| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:QKI-NTRK2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: QKI-NTRK2 | FusionPDB ID: 70986 | FusionGDB2.0 ID: 70986 | Hgene | Tgene | Gene symbol | QKI | NTRK2 | Gene ID | 9444 | 4915 |
| Gene name | QKI, KH domain containing RNA binding | neurotrophic receptor tyrosine kinase 2 | |
| Synonyms | Hqk|QK|QK1|QK3|hqkI | EIEE58|GP145-TrkB|OBHD|TRKB|trk-B | |
| Cytomap | 6q26 | 9q21.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein quakingQKI/LOC100132735 fusionRNA binding protein HQKhomolog of mouse quaking QKI (KH domain RNA binding protein)quaking homolog, KH domain RNA binding | BDNF/NT-3 growth factors receptorBDNF-tropomyosine receptor kinase Bneurotrophic tyrosine kinase receptor type 2tropomyosin-related kinase Btyrosine kinase receptor B | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | Q96PU8 | Q16620 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000275262, ENST00000361195, ENST00000361752, ENST00000392127, ENST00000424802, ENST00000453779, | ENST00000277120, ENST00000304053, ENST00000323115, ENST00000359847, ENST00000376208, ENST00000395866, ENST00000395882, ENST00000376213, ENST00000376214, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 16 X 15 X 11=2640 | 10 X 9 X 7=630 |
| # samples | 23 | 10 | |
| ** MAII score | log2(23/2640*10)=-3.52083216330144 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/630*10)=-2.65535182861255 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: QKI [Title/Abstract] AND NTRK2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | |||
| Anticipated loss of major functional domain due to fusion event. | QKI-NTRK2 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. QKI-NTRK2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. QKI-NTRK2 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. QKI-NTRK2 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across QKI (5'-gene) Fusion gene breakpoints across QKI (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across NTRK2 (3'-gene) Fusion gene breakpoints across NTRK2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | QKI | chr6 | 163984751 | + | NTRK2 | chr9 | 87482157 | + |
| ChimerKB3 | . | . | QKI | chr6 | 163999628 | + | NTRK2 | chr9 | 87482157 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000275262 | QKI | chr6 | 163984751 | + | ENST00000376214 | NTRK2 | chr9 | 87482157 | + | 4711 | 1485 | 551 | 2557 | 668 |
| ENST00000275262 | QKI | chr6 | 163984751 | + | ENST00000376213 | NTRK2 | chr9 | 87482157 | + | 4711 | 1485 | 551 | 2557 | 668 |
| ENST00000392127 | QKI | chr6 | 163984751 | + | ENST00000376214 | NTRK2 | chr9 | 87482157 | + | 10837 | 7611 | 7727 | 8683 | 318 |
| ENST00000392127 | QKI | chr6 | 163984751 | + | ENST00000376213 | NTRK2 | chr9 | 87482157 | + | 10837 | 7611 | 7727 | 8683 | 318 |
| ENST00000453779 | QKI | chr6 | 163984751 | + | ENST00000376214 | NTRK2 | chr9 | 87482157 | + | 4711 | 1485 | 551 | 2557 | 668 |
| ENST00000453779 | QKI | chr6 | 163984751 | + | ENST00000376213 | NTRK2 | chr9 | 87482157 | + | 4711 | 1485 | 551 | 2557 | 668 |
| ENST00000361752 | QKI | chr6 | 163984751 | + | ENST00000376214 | NTRK2 | chr9 | 87482157 | + | 4711 | 1485 | 551 | 2557 | 668 |
