| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:PRKAG2-KMT2C |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: PRKAG2-KMT2C | FusionPDB ID: 68637 | FusionGDB2.0 ID: 68641 | Hgene | Tgene | Gene symbol | PRKAG2 | KMT2C | Gene ID | 51422 | 58508 |
| Gene name | protein kinase AMP-activated non-catalytic subunit gamma 2 | lysine methyltransferase 2C | |
| Synonyms | AAKG|AAKG2|CMH6|H91620p|WPWS | HALR|KLEFS2|MLL3 | |
| Cytomap | 7q36.1 | 7q36.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | 5'-AMP-activated protein kinase subunit gamma-2AMPK subunit gamma-2epididymis secretory sperm binding proteinprotein kinase, AMP-activated, gamma 2 non-catalytic subunit | histone-lysine N-methyltransferase 2CALR-like proteinhistone-lysine N-methyltransferase MLL3histone-lysine N-methyltransferase, H3 lysine-4 specifichomologous to ALR proteinlysine (K)-specific methyltransferase 2Cmyeloid/lymphoid or mixed-lineage le | |
| Modification date | 20200320 | 20200320 | |
| UniProtAcc | . | Q8NEZ4 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000287878, ENST00000392801, ENST00000418337, ENST00000433631, ENST00000492843, ENST00000461529, | ENST00000485241, ENST00000485655, ENST00000262189, ENST00000355193, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 16 X 11 X 10=1760 | 12 X 17 X 8=1632 |
| # samples | 21 | 17 | |
| ** MAII score | log2(21/1760*10)=-3.06711419585854 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(17/1632*10)=-3.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: PRKAG2 [Title/Abstract] AND KMT2C [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | KMT2C(151917608)-PRKAG2(151372723), # samples:1 PRKAG2(151329155)-KMT2C(151834009), # samples:1 PRKAG2(151372506)-KMT2C(151860911), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | PRKAG2-KMT2C seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. KMT2C-PRKAG2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. KMT2C-PRKAG2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Hgene partner, which is a epigenetic factor due to the frame-shifted ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. PRKAG2-KMT2C seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PRKAG2 | GO:0006469 | negative regulation of protein kinase activity | 17255938 |
| Tgene | KMT2C | GO:0097692 | histone H3-K4 monomethylation | 26324722 |
 Fusion gene breakpoints across PRKAG2 (5'-gene) Fusion gene breakpoints across PRKAG2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
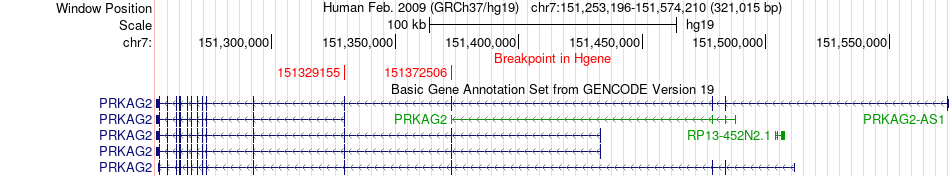 |
 Fusion gene breakpoints across KMT2C (3'-gene) Fusion gene breakpoints across KMT2C (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
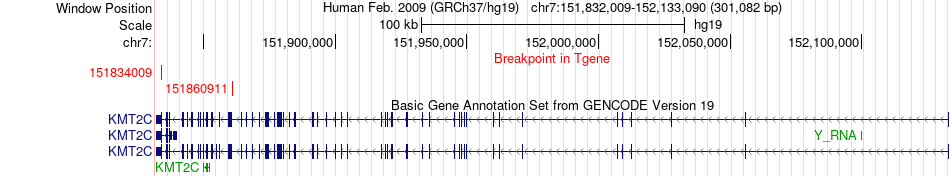 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-AO-A12E-01A | PRKAG2 | chr7 | 151329155 | - | KMT2C | chr7 | 151834009 | - |
| ChimerDB4 | PRAD | TCGA-EJ-A46G-01A | PRKAG2 | chr7 | 151372506 | - | KMT2C | chr7 | 151860911 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000287878 | PRKAG2 | chr7 | 151329155 | - | ENST00000262189 | KMT2C | chr7 | 151834009 | - | 3259 | 1259 | 493 | 1269 | 258 |
| ENST00000287878 | PRKAG2 | chr7 | 151329155 | - | ENST00000355193 | KMT2C | chr7 | 151834009 | - | 3255 | 1259 | 493 | 1269 | 258 |
| ENST00000433631 | PRKAG2 | chr7 | 151329155 | - | ENST00000262189 | KMT2C | chr7 | 151834009 | - | 2652 | 652 | 240 | 662 | 140 |
| ENST00000433631 | PRKAG2 | chr7 | 151329155 | - | ENST00000355193 | KMT2C | chr7 | 151834009 | - | 2648 | 652 | 240 | 662 | 140 |
| ENST00000492843 | PRKAG2 | chr7 | 151329155 | - | ENST00000262189 | KMT2C | chr7 | 151834009 | - | 2703 | 703 | 291 | 713 | 140 |
| ENST00000492843 | PRKAG2 | chr7 | 151329155 | - | ENST00000355193 | KMT2C | chr7 | 151834009 | - | 2699 | 703 | 291 | 713 | 140 |
| ENST00000287878 | PRKAG2 | chr7 | 151372506 | - | ENST00000262189 | KMT2C | chr7 | 151860911 | - | 8082 | 1189 | 493 | 6174 | 1893 |
| ENST00000287878 | PRKAG2 | chr7 | 151372506 | - | ENST00000355193 | KMT2C | chr7 | 151860911 | - | 8249 | 1189 | 493 | 6345 | 1950 |
| ENST00000433631 | PRKAG2 | chr7 | 151372506 | - | ENST00000262189 | KMT2C | chr7 | 151860911 | - | 7475 | 582 | 240 | 5567 | 1775 |
| ENST00000433631 | PRKAG2 | chr7 | 151372506 | - | ENST00000355193 | KMT2C | chr7 | 151860911 | - | 7642 | 582 | 240 | 5738 | 1832 |
| ENST00000492843 | PRKAG2 | chr7 | 151372506 | - | ENST00000262189 | KMT2C | chr7 | 151860911 | - | 7526 | 633 | 291 | 5618 | 1775 |
| ENST00000492843 | PRKAG2 | chr7 | 151372506 | - | ENST00000355193 | KMT2C | chr7 | 151860911 | - | 7693 | 633 | 291 | 5789 | 1832 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000287878 | ENST00000262189 | PRKAG2 | chr7 | 151329155 | - | KMT2C | chr7 | 151834009 | - | 0.042811874 | 0.9571881 |
| ENST00000287878 | ENST00000355193 | PRKAG2 | chr7 | 151329155 | - | KMT2C | chr7 | 151834009 | - | 0.038712412 | 0.9612876 |
