| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:PCCB-KDM4A |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: PCCB-KDM4A | FusionPDB ID: 63171 | FusionGDB2.0 ID: 63171 | Hgene | Tgene | Gene symbol | PCCB | KDM4A | Gene ID | 5096 | 9682 |
| Gene name | propionyl-CoA carboxylase subunit beta | lysine demethylase 4A | |
| Synonyms | - | JHDM3A|JMJD2|JMJD2A|TDRD14A | |
| Cytomap | 3q22.3 | 1p34.2-p34.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | propionyl-CoA carboxylase beta chain, mitochondrialPCCase subunit betapropanoyl-CoA:carbon dioxide ligase subunit betapropionyl CoA carboxylase, beta polypeptidepropionyl Coenzyme A carboxylase, beta polypeptidepropionyl-CoA carboxylase beta subunit | lysine-specific demethylase 4AjmjC domain-containing histone demethylation protein 3Ajumonji C domain-containing histone demethylase 3Ajumonji domain containing 2jumonji domain containing 2Ajumonji domain-containing protein 2Alysine (K)-specific dem | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | O75164 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000474833, ENST00000251654, ENST00000462637, ENST00000466072, ENST00000468777, ENST00000469217, ENST00000471595, ENST00000482086, ENST00000483687, ENST00000490504, ENST00000478469, | ENST00000372396, ENST00000463151, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 8 X 9 X 4=288 | 4 X 4 X 3=48 |
| # samples | 8 | 4 | |
| ** MAII score | log2(8/288*10)=-1.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/48*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: PCCB [Title/Abstract] AND KDM4A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | PCCB(136019953)-KDM4A(44128564), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | PCCB-KDM4A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. PCCB-KDM4A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. PCCB-KDM4A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. PCCB-KDM4A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | KDM4A | GO:0016577 | histone demethylation | 16024779 |
| Tgene | KDM4A | GO:0045892 | negative regulation of transcription, DNA-templated | 16024779 |
| Tgene | KDM4A | GO:0070544 | histone H3-K36 demethylation | 21914792 |
 Fusion gene breakpoints across PCCB (5'-gene) Fusion gene breakpoints across PCCB (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
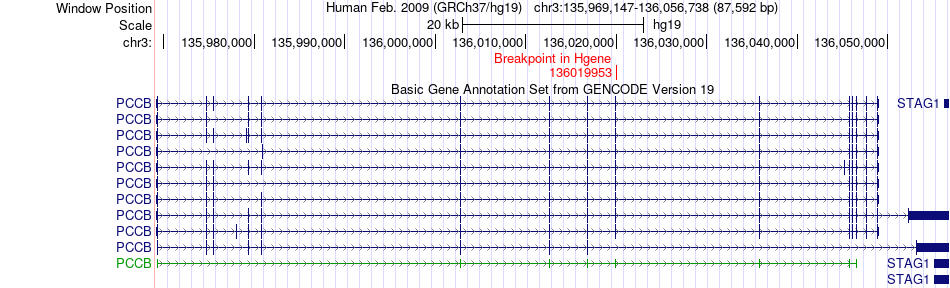 |
 Fusion gene breakpoints across KDM4A (3'-gene) Fusion gene breakpoints across KDM4A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
