| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:RBMS3-RBBP6 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: RBMS3-RBBP6 | FusionPDB ID: 73043 | FusionGDB2.0 ID: 73043 | Hgene | Tgene | Gene symbol | RBMS3 | RBBP6 | Gene ID | 27303 | 5930 |
| Gene name | RNA binding motif single stranded interacting protein 3 | RB binding protein 6, ubiquitin ligase | |
| Synonyms | - | MY038|P2P-R|PACT|RBQ-1|SNAMA | |
| Cytomap | 3p24.1 | 16p12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | RNA-binding motif, single-stranded-interacting protein 3RNA binding motif, single stranded interacting proteinRNA-binding protein | E3 ubiquitin-protein ligase RBBP6RB-binding Q-protein 1RING-type E3 ubiquitin transferase RBBP6p53-associated cellular protein of testisproliferation potential-related proteinprotein P2P-Rretinoblastoma binding protein 6retinoblastoma-binding Q pro | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000273139, ENST00000383766, ENST00000383767, ENST00000396583, ENST00000434693, ENST00000445033, ENST00000452462, ENST00000456853, ENST00000473799, | ENST00000348022, ENST00000381039, ENST00000452655, ENST00000319715, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 12 X 11 X 6=792 | 6 X 6 X 5=180 |
| # samples | 12 | 6 | |
| ** MAII score | log2(12/792*10)=-2.72246602447109 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/180*10)=-1.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: RBMS3 [Title/Abstract] AND RBBP6 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | RBMS3(29323247)-RBBP6(24564834), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | RBMS3-RBBP6 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. RBMS3-RBBP6 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. RBMS3-RBBP6 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. RBMS3-RBBP6 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | RBBP6 | GO:0006511 | ubiquitin-dependent protein catabolic process | 18851979 |
 Fusion gene breakpoints across RBMS3 (5'-gene) Fusion gene breakpoints across RBMS3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
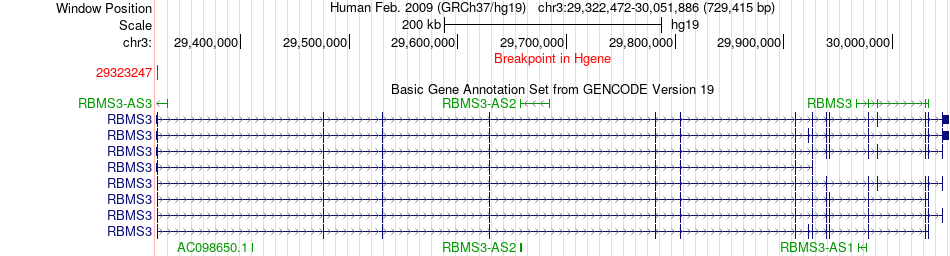 |
