| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:RAB3IL1-NRG1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: RAB3IL1-NRG1 | FusionPDB ID: 71381 | FusionGDB2.0 ID: 71381 | Hgene | Tgene | Gene symbol | RAB3IL1 | NRG1 | Gene ID | 5866 | 3084 |
| Gene name | RAB3A interacting protein like 1 | neuregulin 1 | |
| Synonyms | GRAB | ARIA|GGF|GGF2|HGL|HRG|HRG1|HRGA|MST131|MSTP131|NDF|NRG1-IT2|SMDF | |
| Cytomap | 11q12.2-q12.3 | 8p12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | guanine nucleotide exchange factor for Rab-3ARAB3A interacting protein (rabin3)-like 1rab3A-interacting-like protein 1rabin3-like 1 | pro-neuregulin-1, membrane-bound isoformacetylcholine receptor-inducing activityglial growth factor 2heregulin, alpha (45kD, ERBB2 p185-activator)neu differentiation factorpro-NRG1sensory and motor neuron derived factor | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | . | Q02297 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000301773, ENST00000394836, | ENST00000523681, ENST00000287842, ENST00000287845, ENST00000338921, ENST00000341377, ENST00000356819, ENST00000405005, ENST00000519301, ENST00000520407, ENST00000520502, ENST00000521670, ENST00000523079, ENST00000539990, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 4 X 4 X 2=32 | 25 X 17 X 14=5950 |
| # samples | 4 | 27 | |
| ** MAII score | log2(4/32*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(27/5950*10)=-4.46185835603184 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: RAB3IL1 [Title/Abstract] AND NRG1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | RAB3IL1(61666786)-NRG1(32585467), # samples:2 RAB3IL1(61669914)-NRG1(32585467), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | RAB3IL1-NRG1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. RAB3IL1-NRG1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. RAB3IL1-NRG1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. RAB3IL1-NRG1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. RAB3IL1-NRG1 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | NRG1 | GO:0003222 | ventricular trabecula myocardium morphogenesis | 17336907 |
| Tgene | NRG1 | GO:0031334 | positive regulation of protein complex assembly | 10559227 |
| Tgene | NRG1 | GO:0038127 | ERBB signaling pathway | 11389077 |
| Tgene | NRG1 | GO:0038129 | ERBB3 signaling pathway | 27353365 |
| Tgene | NRG1 | GO:0045892 | negative regulation of transcription, DNA-templated | 15073182 |
| Tgene | NRG1 | GO:0051048 | negative regulation of secretion | 10559227 |
| Tgene | NRG1 | GO:0060379 | cardiac muscle cell myoblast differentiation | 17336907 |
| Tgene | NRG1 | GO:0060956 | endocardial cell differentiation | 17336907 |
 Fusion gene breakpoints across RAB3IL1 (5'-gene) Fusion gene breakpoints across RAB3IL1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
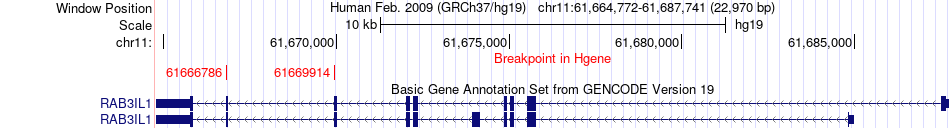 |
 Fusion gene breakpoints across NRG1 (3'-gene) Fusion gene breakpoints across NRG1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
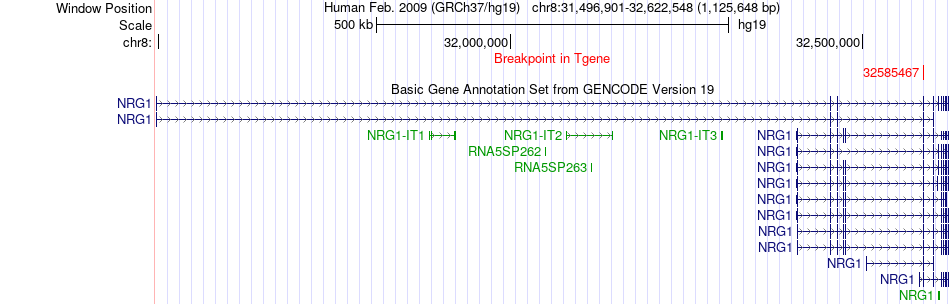 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-04-1362-01A | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + |
| ChimerDB4 | OV | TCGA-04-1362-01A | RAB3IL1 | chr11 | 61669914 | - | NRG1 | chr8 | 32585467 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000519301 | NRG1 | chr8 | 32585467 | + | 2663 | 1224 | 122 | 2659 | 845 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000520407 | NRG1 | chr8 | 32585467 | + | 1904 | 1224 | 122 | 1447 | 441 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000523079 | NRG1 | chr8 | 32585467 | + | 2564 | 1224 | 122 | 1984 | 620 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000338921 | NRG1 | chr8 | 32585467 | + | 3297 | 1224 | 122 | 2668 | 848 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000356819 | NRG1 | chr8 | 32585467 | + | 3288 | 1224 | 122 | 2659 | 845 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000287845 | NRG1 | chr8 | 32585467 | + | 3288 | 1224 | 122 | 2659 | 845 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000341377 | NRG1 | chr8 | 32585467 | + | 3332 | 1224 | 122 | 1465 | 447 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000287842 | NRG1 | chr8 | 32585467 | + | 2745 | 1224 | 122 | 2635 | 837 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000521670 | NRG1 | chr8 | 32585467 | + | 2383 | 1224 | 122 | 2110 | 662 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000405005 | NRG1 | chr8 | 32585467 | + | 2714 | 1224 | 122 | 2644 | 840 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000520502 | NRG1 | chr8 | 32585467 | + | 1887 | 1224 | 122 | 1447 | 441 |