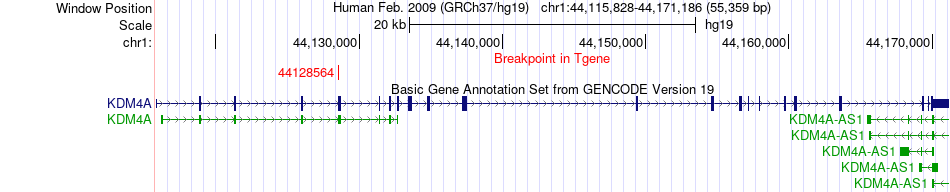 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-13-1403 | PCCB | chr3 | 136019953 | + | KDM4A | chr1 | 44128564 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000471595 | PCCB | chr3 | 136019953 | + | ENST00000372396 | KDM4A | chr1 | 44128564 | + | 4895 | 984 | 18 | 3749 | 1243 |
| ENST00000251654 | PCCB | chr3 | 136019953 | + | ENST00000372396 | KDM4A | chr1 | 44128564 | + | 4947 | 1036 | 70 | 3801 | 1243 |
| ENST00000483687 | PCCB | chr3 | 136019953 | + | ENST00000372396 | KDM4A | chr1 | 44128564 | + | 4841 | 930 | 21 | 3695 | 1224 |
| ENST00000490504 | PCCB | chr3 | 136019953 | + | ENST00000372396 | KDM4A | chr1 | 44128564 | + | 4727 | 816 | 21 | 3581 | 1186 |
| ENST00000466072 | PCCB | chr3 | 136019953 | + | ENST00000372396 | KDM4A | chr1 | 44128564 | + | 4898 | 987 | 21 | 3752 | 1243 |
| ENST00000482086 | PCCB | chr3 | 136019953 | + | ENST00000372396 | KDM4A | chr1 | 44128564 | + | 4550 | 639 | 21 | 3404 | 1127 |
| ENST00000468777 | PCCB | chr3 | 136019953 | + | ENST00000372396 | KDM4A | chr1 | 44128564 | + | 4991 | 1080 | 21 | 3845 | 1274 |
| ENST00000462637 | PCCB | chr3 | 136019953 | + | ENST00000372396 | KDM4A | chr1 | 44128564 | + | 4829 | 918 | 21 | 3683 | 1220 |
| ENST00000469217 | PCCB | chr3 | 136019953 | + | ENST00000372396 | KDM4A | chr1 | 44128564 | + | 4954 | 1043 | 17 | 3808 | 1263 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000471595 | ENST00000372396 | PCCB | chr3 | 136019953 | + | KDM4A | chr1 | 44128564 | + | 0.000674837 | 0.99932516 |
| ENST00000251654 | ENST00000372396 | PCCB | chr3 | 136019953 | + | KDM4A | chr1 | 44128564 | + | 0.000706521 | 0.99929345 |
| ENST00000483687 | ENST00000372396 | PCCB | chr3 | 136019953 | + | KDM4A | chr1 | 44128564 | + | 0.0006907 | 0.9993093 |
| ENST00000490504 | ENST00000372396 | PCCB | chr3 | 136019953 | + | KDM4A | chr1 | 44128564 | + | 0.000625656 | 0.99937433 |
| ENST00000466072 | ENST00000372396 | PCCB | chr3 | 136019953 | + | KDM4A | chr1 | 44128564 | + | 0.000680028 | 0.99932003 |
| ENST00000482086 | ENST00000372396 | PCCB | chr3 | 136019953 | + | KDM4A | chr1 | 44128564 | + | 0.000713133 | 0.99928695 |
| ENST00000468777 | ENST00000372396 | PCCB | chr3 | 136019953 | + | KDM4A | chr1 | 44128564 | + | 0.000673295 | 0.9993267 |
| ENST00000462637 | ENST00000372396 | PCCB | chr3 | 136019953 | + | KDM4A | chr1 | 44128564 | + | 0.000727745 | 0.9992723 |
