLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKRNTHDSEMLGLRRLSSKKNIRKQRMKI LFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILE KLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMD MEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDD EGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEI LLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQP HIVCIERLWEDSAGEKWLYGCWFYRPNETFHLATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSA KTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYND MWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCR PTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSE VIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKK AEYEERAAKVAEQQERERAAQQQQPSASPRAGTPVGALMGVVPPPTPMGMLNQQLTPVAGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPP GQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAVKRFREPKHERRPWRIWFLDTSKQA IGMLFIHFANVYLADLTEEDPCSLYLINFLLDATVGMLLIYVGVRAVSVLVEWQQWESLRFGEYGDPLQCGAWVGQCALYIVIMIFEKSV VFIVLLILQWKKVALLNPIENPDLKLAIVMLIVPFFVNALMFWVVDNFLMRKGKTKAKLEERGANQDSRNGSKVRYRRAASHEESESEIL ISADDEMEESDVEEDLRRLTPLKPVKKKKHRFGLPAGAQEAPIKKKRPPVKDEDLKGARGNLTKNQEIKSKTYQVMRECEQAGSAAPSVF -------------------------------------------------------------- >63012_63012_6_PBRM1-TMEM110-MUSTN1_PBRM1_chr3_52584437_ENST00000409767_TMEM110-MUSTN1_chr3_52889460_ENST00000504329_length(amino acids)=1859AA_BP=1538 MGSKRRRATSPSSSVSGDFDDGHHSVSTPGPSRKRRRLSNLPTVDPIAVCHELYNTIRDYKDEQGRLLCELFIRAPKRRNQPDYYEVVSQ PIDLMKIQQKLKMEEYDDVNLLTADFQLLFNNAKSYYKPDSPEYKAACKLWDLYLRTRNEFVQKGEADDEDDDEDGQDNQGTVTEGSSPA YLKEILEQLLEAIVVATNPSGRLISELFQKLPSKVQYPDYYAIIKEPIDLKTIAQRIQNGSYKSIHAMAKDIDLLAKNAKTYNEPGSQVF KDANSIKKIFYMKKAEIEHHEMAKSSLRMRTPSNLAAARLTGPSHSKGSLGEERNPTSKYYRNKRAVQGGRLSAITMALQYGSESEEDAA LAAARYEEGESEAESITSFMDVSNPFYQLYDTVRSCRNNQGQLIAEPFYHLPSKKKYPDYYQQIKMPISLQQIRTKLKNQEYETLDHLEC DLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKRNTHDSEMLGLRRLSSKKNIRKQRMKI LFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILE KLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMD MEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDD EGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEI LLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQP HIVCIERLWEDSAGEKWLYGCWFYRPNETFHLATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSA KTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYND MWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCR PTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSE VIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKK AEYEGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESN SISKWDQTLAVKRFREPKHERRPWRIWFLDTSKQAIGMLFIHFANVYLADLTEEDPCSLYLINFLLDATVGMLLIYVGVRAVSVLVEWQQ WESLRFGEYGDPLQCGAWVGQCALYIVIMIFEKSVVFIVLLILQWKKVALLNPIENPDLKLAIVMLIVPFFVNALMFWVVDNFLMRKGKT KAKLEERGANQDSRNGSKVRYRRAASHEESESEILISADDEMEESDVEEDLRRLTPLKPVKKKKHRFGLPAGAQEAPIKKKRPPVKDEDL -------------------------------------------------------------- >63012_63012_7_PBRM1-TMEM110-MUSTN1_PBRM1_chr3_52584437_ENST00000410007_TMEM110-MUSTN1_chr3_52889460_ENST00000504329_length(amino acids)=1871AA_BP=1550 MGSKRRRATSPSSSVSGDFDDGHHSVSTPGPSRKRRRLSNLPTVDPIAVCHELYNTIRDYKDEQGRLLCELFIRAPKRRNQPDYYEVVSQ PIDLMKIQQKLKMEEYDDVNLLTADFQLLFNNAKSYYKPDSPEYKAACKLWDLYLRTRNEFVQKGEADDEDDDEDGQDNQGTVTEGSSPA YLKEILEQLLEAIVVATNPSGRLISELFQKLPSKVQYPDYYAIIKEPIDLKTIAQRIQNGSYKSIHAMAKDIDLLAKNAKTYNEPGSQVF KDANSIKKIFYMKKAEIEHHEMAKSSLRMRTPSNLAAARLTGPSHSKGSLGEERNPTSKYYRNKRAVQGGRLSAITMALQYGSESEEDAA LAAARYEEGESEAESITSFMDVSNPFYQLYDTVRSCRNNQGQLIAEPFYHLPSKKKYPDYYQQIKMPISLQQIRTKLKNQEYETLDHLEC DLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGR RLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPD DDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQD IDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAV DPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHN DVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAEK EVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANAD KGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPI FIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVD DEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKGSAKKE GSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGMMGGYPPGLPPLQGPVDGLVSMGSMQPLHPGGPPP HHLPPGVPGLPGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAY LKYIEGLSAESNSISKWDQTLAVKRFREPKHERRPWRIWFLDTSKQAIGMLFIHFANVYLADLTEEDPCSLYLINFLLDATVGMLLIYVG VRAVSVLVEWQQWESLRFGEYGDPLQCGAWVGQCALYIVIMIFEKSVVFIVLLILQWKKVALLNPIENPDLKLAIVMLIVPFFVNALMFW VVDNFLMRKGKTKAKLEERGANQDSRNGSKVRYRRAASHEESESEILISADDEMEESDVEEDLRRLTPLKPVKKKKHRFGLPAGAQEAPI -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:52584437/chr3:52889460) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 1468_1599 | 1632.3333333333333 | 1690.0 | Compositional bias | Note=Pro-rich |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 630_633 | 1632.3333333333333 | 1690.0 | Compositional bias | Note=Poly-Asp |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 630_633 | 1525.3333333333333 | 1583.0 | Compositional bias | Note=Poly-Asp |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 