| ENST00000433631 | ENST00000262189 | PRKAG2 | chr7 | 151329155 | - | KMT2C | chr7 | 151834009 | - | 0.4365546 | 0.5634454 |
| ENST00000433631 | ENST00000355193 | PRKAG2 | chr7 | 151329155 | - | KMT2C | chr7 | 151834009 | - | 0.43175948 | 0.5682405 |
| ENST00000492843 | ENST00000262189 | PRKAG2 | chr7 | 151329155 | - | KMT2C | chr7 | 151834009 | - | 0.43367332 | 0.5663267 |
| ENST00000492843 | ENST00000355193 | PRKAG2 | chr7 | 151329155 | - | KMT2C | chr7 | 151834009 | - | 0.4338613 | 0.5661387 |
| ENST00000287878 | ENST00000262189 | PRKAG2 | chr7 | 151372506 | - | KMT2C | chr7 | 151860911 | - | 0.000700118 | 0.9992999 |
| ENST00000287878 | ENST00000355193 | PRKAG2 | chr7 | 151372506 | - | KMT2C | chr7 | 151860911 | - | 0.000735258 | 0.9992648 |
| ENST00000433631 | ENST00000262189 | PRKAG2 | chr7 | 151372506 | - | KMT2C | chr7 | 151860911 | - | 0.000562269 | 0.99943775 |
| ENST00000433631 | ENST00000355193 | PRKAG2 | chr7 | 151372506 | - | KMT2C | chr7 | 151860911 | - | 0.000594035 | 0.999406 |
| ENST00000492843 | ENST00000262189 | PRKAG2 | chr7 | 151372506 | - | KMT2C | chr7 | 151860911 | - | 0.000610046 | 0.9993899 |
| ENST00000492843 | ENST00000355193 | PRKAG2 | chr7 | 151372506 | - | KMT2C | chr7 | 151860911 | - | 0.000642816 | 0.99935716 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >68637_68637_1_PRKAG2-KMT2C_PRKAG2_chr7_151329155_ENST00000287878_KMT2C_chr7_151834009_ENST00000262189_length(amino acids)=258AA_BP= MVRVMGSAVMDTKKKKDVSSPGGSGGKKNASQKRRSLRVHIPDLSSFAMPLLDGDLEGSGKHSSRKVDSPFGPGSPSKGFFSRGPQPRPS SPMSAPVRPKTSPGSPKTVFPFSYQESPPRSPRRMSFSGIFRSSSKESSPNSNPATSPGGIRFFSRSRKTSGLSSSPSTPTQVTKQHTFP -------------------------------------------------------------- >68637_68637_2_PRKAG2-KMT2C_PRKAG2_chr7_151329155_ENST00000287878_KMT2C_chr7_151834009_ENST00000355193_length(amino acids)=258AA_BP= MVRVMGSAVMDTKKKKDVSSPGGSGGKKNASQKRRSLRVHIPDLSSFAMPLLDGDLEGSGKHSSRKVDSPFGPGSPSKGFFSRGPQPRPS SPMSAPVRPKTSPGSPKTVFPFSYQESPPRSPRRMSFSGIFRSSSKESSPNSNPATSPGGIRFFSRSRKTSGLSSSPSTPTQVTKQHTFP -------------------------------------------------------------- >68637_68637_3_PRKAG2-KMT2C_PRKAG2_chr7_151329155_ENST00000433631_KMT2C_chr7_151834009_ENST00000262189_length(amino acids)=140AA_BP= MSPLLWPQLPMKRFGSLRSNKKHKDQNRSTERRQSEPHGLFASGLSSSPSTPTQVTKQHTFPLESYKHEPERLENRIYASSSPPDTGQRF -------------------------------------------------------------- >68637_68637_4_PRKAG2-KMT2C_PRKAG2_chr7_151329155_ENST00000433631_KMT2C_chr7_151834009_ENST00000355193_length(amino acids)=140AA_BP= MSPLLWPQLPMKRFGSLRSNKKHKDQNRSTERRQSEPHGLFASGLSSSPSTPTQVTKQHTFPLESYKHEPERLENRIYASSSPPDTGQRF -------------------------------------------------------------- >68637_68637_5_PRKAG2-KMT2C_PRKAG2_chr7_151329155_ENST00000492843_KMT2C_chr7_151834009_ENST00000262189_length(amino acids)=140AA_BP= MSPLLWPQLPMKRFGSLRSNKKHKDQNRSTERRQSEPHGLFASGLSSSPSTPTQVTKQHTFPLESYKHEPERLENRIYASSSPPDTGQRF -------------------------------------------------------------- >68637_68637_6_PRKAG2-KMT2C_PRKAG2_chr7_151329155_ENST00000492843_KMT2C_chr7_151834009_ENST00000355193_length(amino acids)=140AA_BP= MSPLLWPQLPMKRFGSLRSNKKHKDQNRSTERRQSEPHGLFASGLSSSPSTPTQVTKQHTFPLESYKHEPERLENRIYASSSPPDTGQRF -------------------------------------------------------------- >68637_68637_7_PRKAG2-KMT2C_PRKAG2_chr7_151372506_ENST00000287878_KMT2C_chr7_151860911_ENST00000262189_length(amino acids)=1893AA_BP=232 MVRVMGSAVMDTKKKKDVSSPGGSGGKKNASQKRRSLRVHIPDLSSFAMPLLDGDLEGSGKHSSRKVDSPFGPGSPSKGFFSRGPQPRPS SPMSAPVRPKTSPGSPKTVFPFSYQESPPRSPRRMSFSGIFRSSSKESSPNSNPATSPGGIRFFSRSRKTSGLSSSPSTPTQVTKQHTFP LESYKHEPERLENRIYASSSPPDTGQRFCPSSFQSPTRPPLASPTHYAPSKAIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMPSVQPQP PLIPGATPPTMSQPTFPMVPQQLQHQQHTTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSNANPQSG PPPRVEFDDNNPFSESFQERERKERLREQQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDFMQPLGP LQQSPQHQQQMGQVLQQQNIQQGSINSPSTQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPSFTPALP AAPPVANSSLPCGQDSTITHGHSYPGSTQSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTSTPAVST PSELPQQADQESVEPVGPSTPNMAAGQLCTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQEEPKLEE QNGSKVEGNAVACPVSSAQSPPHSAGAPAAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQTLGAQMQ GGFGCGNQLPKTDGGSETKKQRSKRTQRTGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQQNNLSNPPTPPASLPPTPPPMACQKMAN GFATTEELAGKAGVLVSHEVTKTLGPKPFQLPFRPQDDLLARALAQGPKTVDVPASLPTPPHNNQEELRIQDHCGDRDTPDSFVPSSSPE SVVGVEVSRYPDLSLVKEEPPEPVPSPIIPILPSTAGKSSESRRNDIKTEPGTLYFASPFGPSPNGPRSGLISVAITLHPTAAENISSVV AAFSDLLHVRIPNSYEVSSAPDVPSMGLVSSHRINPGLEYRQHLLLRGPPPGSANPPRLVSSYRLKQPNVPFPPTSNGLSGYKDSSHGIA ESAALRPQWCCHCKVVILGSGVRKSFKDLTLLNKDSRESTKRVEKDIVFCSNNCFILYSSTAQAKNSENKESIPSLPQSPMRETPSKAFH QYSNNISTLDVHCLPQLPEKASPPASPPIAFPPAFEAAQVEAKPDELKVTVKLKPRLRAVHGGFEDCRPLNKKWRGMKWKKWSIHIVIPK GTFKPPCEDEIDEFLKKLGTSLKPDPVPKDYRKCCFCHEEGDGLTDGPARLLNLDLDLWVHLNCALWSTEVYETQAGALINVELALRRGL QMKCVFCHKTGATSGCHRFRCTNIYHFTCAIKAQCMFFKDKTMLCPMHKPKGIHEQELSYFAVFRRVYVQRDEVRQIASIVQRGERDHTF