| ENST00000469217 | ENST00000372396 | PCCB | chr3 | 136019953 | + | KDM4A | chr1 | 44128564 | + | 0.000677179 | 0.9993229 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >63171_63171_1_PCCB-KDM4A_PCCB_chr3_136019953_ENST00000251654_KDM4A_chr1_44128564_ENST00000372396_length(amino acids)=1243AA_BP=322 MAAALRVAAVGARLSVLASGLRAAVRSLCSQATSVNERIENKRRTALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFVESDMFVEHRC ADFGMAADKNKFPGDSVVTGRGRINGRLVYVFSQDFTVFGGSLSGAHAQKICKIMDQAITVGAPVIGLNDSGGARIQEGVESLAGYADIF LRNVTASGVIPQISLIMGPCAGGAVYSPALTDFTFMVKDTSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTMSGVAHRAFENDVDALCN LRDFFNYLPLSSQDPAPVRECHDPSDRLVPELDTIVPLESTKAYNMVDIIHSHVDEWNIGRLRTILDLVEKESGITIEGVNTPYLYFGMW KTSFAWHTEDMDLYSINYLHFGEPKSWYSVPPEHGKRLERLAKGFFPGSAQSCEAFLRHKMTLISPLMLKKYGIPFDKVTQEAGEFMITF PYGYHAGFNHGFNCAESTNFATRRWIEYGKQAVLCSCRKDMVKISMDVFVRKFQPERYKLWKAGKDNTVIDHTLPTPEAAEFLKESELPP RAGNEEECPEEDMEGVEDGEEGDLKTSLAKHRIGTKRHRVCLEIPQEVSQSELFPKEDLSSEQYEMTECPAALAPVRPTHSSVRQVEDGL TFPDYSDSTEVKFEELKNVKLEEEDEEEEQAAAALDLSVNPASVGGRLVFSGSKKKSSSSLGSGSSRDSISSDSETSEPLSCRAQGQTGV LTVHSYAKGDGRVTVGEPCTRKKGSAARSFSERELAEVADEYMFSLEENKKSKGRRQPLSKLPRHHPLVLQECVSDDETSEQLTPEEEAE ETEAWAKPLSQLWQNRPPNFEAEKEFNETMAQQAPHCAVCMIFQTYHQVEFGGFNQNCGNASDLAPQKQRTKPLIPEMCFTSTGCSTDIN LSTPYLEEDGTSILVSCKKCSVRVHASCYGVPPAKASEDWMCSRCSANALEEDCCLCSLRGGALQRANDDRWVHVSCAVAILEARFVNIA ERSPVDVSKIPLPRFKLKCIFCKKRRKRTAGCCVQCSHGRCPTAFHVSCAQAAGVMMQPDDWPFVVFITCFRHKIPNLERAKGALQSITA GQKVISKHKNGRFYQCEVVRLTTETFYEVNFDDGSFSDNLYPEDIVSQDCLQFGPPAEGEVVQVRWTDGQVYGAKFVASHPIQMYQVEFE -------------------------------------------------------------- >63171_63171_2_PCCB-KDM4A_PCCB_chr3_136019953_ENST00000462637_KDM4A_chr1_44128564_ENST00000372396_length(amino acids)=1220AA_BP=299 MAAALRVAAVGARLSVLASGLRAAVRSLCSQATSVNERIENKRRTALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFVESDMFVEHRC ADFGMAADKNKDFTVFGGSLSGAHAQKICKIMDQAITVGAPVIGLNDSGGARIQEGVESLAGYADIFLRNVTASGVIPQISLIMGPCAGG AVYSPALTDFTFMVKDTSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTMSGVAHRAFENDVDALCNLRDFFNYLPLSSQDPAPVRECHD PSDRLVPELDTIVPLESTKAYNMVDIIHSHVDEWNIGRLRTILDLVEKESGITIEGVNTPYLYFGMWKTSFAWHTEDMDLYSINYLHFGE PKSWYSVPPEHGKRLERLAKGFFPGSAQSCEAFLRHKMTLISPLMLKKYGIPFDKVTQEAGEFMITFPYGYHAGFNHGFNCAESTNFATR RWIEYGKQAVLCSCRKDMVKISMDVFVRKFQPERYKLWKAGKDNTVIDHTLPTPEAAEFLKESELPPRAGNEEECPEEDMEGVEDGEEGD LKTSLAKHRIGTKRHRVCLEIPQEVSQSELFPKEDLSSEQYEMTECPAALAPVRPTHSSVRQVEDGLTFPDYSDSTEVKFEELKNVKLEE EDEEEEQAAAALDLSVNPASVGGRLVFSGSKKKSSSSLGSGSSRDSISSDSETSEPLSCRAQGQTGVLTVHSYAKGDGRVTVGEPCTRKK GSAARSFSERELAEVADEYMFSLEENKKSKGRRQPLSKLPRHHPLVLQECVSDDETSEQLTPEEEAEETEAWAKPLSQLWQNRPPNFEAE KEFNETMAQQAPHCAVCMIFQTYHQVEFGGFNQNCGNASDLAPQKQRTKPLIPEMCFTSTGCSTDINLSTPYLEEDGTSILVSCKKCSVR VHASCYGVPPAKASEDWMCSRCSANALEEDCCLCSLRGGALQRANDDRWVHVSCAVAILEARFVNIAERSPVDVSKIPLPRFKLKCIFCK KRRKRTAGCCVQCSHGRCPTAFHVSCAQAAGVMMQPDDWPFVVFITCFRHKIPNLERAKGALQSITAGQKVISKHKNGRFYQCEVVRLTT ETFYEVNFDDGSFSDNLYPEDIVSQDCLQFGPPAEGEVVQVRWTDGQVYGAKFVASHPIQMYQVEFEDGSQLVVKRDDVYTLDEELPKRV -------------------------------------------------------------- >63171_63171_3_PCCB-KDM4A_PCCB_chr3_136019953_ENST00000466072_KDM4A_chr1_44128564_ENST00000372396_length(amino