| ENST00000394836 | RAB3IL1 | chr11 | 61666786 | - | ENST00000539990 | NRG1 | chr8 | 32585467 | + | 2742 | 1224 | 122 | 2635 | 837 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000519301 | NRG1 | chr8 | 32585467 | + | 2504 | 1065 | 77 | 2500 | 807 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000520407 | NRG1 | chr8 | 32585467 | + | 1745 | 1065 | 77 | 1288 | 403 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000523079 | NRG1 | chr8 | 32585467 | + | 2405 | 1065 | 77 | 1825 | 582 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000338921 | NRG1 | chr8 | 32585467 | + | 3138 | 1065 | 77 | 2509 | 810 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000356819 | NRG1 | chr8 | 32585467 | + | 3129 | 1065 | 77 | 2500 | 807 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000287845 | NRG1 | chr8 | 32585467 | + | 3129 | 1065 | 77 | 2500 | 807 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000341377 | NRG1 | chr8 | 32585467 | + | 3173 | 1065 | 1306 | 2544 | 412 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000287842 | NRG1 | chr8 | 32585467 | + | 2586 | 1065 | 77 | 2476 | 799 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000521670 | NRG1 | chr8 | 32585467 | + | 2224 | 1065 | 77 | 1951 | 624 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000405005 | NRG1 | chr8 | 32585467 | + | 2555 | 1065 | 77 | 2485 | 802 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000520502 | NRG1 | chr8 | 32585467 | + | 1728 | 1065 | 77 | 1288 | 403 |
| ENST00000301773 | RAB3IL1 | chr11 | 61666786 | - | ENST00000539990 | NRG1 | chr8 | 32585467 | + | 2583 | 1065 | 77 | 2476 | 799 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000394836 | ENST00000519301 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.009628521 | 0.9903715 |
| ENST00000394836 | ENST00000520407 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.022910371 | 0.9770897 |
| ENST00000394836 | ENST00000523079 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.022242192 | 0.9777579 |
| ENST00000394836 | ENST00000338921 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.004928527 | 0.99507153 |
| ENST00000394836 | ENST00000356819 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.00437938 | 0.99562067 |
| ENST00000394836 | ENST00000287845 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.00437938 | 0.99562067 |
| ENST00000394836 | ENST00000341377 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.020707669 | 0.9792923 |
| ENST00000394836 | ENST00000287842 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.007423161 | 0.9925768 |
| ENST00000394836 | ENST00000521670 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.044778205 | 0.95522183 |
| ENST00000394836 | ENST00000405005 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.00874865 | 0.9912513 |
| ENST00000394836 | ENST00000520502 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.025658235 | 0.97434175 |
| ENST00000394836 | ENST00000539990 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.007488785 | 0.9925113 |
| ENST00000301773 | ENST00000519301 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.006805878 | 0.9931941 |
| ENST00000301773 | ENST00000520407 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.008710137 | 0.99128985 |
| ENST00000301773 | ENST00000523079 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.015115956 | 0.984884 |
| ENST00000301773 | ENST00000338921 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.003097807 | 0.9969022 |
| ENST00000301773 | ENST00000356819 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.002925247 | 0.9970747 |
| ENST00000301773 | ENST00000287845 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.002925247 | 0.9970747 |
| ENST00000301773 | ENST00000341377 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.010199169 | 0.9898009 |
| ENST00000301773 | ENST00000287842 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.004972085 | 0.99502796 |
| ENST00000301773 | ENST00000521670 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.028741576 | 0.97125846 |
| ENST00000301773 | ENST00000405005 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.006166259 | 0.9938338 |
| ENST00000301773 | ENST00000520502 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.010028931 | 0.9899711 |
| ENST00000301773 | ENST00000539990 | RAB3IL1 | chr11 | 61666786 | - | NRG1 | chr8 | 32585467 | + | 0.005034957 | 0.994965 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >71381_71381_1_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000287842_length(amino acids)=799AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM VKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMN IANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRH SSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPF MEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLE -------------------------------------------------------------- >71381_71381_2_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000287845_length(amino acids)=807AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM VKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLR SERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVM SSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSM PSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPN -------------------------------------------------------------- >71381_71381_3_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000338921_length(amino acids)=810AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM VKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQ SLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSV IVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMT VSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRT KPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARLSSVIANQDPIAV -------------------------------------------------------------- >71381_71381_4_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >71381_71381_5_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000356819_length(amino acids)=807AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM VKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLR SERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVM SSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSM PSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPN -------------------------------------------------------------- >71381_71381_6_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000405005_length(amino acids)=802AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM VKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNN MMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVEN SRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAV SPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIAN -------------------------------------------------------------- >71381_71381_7_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000519301_length(amino acids)=807AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM VKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLR SERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVM SSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSM PSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPN -------------------------------------------------------------- >71381_71381_8_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000520407_length(amino acids)=403AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM -------------------------------------------------------------- >71381_71381_9_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000520502_length(amino acids)=403AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM -------------------------------------------------------------- >71381_71381_10_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000521670_length(amino acids)=624AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM VKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNN MMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVEN -------------------------------------------------------------- >71381_71381_11_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000523079_length(amino acids)=582AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM VKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMN IANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRH -------------------------------------------------------------- >71381_71381_12_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000301773_NRG1_chr8_32585467_ENST00000539990_length(amino acids)=799AA_BP=329 MDSSEEHAGCPARGTCPVFLAMSAGTVRYAPSGLCPVLEGNLREEPWGTDSPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETS AGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEFLKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQ AASEKQLKEARGKVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDFTMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDC SSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQQGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFM VKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMN IANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRH SSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPF MEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLE -------------------------------------------------------------- >71381_71381_13_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000287842_length(amino acids)=837AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ QGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIAL LVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHH STTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPAR MSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDE EYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTN -------------------------------------------------------------- >71381_71381_14_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000287845_length(amino acids)=845AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ QGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLT ITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTS HYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYV SAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPAS PLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAF -------------------------------------------------------------- >71381_71381_15_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000338921_length(amino acids)=848AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ QGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKHSGEPFPEAEELYQKR VLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSF STSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSE RYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSL PASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEAT -------------------------------------------------------------- >71381_71381_16_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000341377_length(amino acids)=447AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ -------------------------------------------------------------- >71381_71381_17_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000356819_length(amino acids)=845AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ QGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLT ITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTS HYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYV SAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPAS PLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAF -------------------------------------------------------------- >71381_71381_18_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000405005_length(amino acids)=840AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ QGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGIC IALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTST AHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTT PARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIV EDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADS -------------------------------------------------------------- >71381_71381_19_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000519301_length(amino acids)=845AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ QGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLT ITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTS HYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYV SAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPAS PLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAF -------------------------------------------------------------- >71381_71381_20_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000520407_length(amino acids)=441AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ -------------------------------------------------------------- >71381_71381_21_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000520502_length(amino acids)=441AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ -------------------------------------------------------------- >71381_71381_22_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000521670_length(amino acids)=662AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ QGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGIC IALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTST AHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERHNLIAEL -------------------------------------------------------------- >71381_71381_23_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000523079_length(amino acids)=620AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ QGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIAL LVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHH -------------------------------------------------------------- >71381_71381_24_RAB3IL1-NRG1_RAB3IL1_chr11_61666786_ENST00000394836_NRG1_chr8_32585467_ENST00000539990_length(amino acids)=837AA_BP=367 MGTDAWAPRGAGMWSGPPQPDQGLPPPLAAVPVPWKSTDPCQGHRESPGALVETSAGEEAQGQEGPAAAQLDVLRLRSSSMEIREKGSEF LKEELHRAQKELKLKDEECERLSKVREQLEQELEELTASLFEEAHKMVREANMKQAASEKQLKEARGKIDMLQAEVTALKTLVITSTPAS PNRELHPQLLSPTKAGPRKGHSRHKSTSSTLCPAVCPAAGHTLTPDREGKEVDTILFAEFQAWRESPTLDKTCPFLERVYREDVGPCLDF TMQELSVLVRAAVEDNTLTIEPVASQTLPTVKVAEVDCSSTNTCALSGLTRTCRHRIRLGDSKSHYYISPSSRARITAVCNFFTYIRYIQ QGLVRQDATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIAL LVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHH STTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPAR MSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDE EYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTN -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:61666786/chr8:32585467) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | NRG1 |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | FUNCTION: Direct ligand for ERBB3 and ERBB4 tyrosine kinase receptors. Concomitantly recruits ERBB1 and ERBB2 coreceptors, resulting in ligand-stimulated tyrosine phosphorylation and activation of the ERBB receptors. The multiple isoforms perform diverse functions such as inducing growth and differentiation of epithelial, glial, neuronal, and skeletal muscle cells; inducing expression of acetylcholine receptor in synaptic vesicles during the formation of the neuromuscular junction; stimulating lobuloalveolar budding and milk production in the mammary gland and inducing differentiation of mammary tumor cells; stimulating Schwann cell proliferation; implication in the development of the myocardium such as trabeculation of the developing heart. Isoform 10 may play a role in motor and sensory neuron development. Binds to ERBB4 (PubMed:10867024, PubMed:7902537). Binds to ERBB3 (PubMed:20682778). Acts as a ligand for integrins and binds (via EGF domain) to integrins ITGAV:ITGB3 or ITGA6:ITGB4. Its binding to integrins and subsequent ternary complex formation with integrins and ERRB3 are essential for NRG1-ERBB signaling. Induces the phosphorylation and activation of MAPK3/ERK1, MAPK1/ERK2 and AKT1 (PubMed:20682778). Ligand-dependent ERBB4 endocytosis is essential for the NRG1-mediated activation of these kinases in neurons (By similarity). {ECO:0000250|UniProtKB:P43322, ECO:0000269|PubMed:10867024, ECO:0000269|PubMed:1348215, ECO:0000269|PubMed:20682778, ECO:0000269|PubMed:7902537}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RAB3IL1 | chr11:61666786 | chr8:32585467 | ENST00000301773 | - | 8 | 9 | 73_161 | 329.3333333333333 | 357.