RVGSLIFHTIGQLLPQQMQAFHSPKALFPVGYEASRLYWSTRYANRRCRYLCSIEEKDGRPVFVIRIVEQGHEDLVLSDISPKGVWDKIL EPVACVRKKSEMLQLFPAYLKGEDLFGLTVSAVARIAESLPGVEACENYTFRYGRNPLMELPLAVNPTGCARSEPKMSAHVKRFVLRPHT LNSTSTSKSFQSTVTGELNAPYSKQFVHSKSSQYRKMKTEWKSNVYLARSRIQGLGLYAARDIEKHTMVIEYIGTIIRNEVANRKEKLYE SQNRGVYMFRMDNDHVIDATLTGGPARYINHSCAPNCVAEVVTFERGHKIIISSSRRIQKGEELCYDYKFDFEDDQHKIPCHCGAVNCRK -------------------------------------------------------------- >68637_68637_8_PRKAG2-KMT2C_PRKAG2_chr7_151372506_ENST00000287878_KMT2C_chr7_151860911_ENST00000355193_length(amino acids)=1950AA_BP=232 MVRVMGSAVMDTKKKKDVSSPGGSGGKKNASQKRRSLRVHIPDLSSFAMPLLDGDLEGSGKHSSRKVDSPFGPGSPSKGFFSRGPQPRPS SPMSAPVRPKTSPGSPKTVFPFSYQESPPRSPRRMSFSGIFRSSSKESSPNSNPATSPGGIRFFSRSRKTSGLSSSPSTPTQVTKQHTFP LESYKHEPERLENRIYASSSPPDTGQRFCPSSFQSPTRPPLASPTHYAPSKAIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMPSVQPQP PLIPGATPPTMSQPTFPMVPQQLQHQQHTTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSNANPQSG PPPRVEFDDNNPFSESFQERERKERLREQQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDFMQPLGP LQQSPQHQQQMGQVLQQQNIQQGSINSPSTQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPSFTPALP AAPPVANSSLPCGQDSTITHGHSYPGSTQSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTSTPAVST PSELPQQADQESVEPVGPSTPNMAAGQLCTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQEEPKLEE QNGSKVEGNAVACPVSSAQSPPHSAGAPAAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQTLGAQMQ GGFGCGNQLPKTDGGSETKKQRSKRTQRTGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQVRQLSLLPLMEPIIGVNFAHFLPYGSGQ FNSGNRLLGTFGSATLEGVSDYYSQLIYKQNNLSNPPTPPASLPPTPPPMACQKMANGFATTEELAGKAGVLVSHEVTKTLGPKPFQLPF RPQDDLLARALAQGPKTVDVPASLPTPPHNNQEELRIQDHCGDRDTPDSFVPSSSPESVVGVEVSRYPDLSLVKEEPPEPVPSPIIPILP STAGKSSESRRNDIKTEPGTLYFASPFGPSPNGPRSGLISVAITLHPTAAENISSVVAAFSDLLHVRIPNSYEVSSAPDVPSMGLVSSHR INPGLEYRQHLLLRGPPPGSANPPRLVSSYRLKQPNVPFPPTSNGLSGYKDSSHGIAESAALRPQWCCHCKVVILGSGVRKSFKDLTLLN KDSRESTKRVEKDIVFCSNNCFILYSSTAQAKNSENKESIPSLPQSPMRETPSKAFHQYSNNISTLDVHCLPQLPEKASPPASPPIAFPP AFEAAQVEAKPDELKVTVKLKPRLRAVHGGFEDCRPLNKKWRGMKWKKWSIHIVIPKGTFKPPCEDEIDEFLKKLGTSLKPDPVPKDYRK CCFCHEEGDGLTDGPARLLNLDLDLWVHLNCALWSTEVYETQAGALINVELALRRGLQMKCVFCHKTGATSGCHRFRCTNIYHFTCAIKA QCMFFKDKTMLCPMHKPKGIHEQELSYFAVFRRVYVQRDEVRQIASIVQRGERDHTFRVGSLIFHTIGQLLPQQMQAFHSPKALFPVGYE ASRLYWSTRYANRRCRYLCSIEEKDGRPVFVIRIVEQGHEDLVLSDISPKGVWDKILEPVACVRKKSEMLQLFPAYLKGEDLFGLTVSAV ARIAESLPGVEACENYTFRYGRNPLMELPLAVNPTGCARSEPKMSAHVKRFVLRPHTLNSTSTSKSFQSTVTGELNAPYSKQFVHSKSSQ YRKMKTEWKSNVYLARSRIQGLGLYAARDIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDHVIDATLTGGPARYINHSC -------------------------------------------------------------- >68637_68637_9_PRKAG2-KMT2C_PRKAG2_chr7_151372506_ENST00000433631_KMT2C_chr7_151860911_ENST00000262189_length(amino acids)=1775AA_BP=114 MSPLLWPQLPMKRFGSLRSNKKHKDQNRSTERRQSEPHGLFASGLSSSPSTPTQVTKQHTFPLESYKHEPERLENRIYASSSPPDTGQRF CPSSFQSPTRPPLASPTHYAPSKAIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMPSVQPQPPLIPGATPPTMSQPTFPMVPQQLQHQQH TTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSNANPQSGPPPRVEFDDNNPFSESFQERERKERLRE QQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDFMQPLGPLQQSPQHQQQMGQVLQQQNIQQGSINSP STQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPSFTPALPAAPPVANSSLPCGQDSTITHGHSYPGST QSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTSTPAVSTPSELPQQADQESVEPVGPSTPNMAAGQL CTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQEEPKLEEQNGSKVEGNAVACPVSSAQSPPHSAGAP AAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQTLGAQMQGGFGCGNQLPKTDGGSETKKQRSKRTQR TGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQQNNLSNPPTPPASLPPTPPPMACQKMANGFATTEELAGKAGVLVSHEVTKTLGPKP FQLPFRPQDDLLARALAQGPKTVDVPASLPTPPHNNQEELRIQDHCGDRDTPDSFVPSSSPESVVGVEVSRYPDLSLVKEEPPEPVPSPI IPILPSTAGKSSESRRNDIKTEPGTLYFASPFGPSPNGPRSGLISVAITLHPTAAENISSVVAAFSDLLHVRIPNSYEVSSAPDVPSMGL VSSHRINPGLEYRQHLLLRGPPPGSANPPRLVSSYRLKQPNVPFPPTSNGLSGYKDSSHGIAESAALRPQWCCHCKVVILGSGVRKSFKD LTLLNKDSRESTKRVEKDIVFCSNNCFILYSSTAQAKNSENKESIPSLPQSPMRETPSKAFHQYSNNISTLDVHCLPQLPEKASPPASPP IAFPPAFEAAQVEAKPDELKVTVKLKPRLRAVHGGFEDCRPLNKKWRGMKWKKWSIHIVIPKGTFKPPCEDEIDEFLKKLGTSLKPDPVP KDYRKCCFCHEEGDGLTDGPARLLNLDLDLWVHLNCALWSTEVYETQAGALINVELALRRGLQMKCVFCHKTGATSGCHRFRCTNIYHFT CAIKAQCMFFKDKTMLCPMHKPKGIHEQELSYFAVFRRVYVQRDEVRQIASIVQRGERDHTFRVGSLIFHTIGQLLPQQMQAFHSPKALF PVGYEASRLYWSTRYANRRCRYLCSIEEKDGRPVFVIRIVEQGHEDLVLSDISPKGVWDKILEPVACVRKKSEMLQLFPAYLKGEDLFGL TVSAVARIAESLPGVEACENYTFRYGRNPLMELPLAVNPTGCARSEPKMSAHVKRFVLRPHTLNSTSTSKSFQSTVTGELNAPYSKQFVH SKSSQYRKMKTEWKSNVYLARSRIQGLGLYAARDIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDHVIDATLTGGPARY -------------------------------------------------------------- >68637_68637_10_PRKAG2-KMT2C_PRKAG2_chr7_151372506_ENST00000433631_KMT2C_chr7_151860911_ENST00000355193_length(amino acids)=1832AA_BP=114 MSPLLWPQLPMKRFGSLRSNKKHKDQNRSTERRQSEPHGLFASGLSSSPSTPTQVTKQHTFPLESYKHEPERLENRIYASSSPPDTGQRF CPSSFQSPTRPPLASPTHYAPSKAIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMPSVQPQPPLIPGATPPTMSQPTFPMVPQQLQHQQH TTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSNANPQSGPPPRVEFDDNNPFSESFQERERKERLRE QQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDFMQPLGPLQQSPQHQQQMGQVLQQQNIQQGSINSP STQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPSFTPALPAAPPVANSSLPCGQDSTITHGHSYPGST QSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTSTPAVSTPSELPQQADQESVEPVGPSTPNMAAGQL CTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQEEPKLEEQNGSKVEGNAVACPVSSAQSPPHSAGAP AAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQTLGAQMQGGFGCGNQLPKTDGGSETKKQRSKRTQR TGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQVRQLSLLPLMEPIIGVNFAHFLPYGSGQFNSGNRLLGTFGSATLEGVSDYYSQLIY KQNNLSNPPTPPASLPPTPPPMACQKMANGFATTEELAGKAGVLVSHEVTKTLGPKPFQLPFRPQDDLLARALAQGPKTVDVPASLPTPP HNNQEELRIQDHCGDRDTPDSFVPSSSPESVVGVEVSRYPDLSLVKEEPPEPVPSPIIPILPSTAGKSSESRRNDIKTEPGTLYFASPFG PSPNGPRSGLISVAITLHPTAAENISSVVAAFSDLLHVRIPNSYEVSSAPDVPSMGLVSSHRINPGLEYRQHLLLRGPPPGSANPPRLVS SYRLKQPNVPFPPTSNGLSGYKDSSHGIAESAALRPQWCCHCKVVILGSGVRKSFKDLTLLNKDSRESTKRVEKDIVFCSNNCFILYSST AQAKNSENKESIPSLPQSPMRETPSKAFHQYSNNISTLDVHCLPQLPEKASPPASPPIAFPPAFEAAQVEAKPDELKVTVKLKPRLRAVH GGFEDCRPLNKKWRGMKWKKWSIHIVIPKGTFKPPCEDEIDEFLKKLGTSLKPDPVPKDYRKCCFCHEEGDGLTDGPARLLNLDLDLWVH LNCALWSTEVYETQAGALINVELALRRGLQMKCVFCHKTGATSGCHRFRCTNIYHFTCAIKAQCMFFKDKTMLCPMHKPKGIHEQELSYF AVFRRVYVQRDEVRQIASIVQRGERDHTFRVGSLIFHTIGQLLPQQMQAFHSPKALFPVGYEASRLYWSTRYANRRCRYLCSIEEKDGRP VFVIRIVEQGHEDLVLSDISPKGVWDKILEPVACVRKKSEMLQLFPAYLKGEDLFGLTVSAVARIAESLPGVEACENYTFRYGRNPLMEL PLAVNPTGCARSEPKMSAHVKRFVLRPHTLNSTSTSKSFQSTVTGELNAPYSKQFVHSKSSQYRKMKTEWKSNVYLARSRIQGLGLYAAR DIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDHVIDATLTGGPARYINHSCAPNCVAEVVTFERGHKIIISSSRRIQKG -------------------------------------------------------------- >68637_68637_11_PRKAG2-KMT2C_PRKAG2_chr7_151372506_ENST00000492843_KMT2C_chr7_151860911_ENST00000262189_length(amino acids)=1775AA_BP=114 MSPLLWPQLPMKRFGSLRSNKKHKDQNRSTERRQSEPHGLFASGLSSSPSTPTQVTKQHTFPLESYKHEPERLENRIYASSSPPDTGQRF CPSSFQSPTRPPLASPTHYAPSKAIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMPSVQPQPPLIPGATPPTMSQPTFPMVPQQLQHQQH TTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSNANPQSGPPPRVEFDDNNPFSESFQERERKERLRE QQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDFMQPLGPLQQSPQHQQQMGQVLQQQNIQQGSINSP STQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPSFTPALPAAPPVANSSLPCGQDSTITHGHSYPGST QSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTSTPAVSTPSELPQQADQESVEPVGPSTPNMAAGQL CTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQEEPKLEEQNGSKVEGNAVACPVSSAQSPPHSAGAP AAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQTLGAQMQGGFGCGNQLPKTDGGSETKKQRSKRTQR TGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQQNNLSNPPTPPASLPPTPPPMACQKMANGFATTEELAGKAGVLVSHEVTKTLGPKP FQLPFRPQDDLLARALAQGPKTVDVPASLPTPPHNNQEELRIQDHCGDRDTPDSFVPSSSPESVVGVEVSRYPDLSLVKEEPPEPVPSPI IPILPSTAGKSSESRRNDIKTEPGTLYFASPFGPSPNGPRSGLISVAITLHPTAAENISSVVAAFSDLLHVRIPNSYEVSSAPDVPSMGL VSSHRINPGLEYRQHLLLRGPPPGSANPPRLVSSYRLKQPNVPFPPTSNGLSGYKDSSHGIAESAALRPQWCCHCKVVILGSGVRKSFKD LTLLNKDSRESTKRVEKDIVFCSNNCFILYSSTAQAKNSENKESIPSLPQSPMRETPSKAFHQYSNNISTLDVHCLPQLPEKASPPASPP IAFPPAFEAAQVEAKPDELKVTVKLKPRLRAVHGGFEDCRPLNKKWRGMKWKKWSIHIVIPKGTFKPPCEDEIDEFLKKLGTSLKPDPVP KDYRKCCFCHEEGDGLTDGPARLLNLDLDLWVHLNCALWSTEVYETQAGALINVELALRRGLQMKCVFCHKTGATSGCHRFRCTNIYHFT CAIKAQCMFFKDKTMLCPMHKPKGIHEQELSYFAVFRRVYVQRDEVRQIASIVQRGERDHTFRVGSLIFHTIGQLLPQQMQAFHSPKALF PVGYEASRLYWSTRYANRRCRYLCSIEEKDGRPVFVIRIVEQGHEDLVLSDISPKGVWDKILEPVACVRKKSEMLQLFPAYLKGEDLFGL TVSAVARIAESLPGVEACENYTFRYGRNPLMELPLAVNPTGCARSEPKMSAHVKRFVLRPHTLNSTSTSKSFQSTVTGELNAPYSKQFVH SKSSQYRKMKTEWKSNVYLARSRIQGLGLYAARDIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDHVIDATLTGGPARY -------------------------------------------------------------- >68637_68637_12_PRKAG2-KMT2C_PRKAG2_chr7_151372506_ENST00000492843_KMT2C_chr7_151860911_ENST00000355193_length(amino acids)=1832AA_BP=114 MSPLLWPQLPMKRFGSLRSNKKHKDQNRSTERRQSEPHGLFASGLSSSPSTPTQVTKQHTFPLESYKHEPERLENRIYASSSPPDTGQRF CPSSFQSPTRPPLASPTHYAPSKAIRKQQKEHAELIEDYRIKQQQQCAMAPPTMMPSVQPQPPLIPGATPPTMSQPTFPMVPQQLQHQQH TTVISGHTSPVRMPSLPGWQPNSAPAHLPLNPPRIQPPIAQLPIKTCTPAPGTVSNANPQSGPPPRVEFDDNNPFSESFQERERKERLRE QQERQRIQLMQEVDRQRALQQRMEMEQHGMVGSEISSSRTSVSQIPFYSSDLPCDFMQPLGPLQQSPQHQQQMGQVLQQQNIQQGSINSP STQTFMQTNERRQVGPPSFVPDSPSIPVGSPNFSSVKQGHGNLSGTSFQQSPVRPSFTPALPAAPPVANSSLPCGQDSTITHGHSYPGST QSLIQLYSDIIPEEKGKKKRTRKKKRDDDAESTKAPSTPHSDITAPPTPGISETTSTPAVSTPSELPQQADQESVEPVGPSTPNMAAGQL CTELENKLPNSDFSQATPNQQTYANSEVDKLSMETPAKTEEIKLEKAETESCPGQEEPKLEEQNGSKVEGNAVACPVSSAQSPPHSAGAP AAKGDSGNELLKHLLKNKKSSSLLNQKPEGSICSEDDCTKDNKLVEKQNPAEGLQTLGAQMQGGFGCGNQLPKTDGGSETKKQRSKRTQR TGEKAAPRSKKRKKDEEEKQAMYSSTDTFTHLKQVRQLSLLPLMEPIIGVNFAHFLPYGSGQFNSGNRLLGTFGSATLEGVSDYYSQLIY KQNNLSNPPTPPASLPPTPPPMACQKMANGFATTEELAGKAGVLVSHEVTKTLGPKPFQLPFRPQDDLLARALAQGPKTVDVPASLPTPP HNNQEELRIQDHCGDRDTPDSFVPSSSPESVVGVEVSRYPDLSLVKEEPPEPVPSPIIPILPSTAGKSSESRRNDIKTEPGTLYFASPFG PSPNGPRSGLISVAITLHPTAAENISSVVAAFSDLLHVRIPNSYEVSSAPDVPSMGLVSSHRINPGLEYRQHLLLRGPPPGSANPPRLVS