acids)=1243AA_BP=322 MAAALRVAAVGARLSVLASGLRAAVRSLCSQATSVNERIENKRRTALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFVESDMFVEHRC ADFGMAADKNKFPGDSVVTGRGRINGRLVYVFSQDFTVFGGSLSGAHAQKICKIMDQAITVGAPVIGLNDSGGARIQEGVESLAGYADIF LRNVTASGVIPQISLIMGPCAGGAVYSPALTDFTFMVKDTSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTMSGVAHRAFENDVDALCN LRDFFNYLPLSSQDPAPVRECHDPSDRLVPELDTIVPLESTKAYNMVDIIHSHVDEWNIGRLRTILDLVEKESGITIEGVNTPYLYFGMW KTSFAWHTEDMDLYSINYLHFGEPKSWYSVPPEHGKRLERLAKGFFPGSAQSCEAFLRHKMTLISPLMLKKYGIPFDKVTQEAGEFMITF PYGYHAGFNHGFNCAESTNFATRRWIEYGKQAVLCSCRKDMVKISMDVFVRKFQPERYKLWKAGKDNTVIDHTLPTPEAAEFLKESELPP RAGNEEECPEEDMEGVEDGEEGDLKTSLAKHRIGTKRHRVCLEIPQEVSQSELFPKEDLSSEQYEMTECPAALAPVRPTHSSVRQVEDGL TFPDYSDSTEVKFEELKNVKLEEEDEEEEQAAAALDLSVNPASVGGRLVFSGSKKKSSSSLGSGSSRDSISSDSETSEPLSCRAQGQTGV LTVHSYAKGDGRVTVGEPCTRKKGSAARSFSERELAEVADEYMFSLEENKKSKGRRQPLSKLPRHHPLVLQECVSDDETSEQLTPEEEAE ETEAWAKPLSQLWQNRPPNFEAEKEFNETMAQQAPHCAVCMIFQTYHQVEFGGFNQNCGNASDLAPQKQRTKPLIPEMCFTSTGCSTDIN LSTPYLEEDGTSILVSCKKCSVRVHASCYGVPPAKASEDWMCSRCSANALEEDCCLCSLRGGALQRANDDRWVHVSCAVAILEARFVNIA ERSPVDVSKIPLPRFKLKCIFCKKRRKRTAGCCVQCSHGRCPTAFHVSCAQAAGVMMQPDDWPFVVFITCFRHKIPNLERAKGALQSITA GQKVISKHKNGRFYQCEVVRLTTETFYEVNFDDGSFSDNLYPEDIVSQDCLQFGPPAEGEVVQVRWTDGQVYGAKFVASHPIQMYQVEFE -------------------------------------------------------------- >63171_63171_4_PCCB-KDM4A_PCCB_chr3_136019953_ENST00000468777_KDM4A_chr1_44128564_ENST00000372396_length(amino acids)=1274AA_BP=132 MAAALRVAAVGARLSVLASGLRAAVRSLCSQATSVNERIENKRRTALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFVESDMFVEHRC ADFGMAADKNKFPGDSVVTGRGRINGRLVYVFSQIRASAMSPRLVSDSWTQAIHLPWPPKVLGLQDFTVFGGSLSGAHAQKICKIMDQAI TVGAPVIGLNDSGGARIQEGVESLAGYADIFLRNVTASGVIPQISLIMGPCAGGAVYSPALTDFTFMVKDTSYLFITGPDVVKSVTNEDV TQEELGGAKTHTTMSGVAHRAFENDVDALCNLRDFFNYLPLSSQDPAPVRECHDPSDRLVPELDTIVPLESTKAYNMVDIIHSHVDEWNI GRLRTILDLVEKESGITIEGVNTPYLYFGMWKTSFAWHTEDMDLYSINYLHFGEPKSWYSVPPEHGKRLERLAKGFFPGSAQSCEAFLRH KMTLISPLMLKKYGIPFDKVTQEAGEFMITFPYGYHAGFNHGFNCAESTNFATRRWIEYGKQAVLCSCRKDMVKISMDVFVRKFQPERYK LWKAGKDNTVIDHTLPTPEAAEFLKESELPPRAGNEEECPEEDMEGVEDGEEGDLKTSLAKHRIGTKRHRVCLEIPQEVSQSELFPKEDL SSEQYEMTECPAALAPVRPTHSSVRQVEDGLTFPDYSDSTEVKFEELKNVKLEEEDEEEEQAAAALDLSVNPASVGGRLVFSGSKKKSSS SLGSGSSRDSISSDSETSEPLSCRAQGQTGVLTVHSYAKGDGRVTVGEPCTRKKGSAARSFSERELAEVADEYMFSLEENKKSKGRRQPL SKLPRHHPLVLQECVSDDETSEQLTPEEEAEETEAWAKPLSQLWQNRPPNFEAEKEFNETMAQQAPHCAVCMIFQTYHQVEFGGFNQNCG NASDLAPQKQRTKPLIPEMCFTSTGCSTDINLSTPYLEEDGTSILVSCKKCSVRVHASCYGVPPAKASEDWMCSRCSANALEEDCCLCSL RGGALQRANDDRWVHVSCAVAILEARFVNIAERSPVDVSKIPLPRFKLKCIFCKKRRKRTAGCCVQCSHGRCPTAFHVSCAQAAGVMMQP