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | RAB3IL1 | chr11:61666786 | chr8:32585467 | ENST00000394836 | - | 9 | 10 | 73_161 | 355.3333333333333 | 383.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000287842 | 4 | 12 | 165_177 | 167.33333333333334 | 638.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000341377 | 4 | 13 | 165_177 | 167.33333333333334 | 525.3333333333334 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000356819 | 4 | 13 | 165_177 | 167.33333333333334 | 646.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000405005 | 4 | 12 | 165_177 | 167.33333333333334 | 641.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000519301 | 2 | 11 | 165_177 | 112.33333333333333 | 591.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000521670 | 4 | 13 | 165_177 | 167.33333333333334 | 477.3333333333333 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000523079 | 4 | 11 | 165_177 | 167.33333333333334 | 421.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000287842 | 4 | 12 | 178_222 | 167.33333333333334 | 638.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000341377 | 4 | 13 | 178_222 | 167.33333333333334 | 525.3333333333334 | Domain | EGF-like | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000356819 | 4 | 13 | 178_222 | 167.33333333333334 | 646.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000405005 | 4 | 12 | 178_222 | 167.33333333333334 | 641.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000519301 | 2 | 11 | 178_222 | 112.33333333333333 | 591.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000521670 | 4 | 13 | 178_222 | 167.33333333333334 | 477.3333333333333 | Domain | EGF-like | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000523079 | 4 | 11 | 178_222 | 167.33333333333334 | 421.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000287842 | 4 | 12 | 266_640 | 167.33333333333334 | 638.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000341377 | 4 | 13 | 266_640 | 167.33333333333334 | 525.3333333333334 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000356819 | 4 | 13 | 266_640 | 167.33333333333334 | 646.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000405005 | 4 | 12 | 266_640 | 167.33333333333334 | 641.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000519301 | 2 | 11 | 266_640 | 112.33333333333333 | 591.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520502 | 0 | 3 | 266_640 | 222.33333333333334 | 297.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000521670 | 4 | 13 | 266_640 | 167.33333333333334 | 477.3333333333333 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000523079 | 4 | 11 | 266_640 | 167.33333333333334 | 421.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000287842 | 4 | 12 | 243_265 | 167.33333333333334 | 638.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000341377 | 4 | 13 | 243_265 | 167.33333333333334 | 525.3333333333334 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000356819 | 4 | 13 | 243_265 | 167.33333333333334 | 646.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000405005 | 4 | 12 | 243_265 | 167.33333333333334 | 641.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000519301 | 2 | 11 | 243_265 | 112.33333333333333 | 591.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520502 | 0 | 3 | 243_265 | 222.33333333333334 | 297.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000521670 | 4 | 13 | 243_265 | 167.33333333333334 | 477.3333333333333 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000523079 | 4 | 11 | 243_265 | 167.33333333333334 | 421.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520407 | 2 | 5 | 165_177 | 348.3333333333333 | 423.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520502 | 0 | 3 | 165_177 | 222.33333333333334 | 297.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000287842 | 4 | 12 | 37_128 | 167.33333333333334 | 638.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000341377 | 4 | 13 | 37_128 | 167.33333333333334 | 525.3333333333334 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000356819 | 4 | 13 | 37_128 | 167.33333333333334 | 646.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000405005 | 4 | 12 | 37_128 | 167.33333333333334 | 641.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000519301 | 2 | 11 | 37_128 | 112.33333333333333 | 591.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520407 | 2 | 5 | 178_222 | 348.3333333333333 | 423.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520407 | 2 | 5 | 37_128 | 348.3333333333333 | 423.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520502 | 0 | 3 | 178_222 | 222.33333333333334 | 297.