SYRLKQPNVPFPPTSNGLSGYKDSSHGIAESAALRPQWCCHCKVVILGSGVRKSFKDLTLLNKDSRESTKRVEKDIVFCSNNCFILYSST AQAKNSENKESIPSLPQSPMRETPSKAFHQYSNNISTLDVHCLPQLPEKASPPASPPIAFPPAFEAAQVEAKPDELKVTVKLKPRLRAVH GGFEDCRPLNKKWRGMKWKKWSIHIVIPKGTFKPPCEDEIDEFLKKLGTSLKPDPVPKDYRKCCFCHEEGDGLTDGPARLLNLDLDLWVH LNCALWSTEVYETQAGALINVELALRRGLQMKCVFCHKTGATSGCHRFRCTNIYHFTCAIKAQCMFFKDKTMLCPMHKPKGIHEQELSYF AVFRRVYVQRDEVRQIASIVQRGERDHTFRVGSLIFHTIGQLLPQQMQAFHSPKALFPVGYEASRLYWSTRYANRRCRYLCSIEEKDGRP VFVIRIVEQGHEDLVLSDISPKGVWDKILEPVACVRKKSEMLQLFPAYLKGEDLFGLTVSAVARIAESLPGVEACENYTFRYGRNPLMEL PLAVNPTGCARSEPKMSAHVKRFVLRPHTLNSTSTSKSFQSTVTGELNAPYSKQFVHSKSSQYRKMKTEWKSNVYLARSRIQGLGLYAAR DIEKHTMVIEYIGTIIRNEVANRKEKLYESQNRGVYMFRMDNDHVIDATLTGGPARYINHSCAPNCVAEVVTFERGHKIIISSSRRIQKG -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr7:151917608/chr7:151372723) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | KMT2C |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | FUNCTION: Histone methyltransferase that methylates 'Lys-4' of histone H3 (PubMed:22266653). H3 'Lys-4' methylation represents a specific tag for epigenetic transcriptional activation. Central component of the MLL2/3 complex, a coactivator complex of nuclear receptors, involved in transcriptional coactivation. KMT2C/MLL3 may be a catalytic subunit of this complex. May be involved in leukemogenesis and developmental disorder. {ECO:0000269|PubMed:17500065, ECO:0000269|PubMed:22266653}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 3391_3433 | 3250.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 3391_3433 | 3250.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 3277_3381 | 3250.0 | 4912.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 3277_3381 | 3250.0 | 4969.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 4895_4911 | 4881.0 | 4912.0 | Domain | Post-SET | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 4545_4605 | 3250.0 | 4912.0 | Domain | FYR N-terminal | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 4606_4691 | 3250.0 | 4912.0 | Domain | FYR C-terminal | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 4771_4887 | 3250.0 | 4912.0 | Domain | SET | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 4895_4911 | 3250.0 | 4912.0 | Domain | Post-SET | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 4545_4605 | 3250.0 | 4969.0 | Domain | FYR N-terminal | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 4606_4691 | 3250.0 | 4969.0 | Domain | FYR C-terminal | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 4771_4887 | 3250.0 | 4969.0 | Domain | SET | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 4895_4911 | 3250.0 | 4969.0 | Domain | Post-SET | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 4848_4849 | 3250.0 | 4912.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 4848_4849 | 3250.0 | 4969.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 4399_4439 | 3250.0 | 4912.0 | Zinc finger | C2HC pre-PHD-type 2 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 4460_4507 | 3250.0 | 4912.0 | Zinc finger | PHD-type 8 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 4399_4439 | 3250.0 | 4969.0 | Zinc finger | C2HC pre-PHD-type 2 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 4460_4507 | 3250.0 | 4969.0 | Zinc finger | PHD-type 8 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 275_335 | 251.33333333333334 | 570.0 | Domain | CBS 1 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 357_415 | 251.33333333333334 | 570.0 | Domain | CBS 2 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 430_492 | 251.33333333333334 | 570.0 | Domain | CBS 3 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 504_562 | 251.33333333333334 | 570.0 | Domain | CBS 4 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 275_335 | 207.33333333333334 | 526.0 | Domain | CBS 1 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 357_415 | 207.33333333333334 | 526.0 | Domain | CBS 2 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 430_492 | 207.33333333333334 | 526.0 | Domain | CBS 3 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 504_562 | 207.33333333333334 | 526.0 | Domain | CBS 4 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 275_335 | 10.333333333333334 | 329.0 | Domain | CBS 1 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 357_415 | 10.333333333333334 | 329.0 | Domain | CBS 2 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 430_492 | 10.333333333333334 | 329.0 | Domain | CBS 3 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 504_562 | 10.333333333333334 | 329.0 | Domain | CBS 4 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 275_335 | 228.0 | 570.0 | Domain | CBS 1 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 357_415 | 228.0 | 570.0 | Domain | CBS 2 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 430_492 | 228.0 | 570.0 | Domain | CBS 3 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 504_562 | 