 Fusion gene breakpoints across RBBP6 (3'-gene) Fusion gene breakpoints across RBBP6 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
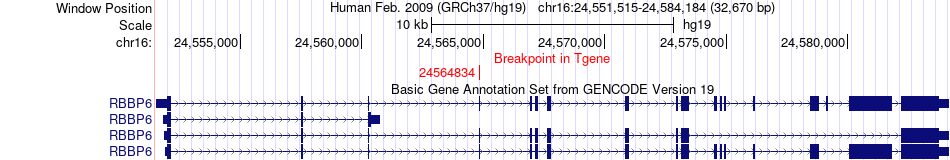 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-FP-8209-01A | RBMS3 | chr3 | 29323247 | + | RBBP6 | chr16 | 24564834 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000434693 | RBMS3 | chr3 | 29323247 | + | ENST00000319715 | RBBP6 | chr16 | 24564834 | + | 6269 | 775 | 352 | 5850 | 1832 |
| ENST00000396583 | RBMS3 | chr3 | 29323247 | + | ENST00000319715 | RBBP6 | chr16 | 24564834 | + | 5939 | 445 | 22 | 5520 | 1832 |
| ENST00000383767 | RBMS3 | chr3 | 29323247 | + | ENST00000319715 | RBBP6 | chr16 | 24564834 | + | 5905 | 411 | 15 | 5486 | 1823 |
| ENST00000445033 | RBMS3 | chr3 | 29323247 | + | ENST00000319715 | RBBP6 | chr16 | 24564834 | + | 5878 | 384 | 117 | 5459 | 1780 |
| ENST00000273139 | RBMS3 | chr3 | 29323247 | + | ENST00000319715 | RBBP6 | chr16 | 24564834 | + | 5799 | 305 | 38 | 5380 | 1780 |
| ENST00000383766 | RBMS3 | chr3 | 29323247 | + | ENST00000319715 | RBBP6 | chr16 | 24564834 | + | 5799 | 305 | 38 | 5380 | 1780 |
| ENST00000452462 | RBMS3 | chr3 | 29323247 | + | ENST00000319715 | RBBP6 | chr16 | 24564834 | + | 5699 | 205 | 130 | 5280 | 1716 |
| ENST00000456853 | RBMS3 | chr3 | 29323247 | + | ENST00000319715 | RBBP6 | chr16 | 24564834 | + | 5635 | 141 | 66 | 5216 | 1716 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000434693 | ENST00000319715 | RBMS3 | chr3 | 29323247 | + | RBBP6 | chr16 | 24564834 | + | 0.000466232 | 0.9995338 |
| ENST00000396583 | ENST00000319715 | RBMS3 | chr3 | 29323247 | + | RBBP6 | chr16 | 24564834 | + | 0.000400572 | 0.9995994 |
| ENST00000383767 | ENST00000319715 | RBMS3 | chr3 | 29323247 | + | RBBP6 | chr16 | 24564834 | + | 0.000392764 | 0.99960726 |
| ENST00000445033 | ENST00000319715 | RBMS3 | chr3 | 29323247 | + | RBBP6 | chr16 | 24564834 | + | 0.000379256 | 0.9996207 |
| ENST00000273139 | ENST00000319715 | RBMS3 | chr3 | 29323247 | + | RBBP6 | chr16 | 24564834 | + | 0.000354368 | 0.9996456 |
| ENST00000383766 | ENST00000319715 | RBMS3 | chr3 | 29323247 | + | RBBP6 | chr16 | 24564834 | + | 0.000354368 | 0.9996456 |