| ENST00000361752 | QKI | chr6 | 163984751 | + | ENST00000376213 | NTRK2 | chr9 | 87482157 | + | 4711 | 1485 | 551 | 2557 | 668 |
| ENST00000361195 | QKI | chr6 | 163984751 | + | ENST00000376214 | NTRK2 | chr9 | 87482157 | + | 4160 | 934 | 24 | 2006 | 660 |
| ENST00000361195 | QKI | chr6 | 163984751 | + | ENST00000376213 | NTRK2 | chr9 | 87482157 | + | 4160 | 934 | 24 | 2006 | 660 |
| ENST00000424802 | QKI | chr6 | 163984751 | + | ENST00000376214 | NTRK2 | chr9 | 87482157 | + | 4136 | 910 | 0 | 1982 | 660 |
| ENST00000424802 | QKI | chr6 | 163984751 | + | ENST00000376213 | NTRK2 | chr9 | 87482157 | + | 4136 | 910 | 0 | 1982 | 660 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >70986_70986_1_QKI-NTRK2_QKI_chr6_163984751_ENST00000275262_NTRK2_chr9_87482157_ENST00000376213_length(amino acids)=668AA_BP=311 MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEISRVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPV KEYPDFNFVGRILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNEDLHVLITVEDAQNRAEIKLKRAVEEVKKLLV PAAEGEDSLKKMQLMELAILNGTYRDANIKSPALAFSLAATAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQIQTAVMPNGTPHP TAAIVPPGPEAGLIYTPYEYPYTLAPATSILEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIENP QYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFY GVCVEGDPLIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGD FGMSRDVYSTDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQRPRTCPQEVYE -------------------------------------------------------------- >70986_70986_2_QKI-NTRK2_QKI_chr6_163984751_ENST00000275262_NTRK2_chr9_87482157_ENST00000376214_length(amino acids)=668AA_BP=311 MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEISRVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPV KEYPDFNFVGRILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNEDLHVLITVEDAQNRAEIKLKRAVEEVKKLLV PAAEGEDSLKKMQLMELAILNGTYRDANIKSPALAFSLAATAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQIQTAVMPNGTPHP TAAIVPPGPEAGLIYTPYEYPYTLAPATSILEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIENP QYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFY GVCVEGDPLIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGD FGMSRDVYSTDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQRPRTCPQEVYE -------------------------------------------------------------- >70986_70986_3_QKI-NTRK2_QKI_chr6_163984751_ENST00000361195_NTRK2_chr9_87482157_ENST00000376213_length(amino acids)=660AA_BP=303 MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEISRVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPV KEYPDFNFVGRILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNEDLHVLITVEDAQNRAEIKLKRAVEEVKKLLV PAAEGEDSLKKMQLMELAILNGTYRDANIKSPTAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQIQTAVMPNGTPHPTAAIVPPG PEAGLIYTPYEYPYTLAPATSILEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIENPQYFGITNS QLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFYGVCVEGDP LIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGDFGMSRDVY STDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQRPRTCPQEVYELMLGCWQR -------------------------------------------------------------- >70986_70986_4_QKI-NTRK2_QKI_chr6_163984751_ENST00000361195_NTRK2_chr9_87482157_ENST00000376214_length(amino acids)=660AA_BP=303 MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEISRVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPV KEYPDFNFVGRILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNEDLHVLITVEDAQNRAEIKLKRAVEEVKKLLV PAAEGEDSLKKMQLMELAILNGTYRDANIKSPTAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQIQTAVMPNGTPHPTAAIVPPG PEAGLIYTPYEYPYTLAPATSILEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIENPQYFGITNS QLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFYGVCVEGDP LIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGDFGMSRDVY STDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQRPRTCPQEVYELMLGCWQR -------------------------------------------------------------- >70986_70986_5_QKI-NTRK2_QKI_chr6_163984751_ENST00000361752_NTRK2_chr9_87482157_ENST00000376213_length(amino acids)=668AA_BP=311 MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEISRVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPV KEYPDFNFVGRILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNEDLHVLITVEDAQNRAEIKLKRAVEEVKKLLV PAAEGEDSLKKMQLMELAILNGTYRDANIKSPALAFSLAATAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQIQTAVMPNGTPHP TAAIVPPGPEAGLIYTPYEYPYTLAPATSILEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIENP QYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFY GVCVEGDPLIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGD FGMSRDVYSTDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQRPRTCPQEVYE -------------------------------------------------------------- >70986_70986_6_QKI-NTRK2_QKI_chr6_163984751_ENST00000361752_NTRK2_chr9_87482157_ENST00000376214_length(amino acids)=668AA_BP=311 MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEISRVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPV KEYPDFNFVGRILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNEDLHVLITVEDAQNRAEIKLKRAVEEVKKLLV PAAEGEDSLKKMQLMELAILNGTYRDANIKSPALAFSLAATAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQIQTAVMPNGTPHP TAAIVPPGPEAGLIYTPYEYPYTLAPATSILEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIENP QYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFY GVCVEGDPLIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGD FGMSRDVYSTDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQRPRTCPQEVYE -------------------------------------------------------------- >70986_70986_7_QKI-NTRK2_QKI_chr6_163984751_ENST00000392127_NTRK2_chr9_87482157_ENST00000376213_length(amino acids)=318AA_BP= MTKIPVIENPQYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTN LQHEHIVKFYGVCVEGDPLIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLV GENLLVKIGDFGMSRDVYSTDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQR -------------------------------------------------------------- >70986_70986_8_QKI-NTRK2_QKI_chr6_163984751_ENST00000392127_NTRK2_chr9_87482157_ENST00000376214_length(amino acids)=318AA_BP= MTKIPVIENPQYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTN LQHEHIVKFYGVCVEGDPLIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLV GENLLVKIGDFGMSRDVYSTDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQR -------------------------------------------------------------- >70986_70986_9_QKI-NTRK2_QKI_chr6_163984751_ENST00000424802_NTRK2_chr9_87482157_ENST00000376213_length(amino acids)=660AA_BP=303 MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEISRVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPV KEYPDFNFVGRILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNEDLHVLITVEDAQNRAEIKLKRAVEEVKKLLV PAAEGEDSLKKMQLMELAILNGTYRDANIKSPTAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQIQTAVMPNGTPHPTAAIVPPG PEAGLIYTPYEYPYTLAPATSILEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIENPQYFGITNS QLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFYGVCVEGDP LIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGDFGMSRDVY STDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQRPRTCPQEVYELMLGCWQR -------------------------------------------------------------- >70986_70986_10_QKI-NTRK2_QKI_chr6_163984751_ENST00000424802_NTRK2_chr9_87482157_ENST00000376214_length(amino acids)=660AA_BP=303 MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEISRVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPV KEYPDFNFVGRILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNEDLHVLITVEDAQNRAEIKLKRAVEEVKKLLV PAAEGEDSLKKMQLMELAILNGTYRDANIKSPTAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQIQTAVMPNGTPHPTAAIVPPG PEAGLIYTPYEYPYTLAPATSILEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIENPQYFGITNS QLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFYGVCVEGDP LIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGDFGMSRDVY STDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQRPRTCPQEVYELMLGCWQR -------------------------------------------------------------- >70986_70986_11_QKI-NTRK2_QKI_chr6_163984751_ENST00000453779_NTRK2_chr9_87482157_ENST00000376213_length(amino acids)=668AA_BP=311 MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEISRVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPV KEYPDFNFVGRILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNEDLHVLITVEDAQNRAEIKLKRAVEEVKKLLV PAAEGEDSLKKMQLMELAILNGTYRDANIKSPALAFSLAATAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQIQTAVMPNGTPHP TAAIVPPGPEAGLIYTPYEYPYTLAPATSILEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIENP QYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFY GVCVEGDPLIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGD FGMSRDVYSTDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQRPRTCPQEVYE -------------------------------------------------------------- >70986_70986_12_QKI-NTRK2_QKI_chr6_163984751_ENST00000453779_NTRK2_chr9_87482157_ENST00000376214_length(amino acids)=668AA_BP=311 MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEISRVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPV KEYPDFNFVGRILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNEDLHVLITVEDAQNRAEIKLKRAVEEVKKLLV PAAEGEDSLKKMQLMELAILNGTYRDANIKSPALAFSLAATAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQIQTAVMPNGTPHP TAAIVPPGPEAGLIYTPYEYPYTLAPATSILEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIENP QYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFY GVCVEGDPLIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGD FGMSRDVYSTDYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEVIECITQGRVLQRPRTCPQEVYE -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr6:/chr9:) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| QKI | NTRK2 |