DDWPFVVFITCFRHKIPNLERAKGALQSITAGQKVISKHKNGRFYQCEVVRLTTETFYEVNFDDGSFSDNLYPEDIVSQDCLQFGPPAEG EVVQVRWTDGQVYGAKFVASHPIQMYQVEFEDGSQLVVKRDDVYTLDEELPKRVKSRLSVASDMRFNEIFTEKEVKQEKKRQRVINSRYR -------------------------------------------------------------- >63171_63171_5_PCCB-KDM4A_PCCB_chr3_136019953_ENST00000469217_KDM4A_chr1_44128564_ENST00000372396_length(amino acids)=1263AA_BP=128 MAAALRVAAVGARLSVLASGLRAAVRSLCSQATSVNERIENKRRTALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFVESDMFVEHRC ADFGMAADKNKFPGDSVVTGRGRINGRLVYVFSQQIIGWAQWLPLVISALWEAEDFTVFGGSLSGAHAQKICKIMDQAITVGAPVIGLND SGGARIQEGVESLAGYADIFLRNVTASGVIPQISLIMGPCAGGAVYSPALTDFTFMVKDTSYLFITGPDVVKSVTNEDVTQEELGGAKTH TTMSGVAHRAFENDVDALCNLRDFFNYLPLSSQDPAPVRECHDPSDRLVPELDTIVPLESTKAYNMVDIIHSHVDEWNIGRLRTILDLVE KESGITIEGVNTPYLYFGMWKTSFAWHTEDMDLYSINYLHFGEPKSWYSVPPEHGKRLERLAKGFFPGSAQSCEAFLRHKMTLISPLMLK KYGIPFDKVTQEAGEFMITFPYGYHAGFNHGFNCAESTNFATRRWIEYGKQAVLCSCRKDMVKISMDVFVRKFQPERYKLWKAGKDNTVI DHTLPTPEAAEFLKESELPPRAGNEEECPEEDMEGVEDGEEGDLKTSLAKHRIGTKRHRVCLEIPQEVSQSELFPKEDLSSEQYEMTECP AALAPVRPTHSSVRQVEDGLTFPDYSDSTEVKFEELKNVKLEEEDEEEEQAAAALDLSVNPASVGGRLVFSGSKKKSSSSLGSGSSRDSI SSDSETSEPLSCRAQGQTGVLTVHSYAKGDGRVTVGEPCTRKKGSAARSFSERELAEVADEYMFSLEENKKSKGRRQPLSKLPRHHPLVL QECVSDDETSEQLTPEEEAEETEAWAKPLSQLWQNRPPNFEAEKEFNETMAQQAPHCAVCMIFQTYHQVEFGGFNQNCGNASDLAPQKQR TKPLIPEMCFTSTGCSTDINLSTPYLEEDGTSILVSCKKCSVRVHASCYGVPPAKASEDWMCSRCSANALEEDCCLCSLRGGALQRANDD RWVHVSCAVAILEARFVNIAERSPVDVSKIPLPRFKLKCIFCKKRRKRTAGCCVQCSHGRCPTAFHVSCAQAAGVMMQPDDWPFVVFITC FRHKIPNLERAKGALQSITAGQKVISKHKNGRFYQCEVVRLTTETFYEVNFDDGSFSDNLYPEDIVSQDCLQFGPPAEGEVVQVRWTDGQ VYGAKFVASHPIQMYQVEFEDGSQLVVKRDDVYTLDEELPKRVKSRLSVASDMRFNEIFTEKEVKQEKKRQRVINSRYREDYIEPALYRA -------------------------------------------------------------- >63171_63171_6_PCCB-KDM4A_PCCB_chr3_136019953_ENST00000471595_KDM4A_chr1_44128564_ENST00000372396_length(amino acids)=1243AA_BP=322 MAAALRVAAVGARLSVLASGLRAAVRSLCSQATSVNERIENKRRTALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFVESDMFVEHRC ADFGMAADKNKFPGDSVVTGRGRINGRLVYVFSQDFTVFGGSLSGAHAQKICKIMDQAITVGAPVIGLNDSGGARIQEGVESLAGYADIF LRNVTASGVIPQISLIMGPCAGGAVYSPALTDFTFMVKDTSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTMSGVAHRAFENDVDALCN LRDFFNYLPLSSQDPAPVRECHDPSDRLVPELDTIVPLESTKAYNMVDIIHSHVDEWNIGRLRTILDLVEKESGITIEGVNTPYLYFGMW KTSFAWHTEDMDLYSINYLHFGEPKSWYSVPPEHGKRLERLAKGFFPGSAQSCEAFLRHKMTLISPLMLKKYGIPFDKVTQEAGEFMITF PYGYHAGFNHGFNCAESTNFATRRWIEYGKQAVLCSCRKDMVKISMDVFVRKFQPERYKLWKAGKDNTVIDHTLPTPEAAEFLKESELPP RAGNEEECPEEDMEGVEDGEEGDLKTSLAKHRIGTKRHRVCLEIPQEVSQSELFPKEDLSSEQYEMTECPAALAPVRPTHSSVRQVEDGL TFPDYSDSTEVKFEELKNVKLEEEDEEEEQAAAALDLSVNPASVGGRLVFSGSKKKSSSSLGSGSSRDSISSDSETSEPLSCRAQGQTGV LTVHSYAKGDGRVTVGEPCTRKKGSAARSFSERELAEVADEYMFSLEENKKSKGRRQPLSKLPRHHPLVLQECVSDDETSEQLTPEEEAE