| ENST00000452462 | ENST00000319715 | RBMS3 | chr3 | 29323247 | + | RBBP6 | chr16 | 24564834 | + | 0.000362763 | 0.99963725 |
| ENST00000456853 | ENST00000319715 | RBMS3 | chr3 | 29323247 | + | RBBP6 | chr16 | 24564834 | + | 0.000361078 | 0.9996389 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >73043_73043_1_RBMS3-RBBP6_RBMS3_chr3_29323247_ENST00000273139_RBBP6_chr16_24564834_ENST00000319715_length(amino acids)=1780AA_BP=1711 MVCVFCFVLFFKKILAVLELASGGVSEASSVTGTVRNSGLRGSSAWGTIPCHPVPCLGDKDSSYMGKRLDQPQMYPQYTYYYPHYLQTKI DDSSASISLAQLTKTANLAEANASEEDKIKAMMSQSGHEYDPINYMKKPLGPPPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKST GIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVIPCCGNSYCDE CIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAVNNFKNETGYTKRLRKQLPPPPPPIPPPRPLIQRNLQPLMRSPISRQQDPLMIP VTSSSTHPAPSISSLTSNQSSLAPPVSGNPSSAPAPVPDITATVSISVHSEKSDGPFRDSDNKILPAAALASEHSKGTSSIAITALMEEK GYQVPVLGTPSLLGQSLLHGQLIPTTGPVRINTARPGGGRPGWEHSNKLGYLVSPPQQIRRGERSCYRSINRGRHHSERSQRTQGPSLPA TPVFVPVPPPPLYPPPPHTLPLPPGVPPPQFSPQFPPGQPPPAGYSVPPPGFPPAPANLSTPWVSSGVQTAHSNTIPTTQAPPLSREEFY REQRRLKEEEKKKSKLDEFTNDFAKELMEYKKIQKERRRSFSRSKSPYSGSSYSRSSYTYSKSRSGSTRSRSYSRSFSRSHSRSYSRSPP YPRRGRGKSRNYRSRSRSHGYHRSRSRSPPYRRYHSRSRSPQAFRGQSPNKRNVPQGETEREYFNRYREVPPPYDMKAYYGRSVDFRDPF EKERYREWERKYREWYEKYYKGYAAGAQPRPSANRENFSPERFLPLNIRNSPFTRGRREDYVGGQSHRSRNIGSNYPEKLSARDGHNQKD NTKSKEKESENAPGDGKGNKHKKHRKRRKGEESEGFLNPELLETSRKSREPTGVEENKTDSLFVLPSRDDATPVRDEPMDAESITFKSVS EKDKRERDKPKAKGDKTKRKNDGSAVSKKENIVKPAKGPQEKVDGERERSPRSEPPIKKAKEETPKTDNTKSSSSSQKDEKITGTPRKAH SKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKAKKPNEKNKPLDNKGEKRKRKTEEKGVDKDFESSSMKISKLEVTEIVKPSPKR KMEPDTEKMDRTPEKDKISLSAPAKKIKLNRETGKKIGSTENISNTKEPSEKLESTSSKVKQEKVKGKVRRKVTGTEGSSSTLVDYTSTS STGGSPVRKSEEKTDTKRTVIKTMEEYNNDNTAPAEDVIIMIQVPQSKWDKDDFESEEEDVKSTQPISSVGKPASVIKNVSTKPSNIVKY PEKESEPSEKIQKFTKDVSHEIIQHEVKSSKNSASSEKGKTKDRDYSVLEKENPEKRKNSTQPEKESNLDRLNEQGNFKSLSQSSKEART SDKHDSTRASSNKDFTPNRDKKTDYDTREYSSSKRRDEKNELTRRKDSPSRNKDSASGQKNKPREERDLPKKGTGDSKKSNSSPSRDRKP HDHKATYDTKRPNEETKSVDKNPCKDREKHVLEARNNKESSGNKLLYILNPPETQVEKEQITGQIDKSTVKPKPQLSHSSRLSSDLTRET DEAAFEPDYNESDSESNVSVKEEESSGNISKDLKDKIVEKAKESLDTAAVVQVGISRNQSHSSPSVSPSRSHSPSGSQTRSHSSSASSAE -------------------------------------------------------------- >73043_73043_2_RBMS3-RBBP6_RBMS3_chr3_29323247_ENST00000383766_RBBP6_chr16_24564834_ENST00000319715_length(amino acids)=1780AA_BP=1711 MVCVFCFVLFFKKILAVLELASGGVSEASSVTGTVRNSGLRGSSAWGTIPCHPVPCLGDKDSSYMGKRLDQPQMYPQYTYYYPHYLQTKI DDSSASISLAQLTKTANLAEANASEEDKIKAMMSQSGHEYDPINYMKKPLGPPPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKST GIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVIPCCGNSYCDE CIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAVNNFKNETGYTKRLRKQLPPPPPPIPPPRPLIQRNLQPLMRSPISRQQDPLMIP VTSSSTHPAPSISSLTSNQSSLAPPVSGNPSSAPAPVPDITATVSISVHSEKSDGPFRDSDNKILPAAALASEHSKGTSSIAITALMEEK GYQVPVLGTPSLLGQSLLHGQLIPTTGPVRINTARPGGGRPGWEHSNKLGYLVSPPQQIRRGERSCYRSINRGRHHSERSQRTQGPSLPA TPVFVPVPPPPLYPPPPHTLPLPPGVPPPQFSPQFPPGQPPPAGYSVPPPGFPPAPANLSTPWVSSGVQTAHSNTIPTTQAPPLSREEFY REQRRLKEEEKKKSKLDEFTNDFAKELMEYKKIQKERRRSFSRSKSPYSGSSYSRSSYTYSKSRSGSTRSRSYSRSFSRSHSRSYSRSPP YPRRGRGKSRNYRSRSRSHGYHRSRSRSPPYRRYHSRSRSPQAFRGQSPNKRNVPQGETEREYFNRYREVPPPYDMKAYYGRSVDFRDPF EKERYREWERKYREWYEKYYKGYAAGAQPRPSANRENFSPERFLPLNIRNSPFTRGRREDYVGGQSHRSRNIGSNYPEKLSARDGHNQKD NTKSKEKESENAPGDGKGNKHKKHRKRRKGEESEGFLNPELLETSRKSREPTGVEENKTDSLFVLPSRDDATPVRDEPMDAESITFKSVS EKDKRERDKPKAKGDKTKRKNDGSAVSKKENIVKPAKGPQEKVDGERERSPRSEPPIKKAKEETPKTDNTKSSSSSQKDEKITGTPRKAH SKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKAKKPNEKNKPLDNKGEKRKRKTEEKGVDKDFESSSMKISKLEVTEIVKPSPKR KMEPDTEKMDRTPEKDKISLSAPAKKIKLNRETGKKIGSTENISNTKEPSEKLESTSSKVKQEKVKGKVRRKVTGTEGSSSTLVDYTSTS STGGSPVRKSEEKTDTKRTVIKTMEEYNNDNTAPAEDVIIMIQVPQSKWDKDDFESEEEDVKSTQPISSVGKPASVIKNVSTKPSNIVKY PEKESEPSEKIQKFTKDVSHEIIQHEVKSSKNSASSEKGKTKDRDYSVLEKENPEKRKNSTQPEKESNLDRLNEQGNFKSLSQSSKEART SDKHDSTRASSNKDFTPNRDKKTDYDTREYSSSKRRDEKNELTRRKDSPSRNKDSASGQKNKPREERDLPKKGTGDSKKSNSSPSRDRKP HDHKATYDTKRPNEETKSVDKNPCKDREKHVLEARNNKESSGNKLLYILNPPETQVEKEQITGQIDKSTVKPKPQLSHSSRLSSDLTRET DEAAFEPDYNESDSESNVSVKEEESSGNISKDLKDKIVEKAKESLDTAAVVQVGISRNQSHSSPSVSPSRSHSPSGSQTRSHSSSASSAE -------------------------------------------------------------- >73043_73043_3_RBMS3-RBBP6_RBMS3_chr3_29323247_ENST00000383767_RBBP6_chr16_24564834_ENST00000319715_length(amino acids)=1823AA_BP=1754 MVREQTASSQVDPGSWGKPGKIWEGCVWVFFLQISLLALFFFPLVCVFCFVLFFKKILAVLELASGGVSEASSVTGTVRNSGLRGSSAWG