228.0 | 570.0 | Domain | CBS 4 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 275_335 | 184.0 | 526.0 | Domain | CBS 1 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 357_415 | 184.0 | 526.0 | Domain | CBS 2 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 430_492 | 184.0 | 526.0 | Domain | CBS 3 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 504_562 | 184.0 | 526.0 | Domain | CBS 4 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 275_335 | 0 | 329.0 | Domain | CBS 1 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 357_415 | 0 | 329.0 | Domain | CBS 2 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 430_492 | 0 | 329.0 | Domain | CBS 3 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 504_562 | 0 | 329.0 | Domain | CBS 4 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 370_391 | 251.33333333333334 | 570.0 | Motif | Note=AMPK pseudosubstrate |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 370_391 | 207.33333333333334 | 526.0 | Motif | Note=AMPK pseudosubstrate |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 370_391 | 10.333333333333334 | 329.0 | Motif | Note=AMPK pseudosubstrate |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 370_391 | 228.0 | 570.0 | Motif | Note=AMPK pseudosubstrate |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 370_391 | 184.0 | 526.0 | Motif | Note=AMPK pseudosubstrate |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 370_391 | 0 | 329.0 | Motif | Note=AMPK pseudosubstrate |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 319_322 | 251.33333333333334 | 570.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 383_384 | 251.33333333333334 | 570.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 458_459 | 251.33333333333334 | 570.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 474_477 | 251.33333333333334 | 570.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 530_531 | 251.33333333333334 | 570.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000287878 | - | 5 | 16 | 546_549 | 251.33333333333334 | 570.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 319_322 | 207.33333333333334 | 526.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 383_384 | 207.33333333333334 | 526.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 458_459 | 207.33333333333334 | 526.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 474_477 | 207.33333333333334 | 526.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 530_531 | 207.33333333333334 | 526.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000392801 | - | 5 | 16 | 546_549 | 207.33333333333334 | 526.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 319_322 | 10.333333333333334 | 329.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 383_384 | 10.333333333333334 | 329.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 458_459 | 10.333333333333334 | 329.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 474_477 | 10.333333333333334 | 329.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 530_531 | 10.333333333333334 | 329.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151329155 | chr7:151834009 | ENST00000418337 | - | 1 | 12 | 546_549 | 10.333333333333334 | 329.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 319_322 | 228.0 | 570.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 383_384 | 228.0 | 570.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 458_459 | 228.0 | 570.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 474_477 | 228.0 | 570.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 530_531 | 228.0 | 570.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000287878 | - | 4 | 16 | 546_549 | 228.0 | 570.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 319_322 | 184.0 | 526.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 383_384 | 184.0 | 526.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 458_459 | 184.0 | 526.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 474_477 | 184.0 | 526.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 530_531 | 184.0 | 526.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000392801 | - | 4 | 16 | 546_549 | 184.0 | 526.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 319_322 | 0 | 329.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 383_384 | 0 | 329.0 | Nucleotide binding | AMP%2C ADP or ATP 1 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 458_459 | 0 | 329.0 | Nucleotide binding | AMP 3 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 474_477 | 0 | 329.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 530_531 | 0 | 329.0 | Nucleotide binding | AMP%2C ADP or ATP 2 |
| Hgene | PRKAG2 | chr7:151372506 | chr7:151860911 | ENST00000418337 | - | 1 | 12 | 546_549 | 0 | 329.0 | Nucleotide binding | AMP 3 |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 1338_1366 | 4881.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 1754_1787 | 4881.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 3054_3081 | 4881.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 3173_3272 | 4881.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 3391_3433 | 4881.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 644_672 | 4881.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 92_112 | 4881.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 1338_1366 | 4938.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 1754_1787 | 4938.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 3054_3081 | 4938.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 3173_3272 | 4938.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 3391_3433 | 4938.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 644_672 | 4938.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 92_112 | 4938.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 1338_1366 | 3250.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 1754_1787 | 3250.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 3054_3081 | 3250.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 3173_3272 | 3250.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 644_672 | 3250.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 92_112 | 3250.0 | 4912.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 1338_1366 | 3250.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 1754_1787 | 3250.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 3054_3081 | 3250.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 3173_3272 | 3250.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 644_672 | 3250.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 92_112 | 3250.0 | 4969.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 1719_1796 | 4881.0 | 4912.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 1834_2281 | 4881.0 | 4912.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 2412_2630 | 4881.0 | 4912.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 2690_2786 | 4881.0 | 4912.0 | Compositional bias | Note=Asp-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 3012_3509 | 4881.0 | 4912.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 3277_3381 | 4881.0 | 4912.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 1719_1796 | 4938.0 | 4969.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 1834_2281 | 4938.0 | 4969.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 2412_2630 | 4938.0 | 4969.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 2690_2786 | 4938.0 | 4969.0 | Compositional bias | Note=Asp-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 3012_3509 | 4938.0 | 4969.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 3277_3381 | 4938.0 | 4969.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 1719_1796 | 3250.0 | 4912.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 1834_2281 | 3250.0 | 4912.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 2412_2630 | 3250.0 | 4912.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 2690_2786 | 3250.0 | 4912.0 | Compositional bias | Note=Asp-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 3012_3509 | 3250.0 | 4912.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 1719_1796 | 3250.0 | 4969.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 1834_2281 | 3250.0 | 4969.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 2412_2630 | 3250.0 | 4969.0 | Compositional bias | Note=Pro-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 2690_2786 | 3250.0 | 4969.0 | Compositional bias | Note=Asp-rich | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 3012_3509 | 3250.0 | 4969.0 | Compositional bias | Note=Gln-rich | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 34_46 | 4881.0 | 4912.0 | DNA binding | Note=A.T hook | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 34_46 | 4938.0 | 4969.0 | DNA binding | Note=A.T hook | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 34_46 | 3250.0 | 4912.0 | DNA binding | Note=A.T hook | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 34_46 | 3250.0 | 4969.0 | DNA binding | Note=A.T hook | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 436_489 | 4881.0 | 4912.0 | Domain | DHHC | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 4545_4605 | 4881.0 | 4912.0 | Domain | FYR N-terminal | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 4606_4691 | 4881.0 | 4912.0 | Domain | FYR C-terminal | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 4771_4887 | 4881.0 | 4912.0 | Domain | SET | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 436_489 | 4938.0 | 4969.0 | Domain | DHHC | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 4545_4605 | 4938.0 | 4969.0 | Domain | FYR N-terminal | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 4606_4691 | 4938.0 | 4969.0 | Domain | FYR C-terminal | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 4771_4887 | 4938.0 | 4969.0 | Domain | SET | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 4895_4911 | 4938.0 | 4969.0 | Domain | Post-SET | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 436_489 | 3250.0 | 4912.0 | Domain | DHHC | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 436_489 | 3250.0 | 4969.0 | Domain | DHHC | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 4848_4849 | 4881.0 | 4912.