| FUNCTION: RNA-binding protein that plays a central role in myelinization (PubMed:16641098). Binds to the 5'-NACUAAY-N(1,20)-UAAY-3' RNA core sequence. Regulates target mRNA stability (PubMed:23630077). In addition, acts by regulating pre-mRNA splicing, mRNA export and protein translation. Required to protect and promote stability of mRNAs such as MBP and CDKN1B. Regulator of oligodendrocyte differentiation and maturation in the brain that may play a role in myelin and oligodendrocyte dysfunction in schizophrenia (PubMed:16641098). Participates in mRNA transport by regulating the nuclear export of MBP mRNA. Also involved in regulation of mRNA splicing of MAG pre-mRNA. Acts as a translational repressor (By similarity). {ECO:0000250|UniProtKB:Q9QYS9, ECO:0000269|PubMed:16641098, ECO:0000269|PubMed:23630077}. | FUNCTION: Receptor tyrosine kinase involved in the development and the maturation of the central and the peripheral nervous systems through regulation of neuron survival, proliferation, migration, differentiation, and synapse formation and plasticity (By similarity). Receptor for BDNF/brain-derived neurotrophic factor and NTF4/neurotrophin-4. Alternatively can also bind NTF3/neurotrophin-3 which is less efficient in activating the receptor but regulates neuron survival through NTRK2 (PubMed:7574684, PubMed:15494731). Upon ligand-binding, undergoes homodimerization, autophosphorylation and activation (PubMed:15494731). Recruits, phosphorylates and/or activates several downstream effectors including SHC1, FRS2, SH2B1, SH2B2 and PLCG1 that regulate distinct overlapping signaling cascades. Through SHC1, FRS2, SH2B1, SH2B2 activates the GRB2-Ras-MAPK cascade that regulates for instance neuronal differentiation including neurite outgrowth. Through the same effectors controls the Ras-PI3 kinase-AKT1 signaling cascade that mainly regulates growth and survival. Through PLCG1 and the downstream protein kinase C-regulated pathways controls synaptic plasticity. Thereby, plays a role in learning and memory by regulating both short term synaptic function and long-term potentiation. PLCG1 also leads to NF-Kappa-B activation and the transcription of genes involved in cell survival. Hence, it is able to suppress anoikis, the apoptosis resulting from loss of cell-matrix interactions. May also play a role in neutrophin-dependent calcium signaling in glial cells and mediate communication between neurons and glia. {ECO:0000250|UniProtKB:P15209, ECO:0000269|PubMed:15494731, ECO:0000269|PubMed:7574684}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
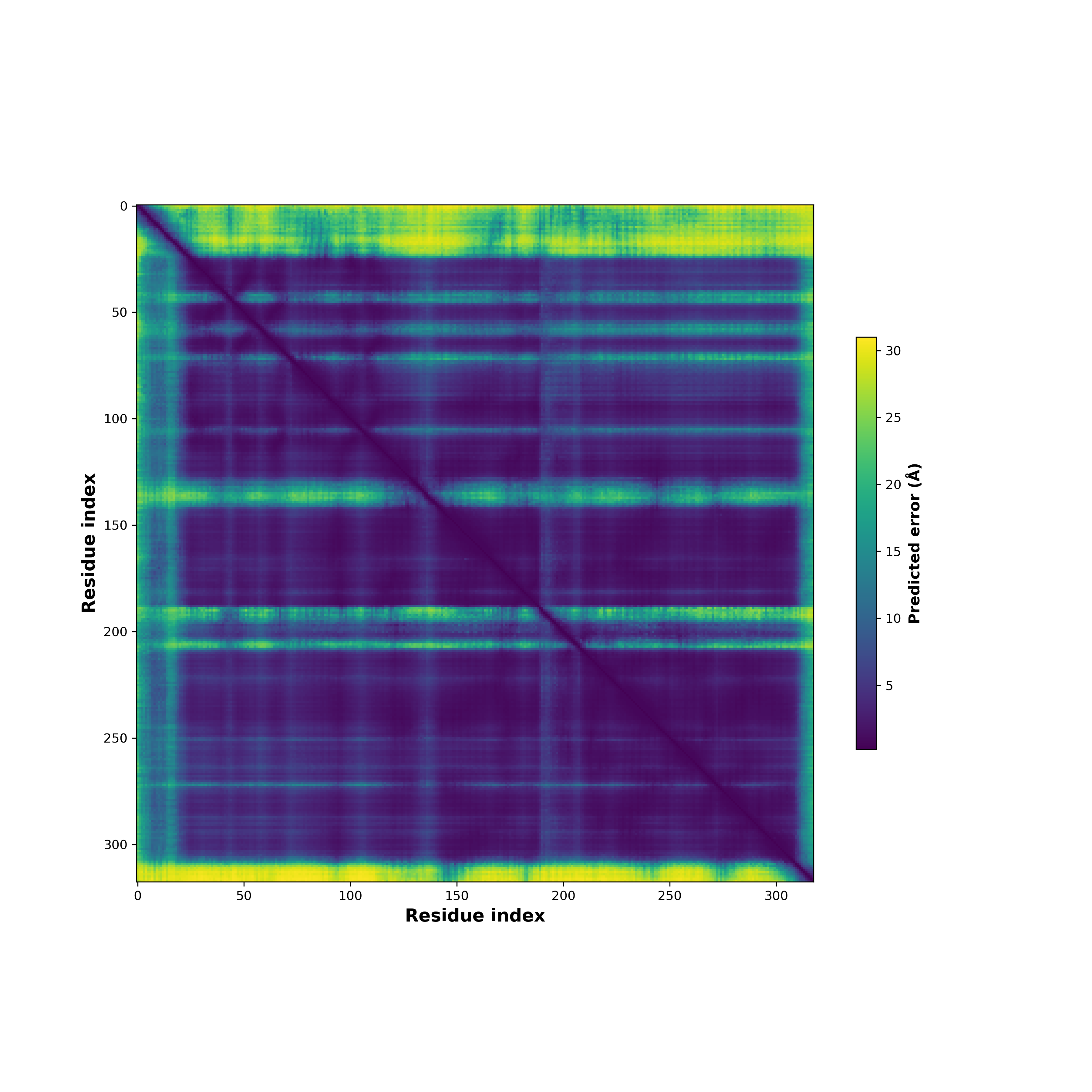
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (78) >>>78.pdbFusion protein BP residue: CIF file (78) >>>78.cif | QKI | chr6 | 163984751 | + | NTRK2 | chr9 | 87482157 | + | MTKIPVIENPQYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFL AECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFY GVCVEGDPLIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQML HIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGDFGMSRDVYST DYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPW YQLSNNEVIECITQGRVLQRPRTCPQEVYELMLGCWQREPHMRKNIKGIH | 318 |