630_633 | 1545.3333333333333 | 1603.0 | Compositional bias | Note=Poly-Asp |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 630_633 | 1525.3333333333333 | 1583.0 | Compositional bias | Note=Poly-Asp |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 630_633 | 1577.3333333333333 | 1635.0 | Compositional bias | Note=Poly-Asp |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 630_633 | 1595.3333333333333 | 1653.0 | Compositional bias | Note=Poly-Asp |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 630_633 | 1540.3333333333333 | 1598.0 | Compositional bias | Note=Poly-Asp |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 630_633 | 1552.3333333333333 | 1610.0 | Compositional bias | Note=Poly-Asp |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 1379_1447 | 1632.3333333333333 | 1690.0 | DNA binding | HMG box |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 1379_1447 | 1525.3333333333333 | 1583.0 | DNA binding | HMG box |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 1379_1447 | 1545.3333333333333 | 1603.0 | DNA binding | HMG box |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 1379_1447 | 1525.3333333333333 | 1583.0 | DNA binding | HMG box |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 1379_1447 | 1577.3333333333333 | 1635.0 | DNA binding | HMG box |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 1379_1447 | 1595.3333333333333 | 1653.0 | DNA binding | HMG box |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 1379_1447 | 1540.3333333333333 | 1598.0 | DNA binding | HMG box |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 1379_1447 | 1552.3333333333333 | 1610.0 | DNA binding | HMG box |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 1156_1272 | 1632.3333333333333 | 1690.0 | Domain | BAH 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 200_270 | 1632.3333333333333 | 1690.0 | Domain | Bromo 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 400_470 | 1632.3333333333333 | 1690.0 | Domain | Bromo 3 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 538_608 | 1632.3333333333333 | 1690.0 | Domain | Bromo 4 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 64_134 | 1632.3333333333333 | 1690.0 | Domain | Bromo 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 676_746 | 1632.3333333333333 | 1690.0 | Domain | Bromo 5 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 792_862 | 1632.3333333333333 | 1690.0 | Domain | Bromo 6 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000296302 | - | 29 | 30 | 956_1074 | 1632.3333333333333 | 1690.0 | Domain | BAH 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 1156_1272 | 1525.3333333333333 | 1583.0 | Domain | BAH 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 200_270 | 1525.3333333333333 | 1583.0 | Domain | Bromo 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 400_470 | 1525.3333333333333 | 1583.0 | Domain | Bromo 3 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 538_608 | 1525.3333333333333 | 1583.0 | Domain | Bromo 4 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 64_134 | 1525.3333333333333 | 1583.0 | Domain | Bromo 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 676_746 | 1525.3333333333333 | 1583.0 | Domain | Bromo 5 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 792_862 | 1525.3333333333333 | 1583.0 | Domain | Bromo 6 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 956_1074 | 1525.3333333333333 | 1583.0 | Domain | BAH 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 1156_1272 | 1545.3333333333333 | 1603.0 | Domain | BAH 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 200_270 | 1545.3333333333333 | 1603.0 | Domain | Bromo 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 400_470 | 1545.3333333333333 | 1603.0 | Domain | Bromo 3 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 538_608 | 1545.3333333333333 | 1603.0 | Domain | Bromo 4 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 64_134 | 1545.3333333333333 | 1603.0 | Domain | Bromo 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 676_746 | 1545.3333333333333 | 1603.0 | Domain | Bromo 5 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 792_862 | 1545.3333333333333 | 1603.0 | Domain | Bromo 6 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 956_1074 | 1545.3333333333333 | 1603.0 | Domain | BAH 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 1156_1272 | 1525.3333333333333 | 1583.0 | Domain | BAH 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 200_270 | 1525.3333333333333 | 1583.0 | Domain | Bromo 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 400_470 | 1525.3333333333333 | 1583.0 | Domain | Bromo 3 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 538_608 | 1525.3333333333333 | 1583.0 | Domain | Bromo 4 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 64_134 | 1525.3333333333333 | 1583.0 | Domain | Bromo 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 676_746 | 1525.3333333333333 | 1583.0 | Domain | Bromo 5 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 