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520502 | 0 | 3 | 37_128 | 222.33333333333334 | 297.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000521670 | 4 | 13 | 37_128 | 167.33333333333334 | 477.3333333333333 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000523079 | 4 | 11 | 37_128 | 167.33333333333334 | 421.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000287842 | 4 | 12 | 20_242 | 167.33333333333334 | 638.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000341377 | 4 | 13 | 20_242 | 167.33333333333334 | 525.3333333333334 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000356819 | 4 | 13 | 20_242 | 167.33333333333334 | 646.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000405005 | 4 | 12 | 20_242 | 167.33333333333334 | 641.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000519301 | 2 | 11 | 20_242 | 112.33333333333333 | 591.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520407 | 2 | 5 | 20_242 | 348.3333333333333 | 423.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520407 | 2 | 5 | 266_640 | 348.3333333333333 | 423.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520502 | 0 | 3 | 20_242 | 222.33333333333334 | 297.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000521670 | 4 | 13 | 20_242 | 167.33333333333334 | 477.3333333333333 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000523079 | 4 | 11 | 20_242 | 167.33333333333334 | 421.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr11:61666786 | chr8:32585467 | ENST00000520407 | 2 | 5 | 243_265 | 348.3333333333333 | 423.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| RAB3IL1 | |
| NRG1 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to RAB3IL1-NRG1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to RAB3IL1-NRG1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | NRG1 | C0036341 | Schizophrenia | 7 | CTD_human |
| Tgene | NRG1 | C0005586 | Bipolar Disorder | 5 | PSYGENET |
| Tgene | NRG1 | C0024809 | Marijuana Abuse | 3 | PSYGENET |
| Tgene | NRG1 | C0011570 | Mental Depression | 2 | PSYGENET |
| Tgene | NRG1 | C0011581 | Depressive disorder | 2 | PSYGENET |
| Tgene | NRG1 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | NRG1 | C0006870 | Cannabis Dependence | 1 | PSYGENET |
| Tgene | NRG1 | C0007621 | Neoplastic Cell Transformation | 1 | CTD_human |
| Tgene | NRG1 | C0011616 | Contact Dermatitis | 1 | CTD_human |
| Tgene | NRG1 | C0018801 | Heart failure | 1 | CTD_human |
| Tgene | NRG1 | C0018802 | Congestive heart failure | 1 | CTD_human |
| Tgene | NRG1 | C0019569 | Hirschsprung Disease | 1 | CTD_human |
| Tgene | NRG1 | C0023212 | Left-Sided Heart Failure | 1 | CTD_human |
| Tgene | NRG1 | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Tgene | NRG1 | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Tgene | NRG1 | C0026650 | Movement Disorders | 1 | CTD_human |
| Tgene | NRG1 | C0027626 | Neoplasm Invasiveness | 1 | CTD_human |
| Tgene | NRG1 | C0030193 | Pain | 1 | CTD_human |
| Tgene | NRG1 | C0032460 | Polycystic Ovary Syndrome | 1 | CTD_human |
| Tgene | NRG1 | C0038358 | Gastric ulcer | 1 | CTD_human |
| Tgene | NRG1 | C0085758 | Aganglionosis, Colonic | 1 | CTD_human |
| Tgene | NRG1 | C0087031 | Juvenile-Onset Still Disease | 1 | CTD_human |
| Tgene | NRG1 | C0162351 | Contact hypersensitivity | 1 | CTD_human |
| Tgene | NRG1 | C0234230 | Pain, Burning | 1 | CTD_human |
| Tgene | NRG1 | C0234238 | Ache | 1 | CTD_human |
| Tgene | NRG1 | C0234254 | Radiating pain | 1 | CTD_human |
| Tgene | NRG1 | C0235527 | Heart Failure, Right-Sided | 1 | CTD_human |
| Tgene | NRG1 | C0236733 | Amphetamine-Related Disorders | 1 | CTD_human |
| Tgene | NRG1 | C0236804 | Amphetamine Addiction | 1 | CTD_human |
| Tgene | NRG1 | C0236807 | Amphetamine Abuse | 1 | CTD_human |
| Tgene | NRG1 | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Tgene | NRG1 | C0266487 | Etat Marbre | 1 | CTD_human |
| Tgene | NRG1 | C0458257 | Pain, Splitting | 1 | CTD_human |
| Tgene | NRG1 | C0458259 | Pain, Crushing | 1 | CTD_human |
| Tgene | NRG1 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | NRG1 | C0751407 | Pain, Migratory | 1 | CTD_human |
| Tgene | NRG1 | C0751408 | Suffering, Physical | 1 | CTD_human |
| Tgene | NRG1 | C0876994 | Cardiotoxicity | 1 | CTD_human |
| Tgene | NRG1 | C1136382 | Sclerocystic Ovaries | 1 | CTD_human |
| Tgene | NRG1 | C1257840 | Aganglionosis, Rectosigmoid Colon | 1 | CTD_human |
| Tgene | NRG1 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | NRG1 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | NRG1 | C1959583 | Myocardial Failure | 1 | CTD_human |
| Tgene | NRG1 | C1961112 | Heart Decompensation | 1 | CTD_human |
| Tgene | NRG1 | C3495559 | Juvenile arthritis | 1 | CTD_human |
| Tgene | NRG1 | C3661523 | Congenital Intestinal Aganglionosis | 1 | CTD_human |
| Tgene | NRG1 | C3714758 | Juvenile psoriatic arthritis | 1 | CTD_human |
| Tgene | NRG1 | C4552091 | Polyarthritis, Juvenile, Rheumatoid Factor Negative | 1 | CTD_human |
| Tgene | NRG1 | C4704862 | Polyarthritis, Juvenile, Rheumatoid Factor Positive | 1 | CTD_human |
| Tgene | NRG1 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |