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 4848_4849 | 4938.0 | 4969.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 1007_1057 | 4881.0 | 4912.0 | Zinc finger | PHD-type 6 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 1084_1139 | 4881.0 | 4912.0 | Zinc finger | PHD-type 7 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 227_262 | 4881.0 | 4912.0 | Zinc finger | C2HC pre-PHD-type 1%3B degenerate | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 283_331 | 4881.0 | 4912.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 341_391 | 4881.0 | 4912.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 344_389 | 4881.0 | 4912.0 | Zinc finger | RING-type | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 388_438 | 4881.0 | 4912.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 4399_4439 | 4881.0 | 4912.0 | Zinc finger | C2HC pre-PHD-type 2 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 4460_4507 | 4881.0 | 4912.0 | Zinc finger | PHD-type 8 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 464_520 | 4881.0 | 4912.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000262189 | 57 | 59 | 957_1010 | 4881.0 | 4912.0 | Zinc finger | PHD-type 5 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 1007_1057 | 4938.0 | 4969.0 | Zinc finger | PHD-type 6 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 1084_1139 | 4938.0 | 4969.0 | Zinc finger | PHD-type 7 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 227_262 | 4938.0 | 4969.0 | Zinc finger | C2HC pre-PHD-type 1%3B degenerate | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 283_331 | 4938.0 | 4969.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 341_391 | 4938.0 | 4969.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 344_389 | 4938.0 | 4969.0 | Zinc finger | RING-type | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 388_438 | 4938.0 | 4969.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 4399_4439 | 4938.0 | 4969.0 | Zinc finger | C2HC pre-PHD-type 2 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 4460_4507 | 4938.0 | 4969.0 | Zinc finger | PHD-type 8 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 464_520 | 4938.0 | 4969.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2C | chr7:151329155 | chr7:151834009 | ENST00000355193 | 58 | 60 | 957_1010 | 4938.0 | 4969.0 | Zinc finger | PHD-type 5 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 1007_1057 | 3250.0 | 4912.0 | Zinc finger | PHD-type 6 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 1084_1139 | 3250.0 | 4912.0 | Zinc finger | PHD-type 7 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 227_262 | 3250.0 | 4912.0 | Zinc finger | C2HC pre-PHD-type 1%3B degenerate | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 283_331 | 3250.0 | 4912.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 341_391 | 3250.0 | 4912.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 344_389 | 3250.0 | 4912.0 | Zinc finger | RING-type | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 388_438 | 3250.0 | 4912.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 464_520 | 3250.0 | 4912.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000262189 | 41 | 59 | 957_1010 | 3250.0 | 4912.0 | Zinc finger | PHD-type 5 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 1007_1057 | 3250.0 | 4969.0 | Zinc finger | PHD-type 6 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 1084_1139 | 3250.0 | 4969.0 | Zinc finger | PHD-type 7 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 227_262 | 3250.0 | 4969.0 | Zinc finger | C2HC pre-PHD-type 1%3B degenerate | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 283_331 | 3250.0 | 4969.0 | Zinc finger | PHD-type 1 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 341_391 | 3250.0 | 4969.0 | Zinc finger | PHD-type 2 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 344_389 | 3250.0 | 4969.0 | Zinc finger | RING-type | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 388_438 | 3250.0 | 4969.0 | Zinc finger | PHD-type 3 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 464_520 | 3250.0 | 4969.0 | Zinc finger | PHD-type 4 | |
| Tgene | KMT2C | chr7:151372506 | chr7:151860911 | ENST00000355193 | 41 | 60 | 957_1010 | 3250.0 | 4969.0 | Zinc finger | PHD-type 5 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| PRKAG2 | |
| KMT2C |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to PRKAG2-KMT2C |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to PRKAG2-KMT2C |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