792_862 | 1525.3333333333333 | 1583.0 | Domain | Bromo 6 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 956_1074 | 1525.3333333333333 | 1583.0 | Domain | BAH 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 1156_1272 | 1577.3333333333333 | 1635.0 | Domain | BAH 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 200_270 | 1577.3333333333333 | 1635.0 | Domain | Bromo 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 400_470 | 1577.3333333333333 | 1635.0 | Domain | Bromo 3 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 538_608 | 1577.3333333333333 | 1635.0 | Domain | Bromo 4 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 64_134 | 1577.3333333333333 | 1635.0 | Domain | Bromo 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 676_746 | 1577.3333333333333 | 1635.0 | Domain | Bromo 5 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 792_862 | 1577.3333333333333 | 1635.0 | Domain | Bromo 6 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 956_1074 | 1577.3333333333333 | 1635.0 | Domain | BAH 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 1156_1272 | 1595.3333333333333 | 1653.0 | Domain | BAH 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 200_270 | 1595.3333333333333 | 1653.0 | Domain | Bromo 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 400_470 | 1595.3333333333333 | 1653.0 | Domain | Bromo 3 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 538_608 | 1595.3333333333333 | 1653.0 | Domain | Bromo 4 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 64_134 | 1595.3333333333333 | 1653.0 | Domain | Bromo 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 676_746 | 1595.3333333333333 | 1653.0 | Domain | Bromo 5 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 792_862 | 1595.3333333333333 | 1653.0 | Domain | Bromo 6 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 956_1074 | 1595.3333333333333 | 1653.0 | Domain | BAH 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 1156_1272 | 1540.3333333333333 | 1598.0 | Domain | BAH 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 200_270 | 1540.3333333333333 | 1598.0 | Domain | Bromo 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 400_470 | 1540.3333333333333 | 1598.0 | Domain | Bromo 3 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 538_608 | 1540.3333333333333 | 1598.0 | Domain | Bromo 4 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 64_134 | 1540.3333333333333 | 1598.0 | Domain | Bromo 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 676_746 | 1540.3333333333333 | 1598.0 | Domain | Bromo 5 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 792_862 | 1540.3333333333333 | 1598.0 | Domain | Bromo 6 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 956_1074 | 1540.3333333333333 | 1598.0 | Domain | BAH 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 1156_1272 | 1552.3333333333333 | 1610.0 | Domain | BAH 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 200_270 | 1552.3333333333333 | 1610.0 | Domain | Bromo 2 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 400_470 | 1552.3333333333333 | 1610.0 | Domain | Bromo 3 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 538_608 | 1552.3333333333333 | 1610.0 | Domain | Bromo 4 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 64_134 | 1552.3333333333333 | 1610.0 | Domain | Bromo 1 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 676_746 | 1552.3333333333333 | 1610.0 | Domain | Bromo 5 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 792_862 | 1552.3333333333333 | 1610.0 | Domain | Bromo 6 |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 956_1074 | 1552.3333333333333 | 1610.0 | Domain | BAH 1 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000337303 | - | 27 | 28 | 1468_1599 | 1525.3333333333333 | 1583.0 | Compositional bias | Note=Pro-rich |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000356770 | - | 27 | 28 | 1468_1599 | 1545.3333333333333 | 1603.0 | Compositional bias | Note=Pro-rich |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000394830 | - | 29 | 30 | 1468_1599 | 1525.3333333333333 | 1583.0 | Compositional bias | Note=Pro-rich |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409057 | - | 28 | 29 | 1468_1599 | 1577.3333333333333 | 1635.0 | Compositional bias | Note=Pro-rich |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409114 | - | 29 | 30 | 1468_1599 | 1595.3333333333333 | 1653.0 | Compositional bias | Note=Pro-rich |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000409767 | - | 28 | 29 | 1468_1599 | 1540.3333333333333 | 1598.0 | Compositional bias | Note=Pro-rich |
| Hgene | PBRM1 | chr3:52584437 | chr3:52889460 | ENST00000410007 | - | 28 | 29 | 1468_1599 | 1552.3333333333333 | 1610.0 | Compositional bias | Note=Pro-rich |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| PBRM1 | |
| TMEM110-MUSTN1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to PBRM1-TMEM110-MUSTN1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to PBRM1-TMEM110-MUSTN1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