| 3D view using mol* of 78 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (537) >>>537.pdbFusion protein BP residue: 303 CIF file (537) >>>537.cif | QKI | chr6 | 163984751 | + | NTRK2 | chr9 | 87482157 | + | MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEIS RVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPVKEYPDFNFVG RILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNED LHVLITVEDAQNRAEIKLKRAVEEVKKLLVPAAEGEDSLKKMQLMELAIL NGTYRDANIKSPTAQAAPRIITGPAPVLPPAALRTPTPAGPTIMPLIRQI QTAVMPNGTPHPTAAIVPPGPEAGLIYTPYEYPYTLAPATSILEYPIEPS GVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIGMTKIPVIE NPQYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFLAECYNLCP EQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFYGVCVEGDP LIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQMLHIAQQIAA GMVYLASQHFVHRDLATRNCLVGENLLVKIGDFGMSRDVYSTDYYRVGGH TMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPWYQLSNNEV IECITQGRVLQRPRTCPQEVYELMLGCWQREPHMRKNIKGIHTLLQNLAK | 660 |
| 3D view using mol* of 537 (AA BP:303) | ||||||||||
 | ||||||||||
| PDB file (543) >>>543.pdbFusion protein BP residue: 311 CIF file (543) >>>543.cif | QKI | chr6 | 163984751 | + | NTRK2 | chr9 | 87482157 | + | MVGEMETKEKPKPTPDYLMQLMNDKKLMSSLPNFCGIFNHLERLLDEEIS RVRKDMYNDTLNGSTEKRSAELPDAVGPIVQLQEKLYVPVKEYPDFNFVG RILGPRGLTAKQLEAETGCKIMVRGKGSMRDKKKEEQNRGKPNWEHLNED LHVLITVEDAQNRAEIKLKRAVEEVKKLLVPAAEGEDSLKKMQLMELAIL NGTYRDANIKSPALAFSLAATAQAAPRIITGPAPVLPPAALRTPTPAGPT IMPLIRQIQTAVMPNGTPHPTAAIVPPGPEAGLIYTPYEYPYTLAPATSI LEYPIEPSGVLGPASVISNDDDSASPLHHISNGSNTPSSSEGGPDAVIIG MTKIPVIENPQYFGITNSQLKPDTFVQHIKRHNIVLKRELGEGAFGKVFL AECYNLCPEQDKILVAVKTLKDASDNARKDFHREAELLTNLQHEHIVKFY GVCVEGDPLIMVFEYMKHGDLNKFLRAHGPDAVLMAEGNPPTELTQSQML HIAQQIAAGMVYLASQHFVHRDLATRNCLVGENLLVKIGDFGMSRDVYST DYYRVGGHTMLPIRWMPPESIMYRKFTTESDVWSLGVVLWEIFTYGKQPW YQLSNNEVIECITQGRVLQRPRTCPQEVYELMLGCWQREPHMRKNIKGIH | 668 |
| 3D view using mol* of 543 (AA BP:311) | ||||||||||
 | ||||||||||
Top |
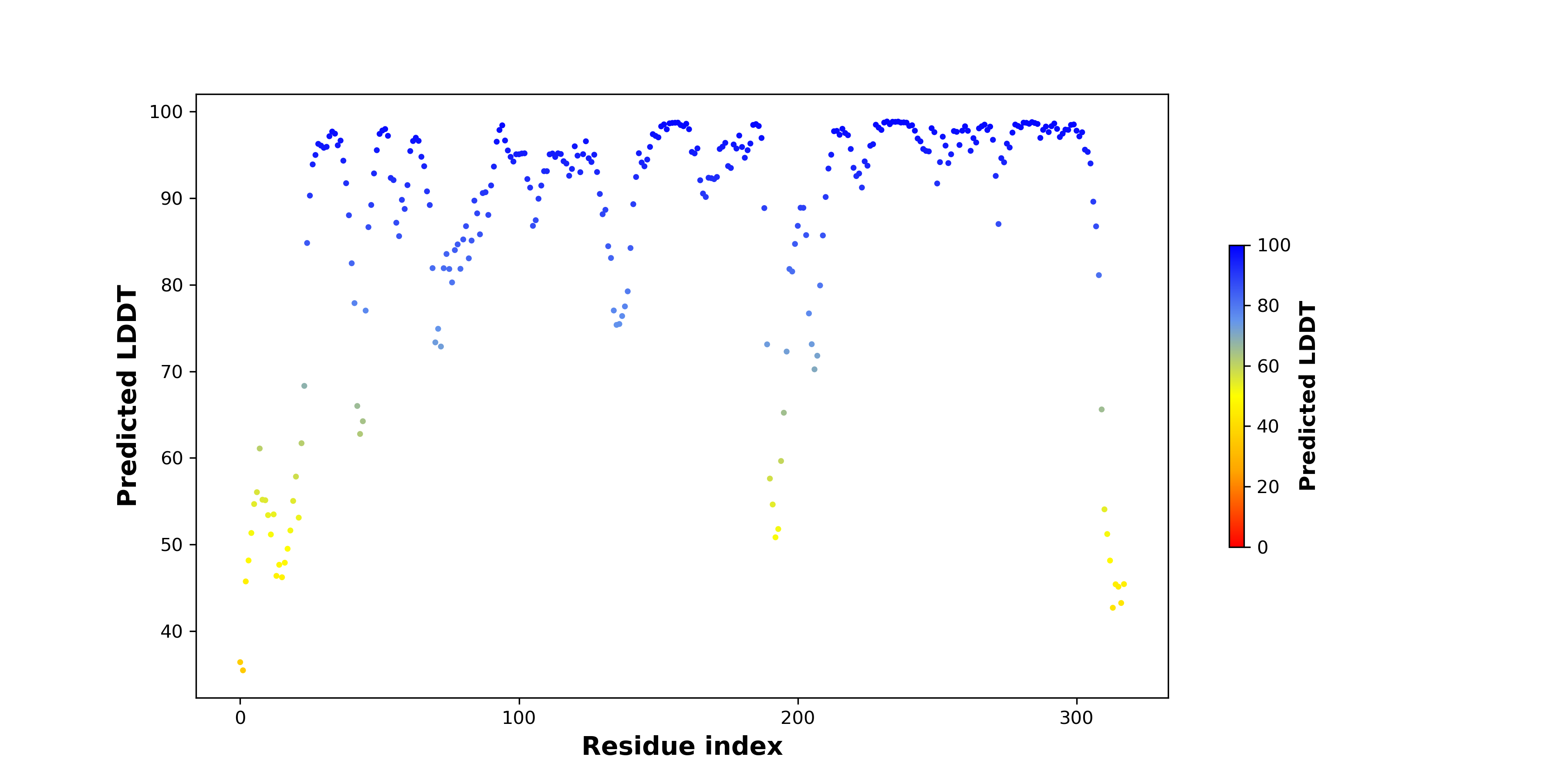
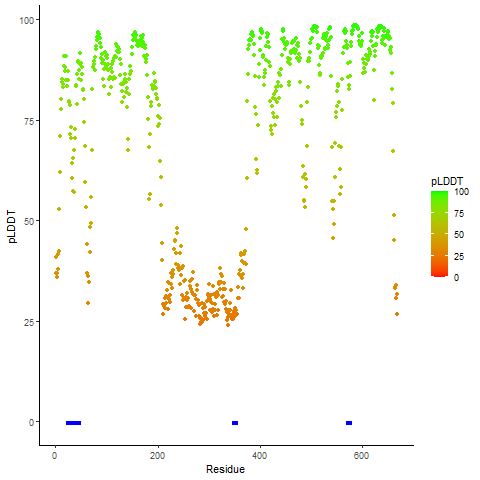
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
QKI_pLDDT.png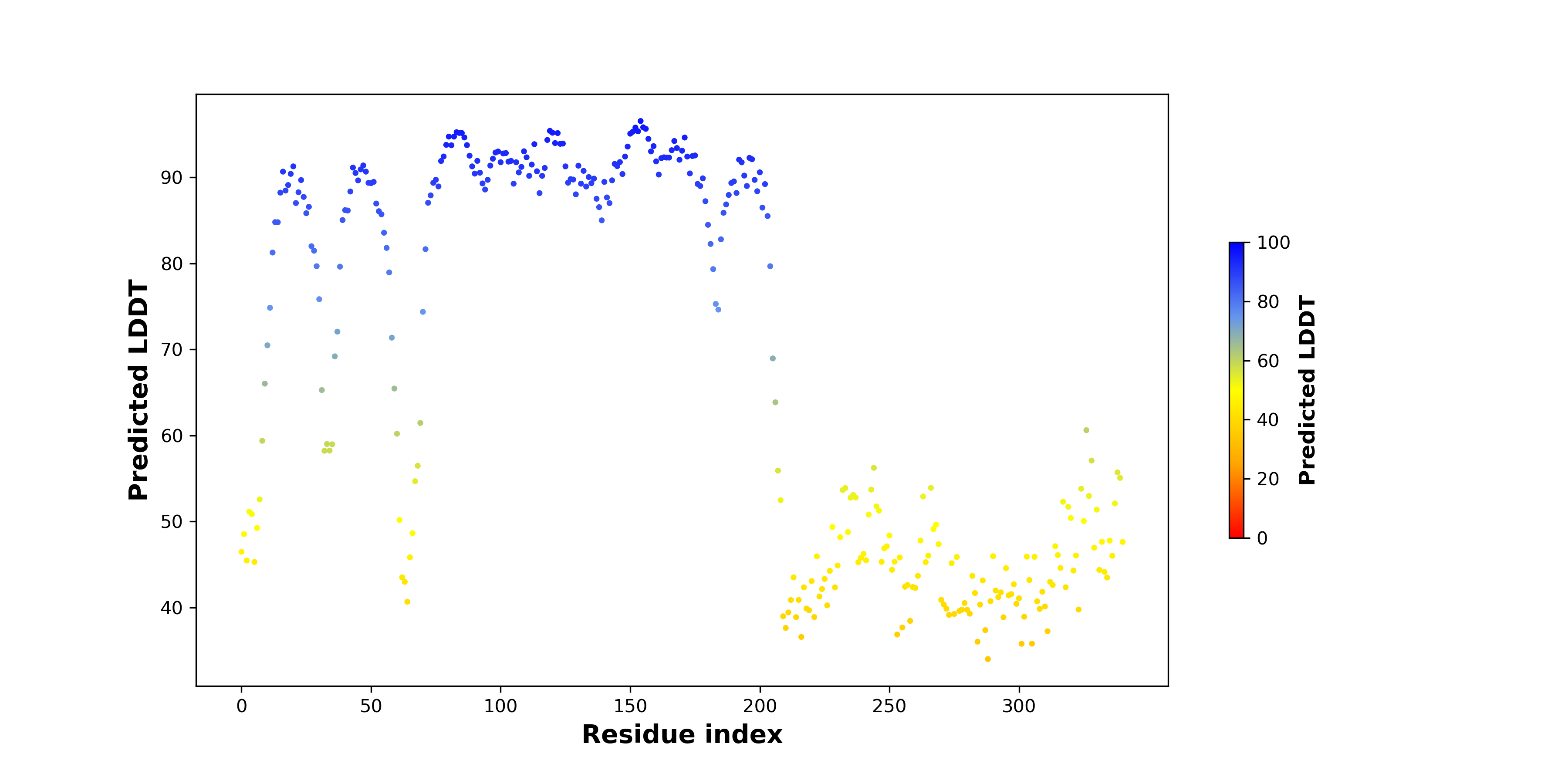 |
NTRK2_pLDDT.png |
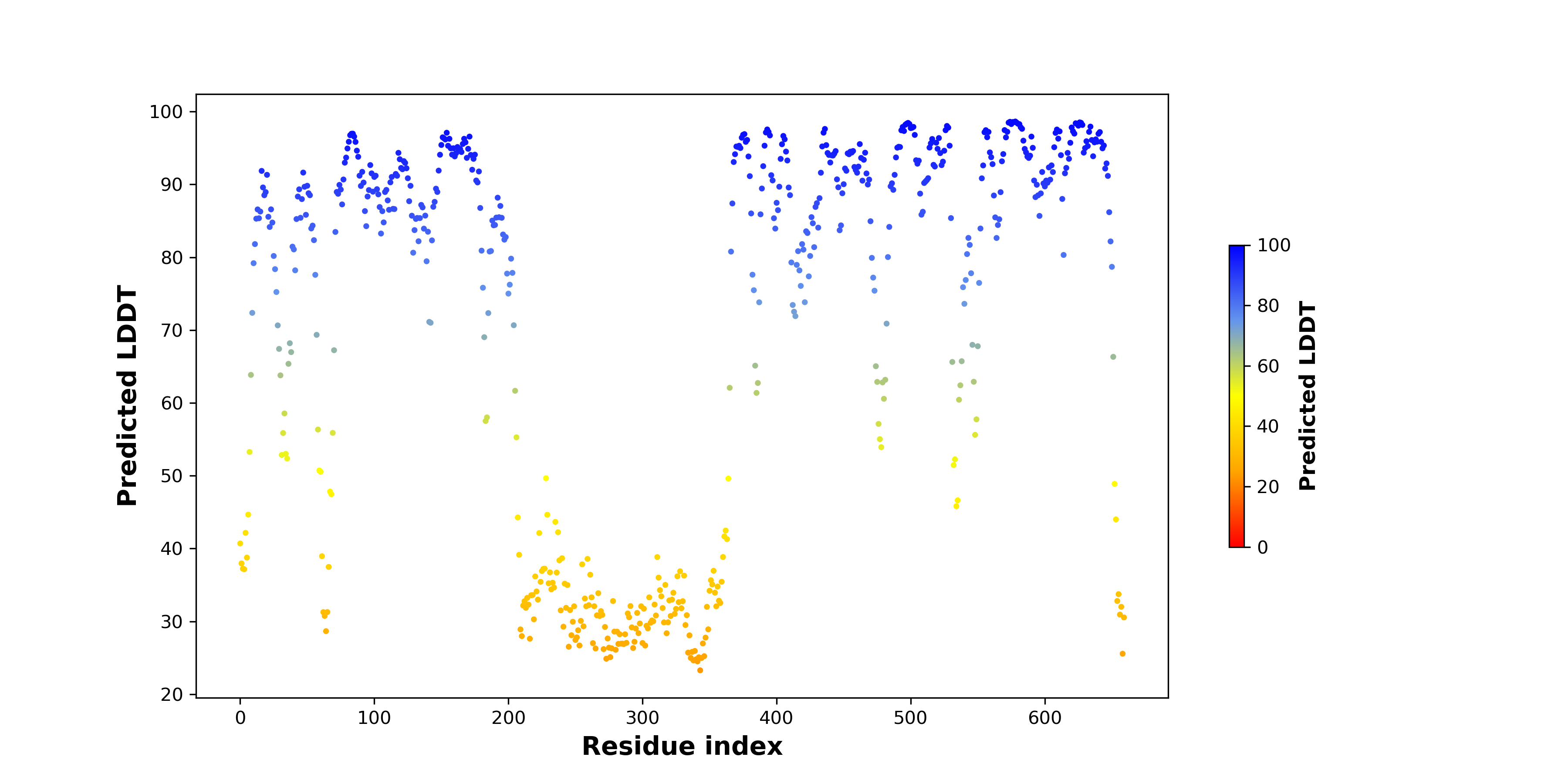
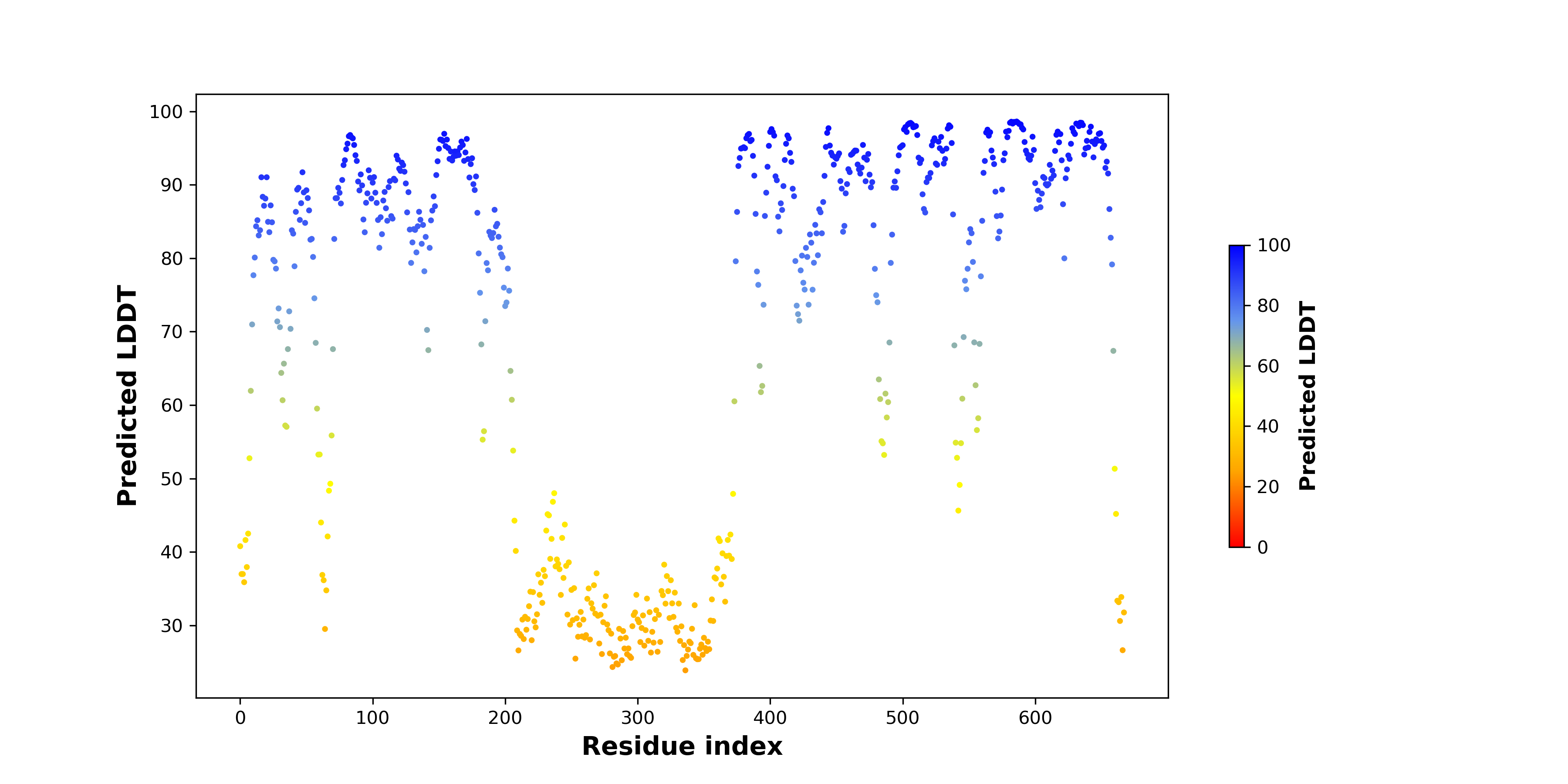
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
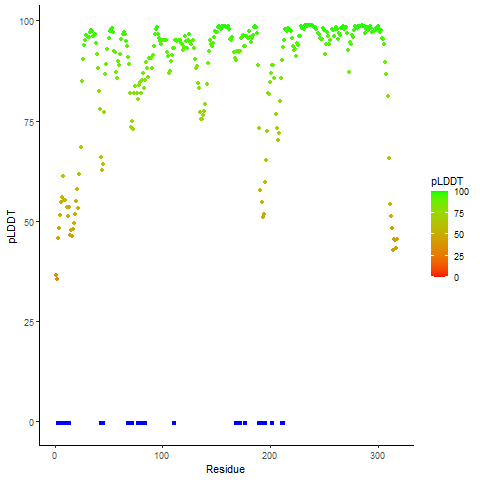
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| QKI_NTRK2_537.png |
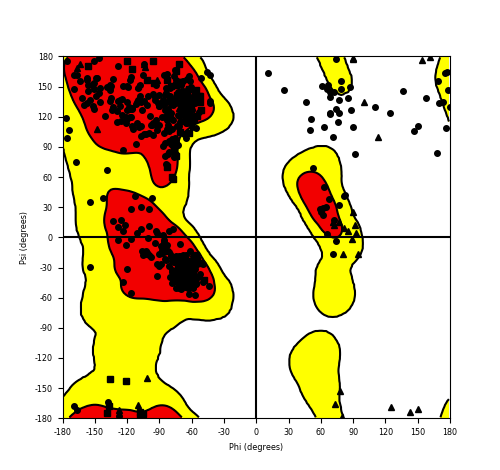 |
Top |
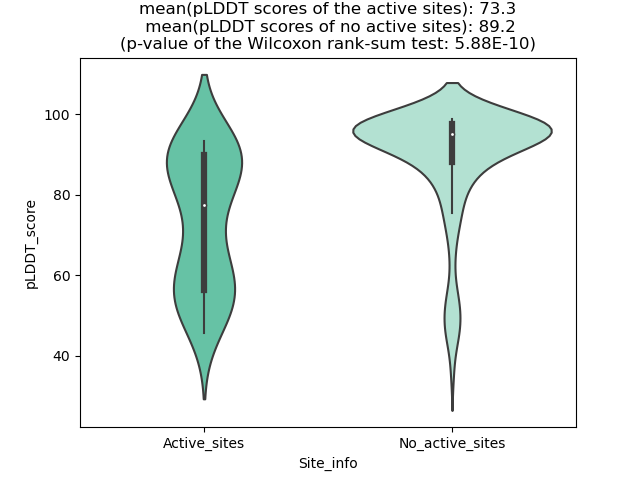
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 78 | 1.107 | 146 | 0.998 | 341.971 | 0.493 | 0.858 | 1.161 | 0.446 | 1.403 | 0.318 | 1.091 | Chain A: 3,4,5,6,7,8,9,12,13,43,44,45,68,69,70,72, 77,80,81,83,84,111,168,170,171,172,177,190,191,192 ,195,202,211,212 |
| 537 | 1.074 | 305 | 1.12 | 1013.565 | 0.574 | 0.744 | 0.915 | 0.896 | 0.819 | 1.094 | 1.226 | Chain A: 288,289,290,291,292,293,294,295,349,350,3 51,354,355,357,358,361,362,363,366,367,368,386,387 ,410,412,413,414,419,422,423,425,426,428,429,432,4 33,434,453,508,509,510,511,512,513,514,515,532,533 ,534,551,552,553,554,557,558,566,568,576 |