ETEAWAKPLSQLWQNRPPNFEAEKEFNETMAQQAPHCAVCMIFQTYHQVEFGGFNQNCGNASDLAPQKQRTKPLIPEMCFTSTGCSTDIN LSTPYLEEDGTSILVSCKKCSVRVHASCYGVPPAKASEDWMCSRCSANALEEDCCLCSLRGGALQRANDDRWVHVSCAVAILEARFVNIA ERSPVDVSKIPLPRFKLKCIFCKKRRKRTAGCCVQCSHGRCPTAFHVSCAQAAGVMMQPDDWPFVVFITCFRHKIPNLERAKGALQSITA GQKVISKHKNGRFYQCEVVRLTTETFYEVNFDDGSFSDNLYPEDIVSQDCLQFGPPAEGEVVQVRWTDGQVYGAKFVASHPIQMYQVEFE -------------------------------------------------------------- >63171_63171_7_PCCB-KDM4A_PCCB_chr3_136019953_ENST00000482086_KDM4A_chr1_44128564_ENST00000372396_length(amino acids)=1127AA_BP=206 MAAALRVAAVGARLSVLASGLRAAVRSLCSQAITVGAPVIGLNDSGGARIQEGVESLAGYADIFLRNVTASGVIPQISLIMGPCAGGAVY SPALTDFTFMVKDTSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTMSGVAHRAFENDVDALCNLRDFFNYLPLSSQDPAPVRECHDPSD RLVPELDTIVPLESTKAYNMVDIIHSHVDEWNIGRLRTILDLVEKESGITIEGVNTPYLYFGMWKTSFAWHTEDMDLYSINYLHFGEPKS WYSVPPEHGKRLERLAKGFFPGSAQSCEAFLRHKMTLISPLMLKKYGIPFDKVTQEAGEFMITFPYGYHAGFNHGFNCAESTNFATRRWI EYGKQAVLCSCRKDMVKISMDVFVRKFQPERYKLWKAGKDNTVIDHTLPTPEAAEFLKESELPPRAGNEEECPEEDMEGVEDGEEGDLKT SLAKHRIGTKRHRVCLEIPQEVSQSELFPKEDLSSEQYEMTECPAALAPVRPTHSSVRQVEDGLTFPDYSDSTEVKFEELKNVKLEEEDE EEEQAAAALDLSVNPASVGGRLVFSGSKKKSSSSLGSGSSRDSISSDSETSEPLSCRAQGQTGVLTVHSYAKGDGRVTVGEPCTRKKGSA ARSFSERELAEVADEYMFSLEENKKSKGRRQPLSKLPRHHPLVLQECVSDDETSEQLTPEEEAEETEAWAKPLSQLWQNRPPNFEAEKEF NETMAQQAPHCAVCMIFQTYHQVEFGGFNQNCGNASDLAPQKQRTKPLIPEMCFTSTGCSTDINLSTPYLEEDGTSILVSCKKCSVRVHA SCYGVPPAKASEDWMCSRCSANALEEDCCLCSLRGGALQRANDDRWVHVSCAVAILEARFVNIAERSPVDVSKIPLPRFKLKCIFCKKRR KRTAGCCVQCSHGRCPTAFHVSCAQAAGVMMQPDDWPFVVFITCFRHKIPNLERAKGALQSITAGQKVISKHKNGRFYQCEVVRLTTETF YEVNFDDGSFSDNLYPEDIVSQDCLQFGPPAEGEVVQVRWTDGQVYGAKFVASHPIQMYQVEFEDGSQLVVKRDDVYTLDEELPKRVKSR -------------------------------------------------------------- >63171_63171_8_PCCB-KDM4A_PCCB_chr3_136019953_ENST00000483687_KDM4A_chr1_44128564_ENST00000372396_length(amino acids)=1224AA_BP=303 MAAALRVAAVGARLSVLASGLRAAVRSLCSQATSVNERIENKRRTALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFVESDMFVEHRC ADFGMAADKNKFPGDSVVTGRGRINGRLVYVFSQIMDQAITVGAPVIGLNDSGGARIQEGVESLAGYADIFLRNVTASGVIPQISLIMGP CAGGAVYSPALTDFTFMVKDTSYLFITGPDVVKSVTNEDVTQEELGGAKTHTTMSGVAHRAFENDVDALCNLRDFFNYLPLSSQDPAPVR ECHDPSDRLVPELDTIVPLESTKAYNMVDIIHSHVDEWNIGRLRTILDLVEKESGITIEGVNTPYLYFGMWKTSFAWHTEDMDLYSINYL HFGEPKSWYSVPPEHGKRLERLAKGFFPGSAQSCEAFLRHKMTLISPLMLKKYGIPFDKVTQEAGEFMITFPYGYHAGFNHGFNCAESTN FATRRWIEYGKQAVLCSCRKDMVKISMDVFVRKFQPERYKLWKAGKDNTVIDHTLPTPEAAEFLKESELPPRAGNEEECPEEDMEGVEDG EEGDLKTSLAKHRIGTKRHRVCLEIPQEVSQSELFPKEDLSSEQYEMTECPAALAPVRPTHSSVRQVEDGLTFPDYSDSTEVKFEELKNV KLEEEDEEEEQAAAALDLSVNPASVGGRLVFSGSKKKSSSSLGSGSSRDSISSDSETSEPLSCRAQGQTGVLTVHSYAKGDGRVTVGEPC TRKKGSAARSFSERELAEVADEYMFSLEENKKSKGRRQPLSKLPRHHPLVLQECVSDDETSEQLTPEEEAEETEAWAKPLSQLWQNRPPN FEAEKEFNETMAQQAPHCAVCMIFQTYHQVEFGGFNQNCGNASDLAPQKQRTKPLIPEMCFTSTGCSTDINLSTPYLEEDGTSILVSCKK CSVRVHASCYGVPPAKASEDWMCSRCSANALEEDCCLCSLRGGALQRANDDRWVHVSCAVAILEARFVNIAERSPVDVSKIPLPRFKLKC IFCKKRRKRTAGCCVQCSHGRCPTAFHVSCAQAAGVMMQPDDWPFVVFITCFRHKIPNLERAKGALQSITAGQKVISKHKNGRFYQCEVV RLTTETFYEVNFDDGSFSDNLYPEDIVSQDCLQFGPPAEGEVVQVRWTDGQVYGAKFVASHPIQMYQVEFEDGSQLVVKRDDVYTLDEEL -------------------------------------------------------------- >63171_63171_9_PCCB-KDM4A_PCCB_chr3_136019953_ENST00000490504_KDM4A_chr1_44128564_ENST00000372396_length(amino acids)=1186AA_BP=265 MAAALRVAAVGARLSVLASGLRAAVRSLCSQATSVNERIENKRRTALLGGGQRRIDAQHKRGKLTARERISLLLDPGSFVESDMFVEHRC ADFGMAADKNKFPGDSVVTGRGRINGRLVYVFSQRNVTASGVIPQISLIMGPCAGGAVYSPALTDFTFMVKDTSYLFITGPDVVKSVTNE DVTQEELGGAKTHTTMSGVAHRAFENDVDALCNLRDFFNYLPLSSQDPAPVRECHDPSDRLVPELDTIVPLESTKAYNMVDIIHSHVDEW NIGRLRTILDLVEKESGITIEGVNTPYLYFGMWKTSFAWHTEDMDLYSINYLHFGEPKSWYSVPPEHGKRLERLAKGFFPGSAQSCEAFL RHKMTLISPLMLKKYGIPFDKVTQEAGEFMITFPYGYHAGFNHGFNCAESTNFATRRWIEYGKQAVLCSCRKDMVKISMDVFVRKFQPER YKLWKAGKDNTVIDHTLPTPEAAEFLKESELPPRAGNEEECPEEDMEGVEDGEEGDLKTSLAKHRIGTKRHRVCLEIPQEVSQSELFPKE DLSSEQYEMTECPAALAPVRPTHSSVRQVEDGLTFPDYSDSTEVKFEELKNVKLEEEDEEEEQAAAALDLSVNPASVGGRLVFSGSKKKS SSSLGSGSSRDSISSDSETSEPLSCRAQGQTGVLTVHSYAKGDGRVTVGEPCTRKKGSAARSFSERELAEVADEYMFSLEENKKSKGRRQ PLSKLPRHHPLVLQECVSDDETSEQLTPEEEAEETEAWAKPLSQLWQNRPPNFEAEKEFNETMAQQAPHCAVCMIFQTYHQVEFGGFNQN CGNASDLAPQKQRTKPLIPEMCFTSTGCSTDINLSTPYLEEDGTSILVSCKKCSVRVHASCYGVPPAKASEDWMCSRCSANALEEDCCLC SLRGGALQRANDDRWVHVSCAVAILEARFVNIAERSPVDVSKIPLPRFKLKCIFCKKRRKRTAGCCVQCSHGRCPTAFHVSCAQAAGVMM QPDDWPFVVFITCFRHKIPNLERAKGALQSITAGQKVISKHKNGRFYQCEVVRLTTETFYEVNFDDGSFSDNLYPEDIVSQDCLQFGPPA EGEVVQVRWTDGQVYGAKFVASHPIQMYQVEFEDGSQLVVKRDDVYTLDEELPKRVKSRLSVASDMRFNEIFTEKEVKQEKKRQRVINSR -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:136019953/chr1:44128564) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | KDM4A |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | FUNCTION: Histone demethylase that specifically demethylates 'Lys-9' and 'Lys-36' residues of histone H3, thereby playing a central role in histone code (PubMed:26741168). Does not demethylate histone H3 'Lys-4', H3 'Lys-27' nor H4 'Lys-20'. Demethylates trimethylated H3 'Lys-9' and H3 'Lys-36' residue, while it has no activity on mono- and dimethylated residues. Demethylation of Lys residue generates formaldehyde and succinate. Participates in transcriptional repression of ASCL2 and E2F-responsive promoters via the recruitment of histone deacetylases and NCOR1, respectively. {ECO:0000269|PubMed:16024779, ECO:0000269|PubMed:16603238, ECO:0000269|PubMed:26741168}.; FUNCTION: [Isoform 2]: Crucial for muscle differentiation, promotes transcriptional activation of the Myog gene by directing the removal of repressive chromatin marks at its promoter. Lacks the N-terminal demethylase domain. {ECO:0000269|PubMed:21694756}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PCCB | chr3:136019953 | chr1:44128564 | ENST00000251654 | + | 9 | 15 | 32_290 | 322.0 | 540.0 | Domain | CoA carboxyltransferase N-terminal |
| Hgene | PCCB | chr3:136019953 | chr1:44128564 | ENST00000469217 | + | 10 | 16 | 32_290 | 342.0 | 560.0 | Domain | CoA carboxyltransferase N-terminal |
| Tgene | KDM4A | chr3:136019953 | chr1:44128564 | ENST00000372396 | 3 | 22 | 142_308 | 143.0 | 1065.0 | Domain | JmjC | |
| Tgene | KDM4A | chr3:136019953 | chr1:44128564 | ENST00000372396 | 3 | 22 | 897_954 | 143.0 | 1065.0 | Domain | Note=Tudor 1 | |
| Tgene | KDM4A | chr3:136019953 | chr1:44128564 | ENST00000372396 | 3 | 22 | 955_1011 | 143.0 | 1065.0 | Domain | Note=Tudor 2 | |
| Tgene | KDM4A | chr3:136019953 | chr1:44128564 | ENST00000372396 | 3 | 22 | 709_767 | 143.0 | 1065.0 | Zinc finger | Note=PHD-type 1 | |
| Tgene | KDM4A | chr3:136019953 | chr1:44128564 | ENST00000372396 | 3 | 22 | 772_805 | 143.0 | 1065.0 | Zinc finger | C2HC pre-PHD-type | |
| Tgene | KDM4A | chr3:136019953 | chr1:44128564 | ENST00000372396 | 3 | 22 | 828_885 | 143.0 | 1065.0 | Zinc finger | PHD-type 2 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PCCB | chr3:136019953 | chr1:44128564 | ENST00000251654 | + | 9 | 15 | 294_533 | 322.0 | 540.0 | Domain | CoA carboxyltransferase C-terminal |
| Hgene | PCCB | chr3:136019953 | chr1:44128564 | ENST00000469217 | + | 10 | 16 | 294_533 | 342.0 | 560.0 | Domain | CoA carboxyltransferase C-terminal |
| Hgene | PCCB | chr3:136019953 | chr1:44128564 | ENST00000251654 | + | 9 | 15 | 325_358 | 322.0 | 540.0 | Region | Acyl-CoA binding |
| Hgene | PCCB | chr3:136019953 | chr1:44128564 | ENST00000251654 | + | 9 | 15 | 32_533 | 322.0 | 540.0 | Region | Carboxyltransferase |
| Hgene | PCCB | chr3:136019953 | chr1:44128564 | ENST00000469217 | + | 10 | 16 | 325_358 | 342.0 | 560.0 | Region | Acyl-CoA binding |
| Hgene | PCCB | chr3:136019953 | chr1:44128564 | ENST00000469217 | + | 10 | 16 | 32_533 | 342.0 | 560.0 | Region | Carboxyltransferase |
| Tgene | KDM4A | chr3:136019953 | chr1:44128564 | ENST00000372396 | 3 | 22 | 14_56 | 143.0 | 1065.0 | Domain | JmjN |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| PCCB | |
| KDM4A |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to PCCB-KDM4A |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to PCCB-KDM4A |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