| 543 | 1.029 | 140 | 1.056 | 457.905 | 0.61 | 0.719 | 0.891 | 0.54 | 0.967 | 0.558 | 0.965 | Chain A: 25,28,29,30,31,32,33,34,35,36,38,39,40,41 ,42,43,46,349,350,351,352,354,572,573,574,575 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| QKI | HNRNPK, USP30, MED12, MED13, MED25, MED16, MED17, MED1, MED24, MED23, MED4, MED29, MED31, MED15, MED14, MED26, MED10, POLR2A, MED27, ELAVL1, PSMF1, C10orf10, FUBP3, HCLS1, PCBP2, PTBP1, PTBP2, RBM14, RBFOX2, RBPMS, RNF138, SERF2, THAP1, USH1C, RBFOX1, PCBP1, PCBP4, APP, NDUFA7, CDV3, ATXN1, FN1, U2AF2, RBM23, RALY, WDR77, HNRNPH2, QKI, DAZ1, IL7R, CARM1, SNRPA, RBM11, NABP1, HNRNPLL, OIP5, RBMX, ZACN, CYP1A1, NTRK1, EWSR1, HYPK, DUSP22, DUSP4, ATE1, RBM42, FBXW11, SLC39A12, VAC14, TRIM11, SYNCRIP, SNRNP27, TAE1, PBP2, FBXO7, DMRTB1, CDKN2D, CCND3, TSG101, LSM3, TEX11, PIAS1, FHL3, ESR2, RAF1, APEX1, SNRNP70, DUSP14, LMBR1L, PRKAB2, TIAL1, E7, MIR7-1, MIR19B2, MIR34B, MIR34C, MIR141, MIR200A, CELF1, RBM47, RBMS1, C8orf82, RBM45, UFL1, DDRGK1, HNRNPA1, FZR1, HSPA2, HSPA1A, RBMS2, CYB5B, PRR3, CENPM, HNRNPUL2, FAM120A, HNRNPC, BAG1, IL7, TGM1, ELAVL2, BAG2, HSPA8, HNRNPCL2, HNRNPCL1, EIF4ENIF1, MBNL1, PCBP3, ALG13, YTHDF1, SNRPC, UNKL, BAG4, PRRC2B, MSI2, TNRC6A, ANKHD1, TXNL4A, CALD1, RBM7, ZMAT5, CCDC59, BCAS2, NXF1, TRNAU1AP, YTHDF3, NIP7, ZC3H10, CPSF7, SLAIN1, SECISBP2, RPL9, PRPF6, PRPF39, MBNL2, PCGF5, KIF21B, MRPL44, LSM5, UTP6, Pcbp1, Hnrnpk, Rbm38, Rbm47, Dusp14, Mrpl13, Ywhag, Pum2, Pef1, Hsp90aa1, GATA3, PAX9, SP7, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| QKI | 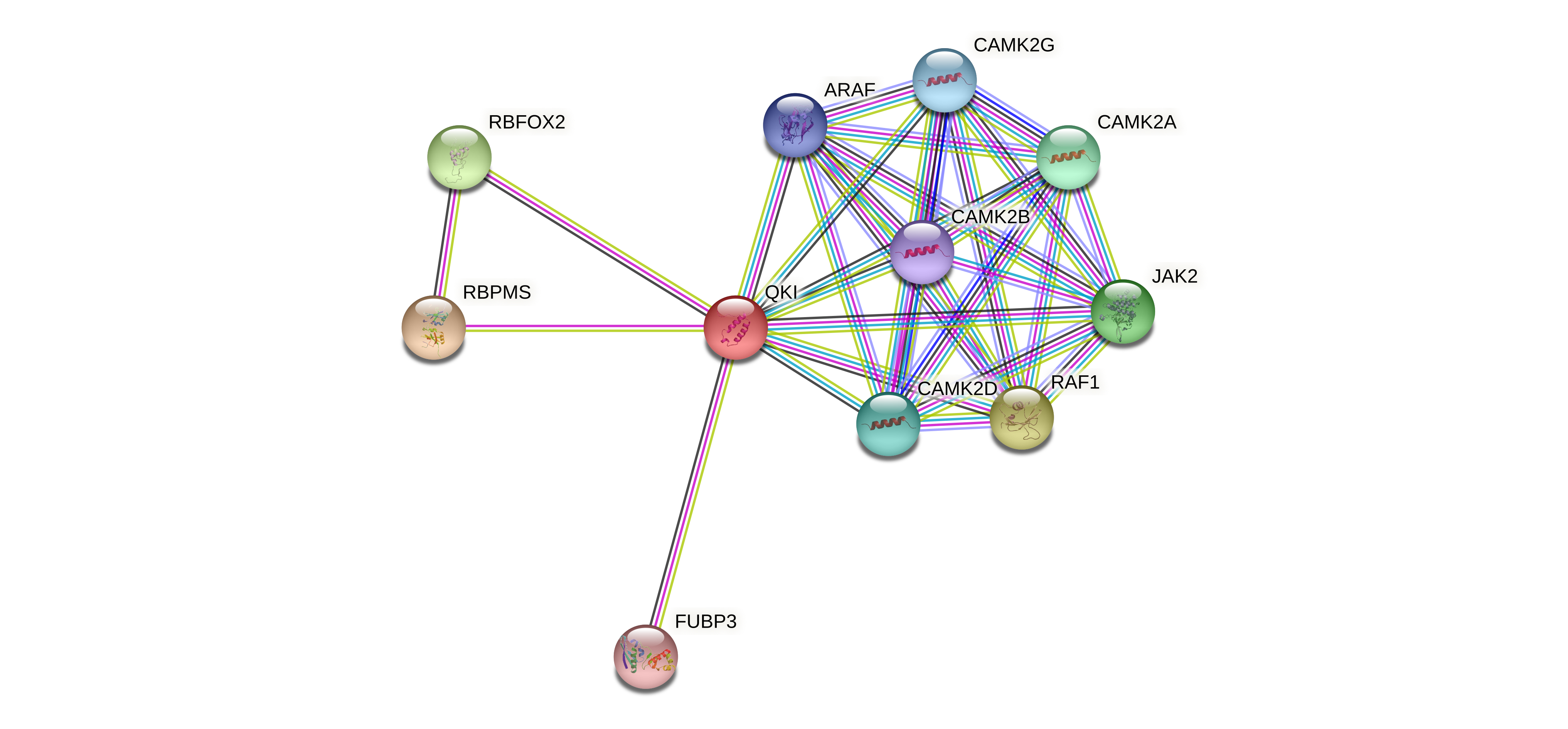 |
| NTRK2 | 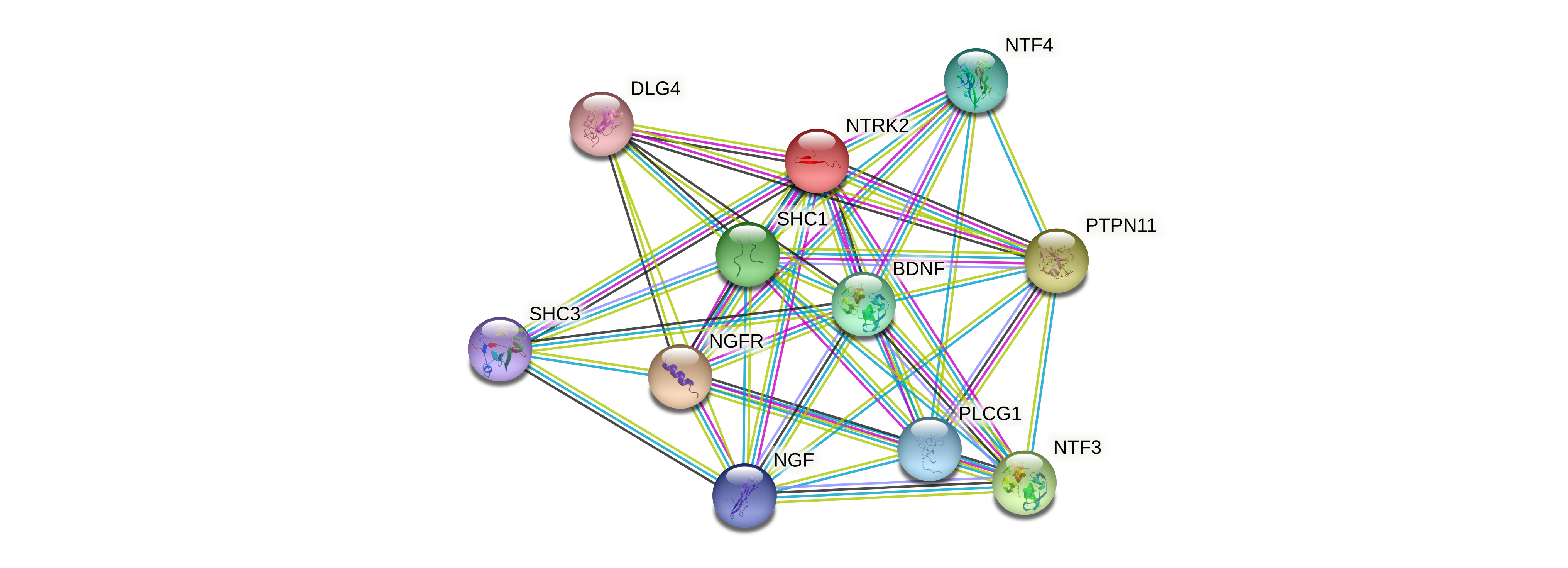 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to QKI-NTRK2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to QKI-NTRK2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| QKI | NTRK2 | Prostate Adenocarcinoma | MyCancerGenome | |
| QKI | NTRK2 | Anal Squamous Cell Carcinoma | MyCancerGenome | |
| QKI | NTRK2 | Cancer Of Unknown Primary | MyCancerGenome | |
| QKI | NTRK2 | Nos | MyCancerGenome | |
| QKI | NTRK2 | Cutaneous Melanoma | MyCancerGenome | |
| QKI | NTRK2 | Esophagogastric Carcinoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | QKI | C0036341 | Schizophrenia | 5 | PSYGENET |
| Hgene | QKI | C0017638 | Glioma | 1 | CTD_human |
| Hgene | QKI | C0041696 | Unipolar Depression | 1 | PSYGENET |
| Hgene | QKI | C0259783 | mixed gliomas | 1 | CTD_human |
| Hgene | QKI | C0555198 | Malignant Glioma | 1 | CTD_human |
| Hgene | QKI | C1269683 | Major Depressive Disorder | 1 | PSYGENET |
| Hgene | QKI | C2363903 | Angiocentric glioma | 1 | ORPHANET |
| Hgene | QKI | C3714756 | Intellectual Disability | 1 | GENOMICS_ENGLAND |
| Tgene | NTRK2 | C0011570 | Mental Depression | 5 | PSYGENET |
| Tgene | NTRK2 | C0011581 | Depressive disorder | 5 | PSYGENET |
| Tgene | NTRK2 | C0041696 | Unipolar Depression | 5 | PSYGENET |
| Tgene | NTRK2 | C0525045 | Mood Disorders | 5 | PSYGENET |
| Tgene | NTRK2 | C1269683 | Major Depressive Disorder | 5 | PSYGENET |
| Tgene | NTRK2 | C3151303 | Obesity, Hyperphagia, and Developmental Delay | 4 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | NTRK2 | C0005586 | Bipolar Disorder | 3 | CTD_human;PSYGENET |
| Tgene | NTRK2 | C4693367 | EPILEPTIC ENCEPHALOPATHY, EARLY INFANTILE, 58 | 3 | GENOMICS_ENGLAND;UNIPROT |
| Tgene | NTRK2 | C0009171 | Cocaine Abuse | 2 | CTD_human |
| Tgene | NTRK2 | C0036341 | Schizophrenia | 2 | PSYGENET |
| Tgene | NTRK2 | C0038220 | Status Epilepticus | 2 | CTD_human |
| Tgene | NTRK2 | C0236736 | Cocaine-Related Disorders | 2 | CTD_human |
| Tgene | NTRK2 | C0270823 | Petit mal status | 2 | CTD_human |
| Tgene | NTRK2 | C0311335 | Grand Mal Status Epilepticus | 2 | CTD_human |
| Tgene | NTRK2 | C0393734 | Complex Partial Status Epilepticus | 2 | CTD_human |
| Tgene | NTRK2 | C0600427 | Cocaine Dependence | 2 | CTD_human |
| Tgene | NTRK2 | C0751217 | Hyperkinesia, Generalized | 2 | CTD_human |
| Tgene | NTRK2 | C0751522 | Status Epilepticus, Subclinical | 2 | CTD_human |
| Tgene | NTRK2 | C0751523 | Non-Convulsive Status Epilepticus | 2 | CTD_human |
| Tgene | NTRK2 | C0751524 | Simple Partial Status Epilepticus | 2 | CTD_human |
| Tgene | NTRK2 | C3887506 | Hyperkinesia | 2 | CTD_human |
| Tgene | NTRK2 | C0001973 | Alcoholic Intoxication, Chronic | 1 | PSYGENET |
| Tgene | NTRK2 | C0004114 | Astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0004352 | Autistic Disorder | 1 | CTD_human |
| Tgene | NTRK2 | C0005587 | Depression, Bipolar | 1 | CTD_human |
| Tgene | NTRK2 | C0008073 | Developmental Disabilities | 1 | CTD_human |
| Tgene | NTRK2 | C0013415 | Dysthymic Disorder | 1 | PSYGENET |
| Tgene | NTRK2 | C0017638 | Glioma | 1 | CTD_human |
| Tgene | NTRK2 | C0020505 | Hyperphagia | 1 | CTD_human |
| Tgene | NTRK2 | C0024713 | Manic Disorder | 1 | CTD_human |
| Tgene | NTRK2 | C0027819 | Neuroblastoma | 1 | CTD_human |
| Tgene | NTRK2 | C0028754 | Obesity | 1 | CTD_human |
| Tgene | NTRK2 | C0036349 | Paranoid Schizophrenia | 1 | PSYGENET |
| Tgene | NTRK2 | C0037769 | West Syndrome | 1 | ORPHANET |
| Tgene | NTRK2 | C0085996 | Child Development Deviations | 1 | CTD_human |
| Tgene | NTRK2 | C0085997 | Child Development Disorders, Specific | 1 | CTD_human |
| Tgene | NTRK2 | C0205768 | Subependymal Giant Cell Astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0259783 | mixed gliomas | 1 | CTD_human |
| Tgene | NTRK2 | C0280783 | Juvenile Pilocytic Astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0280785 | Diffuse Astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0334579 | Anaplastic astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0334580 | Protoplasmic astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0334581 | Gemistocytic astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0334582 | Fibrillary Astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0334583 | Pilocytic Astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0338070 | Childhood Cerebral Astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0338831 | Manic | 1 | CTD_human |
| Tgene | NTRK2 | C0547065 | Mixed oligoastrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0555198 | Malignant Glioma | 1 | CTD_human |
| Tgene | NTRK2 | C0678807 | prenatal alcohol exposure | 1 | PSYGENET |
| Tgene | NTRK2 | C0750935 | Cerebral Astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0750936 | Intracranial Astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C0752347 | Lewy Body Disease | 1 | CTD_human |
| Tgene | NTRK2 | C1519086 | Pilomyxoid astrocytoma | 1 | ORPHANET |
| Tgene | NTRK2 | C1704230 | Grade I Astrocytoma | 1 | CTD_human |
| Tgene | NTRK2 | C3146244 | Alcohol Related Birth Defect | 1 | PSYGENET |