TIPCHPVPCLGDKDSSYMGKRLDQPQMYPQYTYYYPHYLQTKIDDSSASISLAQLTKTANLAEANASEEDKIKAMMSQSGHEYDPINYMK KPLGPPPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAIGKKEKPPFL PEEPSSSSEEDDPIPDELLCLICKDIMTDAVVIPCCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAVNNFKNETGYT KRLRKQLPPPPPPIPPPRPLIQRNLQPLMRSPISRQQDPLMIPVTSSSTHPAPSISSLTSNQSSLAPPVSGNPSSAPAPVPDITATVSIS VHSEKSDGPFRDSDNKILPAAALASEHSKGTSSIAITALMEEKGYQVPVLGTPSLLGQSLLHGQLIPTTGPVRINTARPGGGRPGWEHSN KLGYLVSPPQQIRRGERSCYRSINRGRHHSERSQRTQGPSLPATPVFVPVPPPPLYPPPPHTLPLPPGVPPPQFSPQFPPGQPPPAGYSV PPPGFPPAPANLSTPWVSSGVQTAHSNTIPTTQAPPLSREEFYREQRRLKEEEKKKSKLDEFTNDFAKELMEYKKIQKERRRSFSRSKSP YSGSSYSRSSYTYSKSRSGSTRSRSYSRSFSRSHSRSYSRSPPYPRRGRGKSRNYRSRSRSHGYHRSRSRSPPYRRYHSRSRSPQAFRGQ SPNKRNVPQGETEREYFNRYREVPPPYDMKAYYGRSVDFRDPFEKERYREWERKYREWYEKYYKGYAAGAQPRPSANRENFSPERFLPLN IRNSPFTRGRREDYVGGQSHRSRNIGSNYPEKLSARDGHNQKDNTKSKEKESENAPGDGKGNKHKKHRKRRKGEESEGFLNPELLETSRK SREPTGVEENKTDSLFVLPSRDDATPVRDEPMDAESITFKSVSEKDKRERDKPKAKGDKTKRKNDGSAVSKKENIVKPAKGPQEKVDGER ERSPRSEPPIKKAKEETPKTDNTKSSSSSQKDEKITGTPRKAHSKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKAKKPNEKNKP LDNKGEKRKRKTEEKGVDKDFESSSMKISKLEVTEIVKPSPKRKMEPDTEKMDRTPEKDKISLSAPAKKIKLNRETGKKIGSTENISNTK EPSEKLESTSSKVKQEKVKGKVRRKVTGTEGSSSTLVDYTSTSSTGGSPVRKSEEKTDTKRTVIKTMEEYNNDNTAPAEDVIIMIQVPQS KWDKDDFESEEEDVKSTQPISSVGKPASVIKNVSTKPSNIVKYPEKESEPSEKIQKFTKDVSHEIIQHEVKSSKNSASSEKGKTKDRDYS VLEKENPEKRKNSTQPEKESNLDRLNEQGNFKSLSQSSKEARTSDKHDSTRASSNKDFTPNRDKKTDYDTREYSSSKRRDEKNELTRRKD SPSRNKDSASGQKNKPREERDLPKKGTGDSKKSNSSPSRDRKPHDHKATYDTKRPNEETKSVDKNPCKDREKHVLEARNNKESSGNKLLY ILNPPETQVEKEQITGQIDKSTVKPKPQLSHSSRLSSDLTRETDEAAFEPDYNESDSESNVSVKEEESSGNISKDLKDKIVEKAKESLDT AAVVQVGISRNQSHSSPSVSPSRSHSPSGSQTRSHSSSASSAESQDSKKKKKKKEKKKHKKHKKHKKHKKHAGTEVELEKSQKHKHKKKK -------------------------------------------------------------- >73043_73043_4_RBMS3-RBBP6_RBMS3_chr3_29323247_ENST00000396583_RBBP6_chr16_24564834_ENST00000319715_length(amino acids)=1832AA_BP=1763 MAIDITTDSLVREQTASSQVDPGSWGKPGKIWEGCVWVFFLQISLLALFFFPLVCVFCFVLFFKKILAVLELASGGVSEASSVTGTVRNS GLRGSSAWGTIPCHPVPCLGDKDSSYMGKRLDQPQMYPQYTYYYPHYLQTKIDDSSASISLAQLTKTANLAEANASEEDKIKAMMSQSGH EYDPINYMKKPLGPPPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAI GKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVIPCCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAVN NFKNETGYTKRLRKQLPPPPPPIPPPRPLIQRNLQPLMRSPISRQQDPLMIPVTSSSTHPAPSISSLTSNQSSLAPPVSGNPSSAPAPVP DITATVSISVHSEKSDGPFRDSDNKILPAAALASEHSKGTSSIAITALMEEKGYQVPVLGTPSLLGQSLLHGQLIPTTGPVRINTARPGG GRPGWEHSNKLGYLVSPPQQIRRGERSCYRSINRGRHHSERSQRTQGPSLPATPVFVPVPPPPLYPPPPHTLPLPPGVPPPQFSPQFPPG QPPPAGYSVPPPGFPPAPANLSTPWVSSGVQTAHSNTIPTTQAPPLSREEFYREQRRLKEEEKKKSKLDEFTNDFAKELMEYKKIQKERR RSFSRSKSPYSGSSYSRSSYTYSKSRSGSTRSRSYSRSFSRSHSRSYSRSPPYPRRGRGKSRNYRSRSRSHGYHRSRSRSPPYRRYHSRS RSPQAFRGQSPNKRNVPQGETEREYFNRYREVPPPYDMKAYYGRSVDFRDPFEKERYREWERKYREWYEKYYKGYAAGAQPRPSANRENF SPERFLPLNIRNSPFTRGRREDYVGGQSHRSRNIGSNYPEKLSARDGHNQKDNTKSKEKESENAPGDGKGNKHKKHRKRRKGEESEGFLN PELLETSRKSREPTGVEENKTDSLFVLPSRDDATPVRDEPMDAESITFKSVSEKDKRERDKPKAKGDKTKRKNDGSAVSKKENIVKPAKG PQEKVDGERERSPRSEPPIKKAKEETPKTDNTKSSSSSQKDEKITGTPRKAHSKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKA KKPNEKNKPLDNKGEKRKRKTEEKGVDKDFESSSMKISKLEVTEIVKPSPKRKMEPDTEKMDRTPEKDKISLSAPAKKIKLNRETGKKIG STENISNTKEPSEKLESTSSKVKQEKVKGKVRRKVTGTEGSSSTLVDYTSTSSTGGSPVRKSEEKTDTKRTVIKTMEEYNNDNTAPAEDV IIMIQVPQSKWDKDDFESEEEDVKSTQPISSVGKPASVIKNVSTKPSNIVKYPEKESEPSEKIQKFTKDVSHEIIQHEVKSSKNSASSEK GKTKDRDYSVLEKENPEKRKNSTQPEKESNLDRLNEQGNFKSLSQSSKEARTSDKHDSTRASSNKDFTPNRDKKTDYDTREYSSSKRRDE KNELTRRKDSPSRNKDSASGQKNKPREERDLPKKGTGDSKKSNSSPSRDRKPHDHKATYDTKRPNEETKSVDKNPCKDREKHVLEARNNK ESSGNKLLYILNPPETQVEKEQITGQIDKSTVKPKPQLSHSSRLSSDLTRETDEAAFEPDYNESDSESNVSVKEEESSGNISKDLKDKIV EKAKESLDTAAVVQVGISRNQSHSSPSVSPSRSHSPSGSQTRSHSSSASSAESQDSKKKKKKKEKKKHKKHKKHKKHKKHAGTEVELEKS -------------------------------------------------------------- >73043_73043_5_RBMS3-RBBP6_RBMS3_chr3_29323247_ENST00000434693_RBBP6_chr16_24564834_ENST00000319715_length(amino acids)=1832AA_BP=1763 MAIDITTDSLVREQTASSQVDPGSWGKPGKIWEGCVWVFFLQISLLALFFFPLVCVFCFVLFFKKILAVLELASGGVSEASSVTGTVRNS GLRGSSAWGTIPCHPVPCLGDKDSSYMGKRLDQPQMYPQYTYYYPHYLQTKIDDSSASISLAQLTKTANLAEANASEEDKIKAMMSQSGH EYDPINYMKKPLGPPPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAI GKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVIPCCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAVN NFKNETGYTKRLRKQLPPPPPPIPPPRPLIQRNLQPLMRSPISRQQDPLMIPVTSSSTHPAPSISSLTSNQSSLAPPVSGNPSSAPAPVP DITATVSISVHSEKSDGPFRDSDNKILPAAALASEHSKGTSSIAITALMEEKGYQVPVLGTPSLLGQSLLHGQLIPTTGPVRINTARPGG GRPGWEHSNKLGYLVSPPQQIRRGERSCYRSINRGRHHSERSQRTQGPSLPATPVFVPVPPPPLYPPPPHTLPLPPGVPPPQFSPQFPPG QPPPAGYSVPPPGFPPAPANLSTPWVSSGVQTAHSNTIPTTQAPPLSREEFYREQRRLKEEEKKKSKLDEFTNDFAKELMEYKKIQKERR RSFSRSKSPYSGSSYSRSSYTYSKSRSGSTRSRSYSRSFSRSHSRSYSRSPPYPRRGRGKSRNYRSRSRSHGYHRSRSRSPPYRRYHSRS RSPQAFRGQSPNKRNVPQGETEREYFNRYREVPPPYDMKAYYGRSVDFRDPFEKERYREWERKYREWYEKYYKGYAAGAQPRPSANRENF SPERFLPLNIRNSPFTRGRREDYVGGQSHRSRNIGSNYPEKLSARDGHNQKDNTKSKEKESENAPGDGKGNKHKKHRKRRKGEESEGFLN PELLETSRKSREPTGVEENKTDSLFVLPSRDDATPVRDEPMDAESITFKSVSEKDKRERDKPKAKGDKTKRKNDGSAVSKKENIVKPAKG PQEKVDGERERSPRSEPPIKKAKEETPKTDNTKSSSSSQKDEKITGTPRKAHSKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKA KKPNEKNKPLDNKGEKRKRKTEEKGVDKDFESSSMKISKLEVTEIVKPSPKRKMEPDTEKMDRTPEKDKISLSAPAKKIKLNRETGKKIG STENISNTKEPSEKLESTSSKVKQEKVKGKVRRKVTGTEGSSSTLVDYTSTSSTGGSPVRKSEEKTDTKRTVIKTMEEYNNDNTAPAEDV IIMIQVPQSKWDKDDFESEEEDVKSTQPISSVGKPASVIKNVSTKPSNIVKYPEKESEPSEKIQKFTKDVSHEIIQHEVKSSKNSASSEK GKTKDRDYSVLEKENPEKRKNSTQPEKESNLDRLNEQGNFKSLSQSSKEARTSDKHDSTRASSNKDFTPNRDKKTDYDTREYSSSKRRDE KNELTRRKDSPSRNKDSASGQKNKPREERDLPKKGTGDSKKSNSSPSRDRKPHDHKATYDTKRPNEETKSVDKNPCKDREKHVLEARNNK ESSGNKLLYILNPPETQVEKEQITGQIDKSTVKPKPQLSHSSRLSSDLTRETDEAAFEPDYNESDSESNVSVKEEESSGNISKDLKDKIV EKAKESLDTAAVVQVGISRNQSHSSPSVSPSRSHSPSGSQTRSHSSSASSAESQDSKKKKKKKEKKKHKKHKKHKKHKKHAGTEVELEKS -------------------------------------------------------------- >73043_73043_6_RBMS3-RBBP6_RBMS3_chr3_29323247_ENST00000445033_RBBP6_chr16_24564834_ENST00000319715_length(amino acids)=1780AA_BP=1711 MVCVFCFVLFFKKILAVLELASGGVSEASSVTGTVRNSGLRGSSAWGTIPCHPVPCLGDKDSSYMGKRLDQPQMYPQYTYYYPHYLQTKI DDSSASISLAQLTKTANLAEANASEEDKIKAMMSQSGHEYDPINYMKKPLGPPPPSYTCFRCGKPGHYIKNCPTNGDKNFESGPRIKKST GIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDELLCLICKDIMTDAVVIPCCGNSYCDE CIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAVNNFKNETGYTKRLRKQLPPPPPPIPPPRPLIQRNLQPLMRSPISRQQDPLMIP VTSSSTHPAPSISSLTSNQSSLAPPVSGNPSSAPAPVPDITATVSISVHSEKSDGPFRDSDNKILPAAALASEHSKGTSSIAITALMEEK GYQVPVLGTPSLLGQSLLHGQLIPTTGPVRINTARPGGGRPGWEHSNKLGYLVSPPQQIRRGERSCYRSINRGRHHSERSQRTQGPSLPA TPVFVPVPPPPLYPPPPHTLPLPPGVPPPQFSPQFPPGQPPPAGYSVPPPGFPPAPANLSTPWVSSGVQTAHSNTIPTTQAPPLSREEFY REQRRLKEEEKKKSKLDEFTNDFAKELMEYKKIQKERRRSFSRSKSPYSGSSYSRSSYTYSKSRSGSTRSRSYSRSFSRSHSRSYSRSPP YPRRGRGKSRNYRSRSRSHGYHRSRSRSPPYRRYHSRSRSPQAFRGQSPNKRNVPQGETEREYFNRYREVPPPYDMKAYYGRSVDFRDPF EKERYREWERKYREWYEKYYKGYAAGAQPRPSANRENFSPERFLPLNIRNSPFTRGRREDYVGGQSHRSRNIGSNYPEKLSARDGHNQKD NTKSKEKESENAPGDGKGNKHKKHRKRRKGEESEGFLNPELLETSRKSREPTGVEENKTDSLFVLPSRDDATPVRDEPMDAESITFKSVS EKDKRERDKPKAKGDKTKRKNDGSAVSKKENIVKPAKGPQEKVDGERERSPRSEPPIKKAKEETPKTDNTKSSSSSQKDEKITGTPRKAH SKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKAKKPNEKNKPLDNKGEKRKRKTEEKGVDKDFESSSMKISKLEVTEIVKPSPKR KMEPDTEKMDRTPEKDKISLSAPAKKIKLNRETGKKIGSTENISNTKEPSEKLESTSSKVKQEKVKGKVRRKVTGTEGSSSTLVDYTSTS STGGSPVRKSEEKTDTKRTVIKTMEEYNNDNTAPAEDVIIMIQVPQSKWDKDDFESEEEDVKSTQPISSVGKPASVIKNVSTKPSNIVKY PEKESEPSEKIQKFTKDVSHEIIQHEVKSSKNSASSEKGKTKDRDYSVLEKENPEKRKNSTQPEKESNLDRLNEQGNFKSLSQSSKEART SDKHDSTRASSNKDFTPNRDKKTDYDTREYSSSKRRDEKNELTRRKDSPSRNKDSASGQKNKPREERDLPKKGTGDSKKSNSSPSRDRKP HDHKATYDTKRPNEETKSVDKNPCKDREKHVLEARNNKESSGNKLLYILNPPETQVEKEQITGQIDKSTVKPKPQLSHSSRLSSDLTRET DEAAFEPDYNESDSESNVSVKEEESSGNISKDLKDKIVEKAKESLDTAAVVQVGISRNQSHSSPSVSPSRSHSPSGSQTRSHSSSASSAE -------------------------------------------------------------- >73043_73043_7_RBMS3-RBBP6_RBMS3_chr3_29323247_ENST00000452462_RBBP6_chr16_24564834_ENST00000319715_length(amino acids)=1716AA_BP=1647 MGKRLDQPQMYPQYTYYYPHYLQTKIDDSSASISLAQLTKTANLAEANASEEDKIKAMMSQSGHEYDPINYMKKPLGPPPPSYTCFRCGK PGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDE LLCLICKDIMTDAVVIPCCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAVNNFKNETGYTKRLRKQLPPPPPPIPPP RPLIQRNLQPLMRSPISRQQDPLMIPVTSSSTHPAPSISSLTSNQSSLAPPVSGNPSSAPAPVPDITATVSISVHSEKSDGPFRDSDNKI LPAAALASEHSKGTSSIAITALMEEKGYQVPVLGTPSLLGQSLLHGQLIPTTGPVRINTARPGGGRPGWEHSNKLGYLVSPPQQIRRGER SCYRSINRGRHHSERSQRTQGPSLPATPVFVPVPPPPLYPPPPHTLPLPPGVPPPQFSPQFPPGQPPPAGYSVPPPGFPPAPANLSTPWV SSGVQTAHSNTIPTTQAPPLSREEFYREQRRLKEEEKKKSKLDEFTNDFAKELMEYKKIQKERRRSFSRSKSPYSGSSYSRSSYTYSKSR SGSTRSRSYSRSFSRSHSRSYSRSPPYPRRGRGKSRNYRSRSRSHGYHRSRSRSPPYRRYHSRSRSPQAFRGQSPNKRNVPQGETEREYF NRYREVPPPYDMKAYYGRSVDFRDPFEKERYREWERKYREWYEKYYKGYAAGAQPRPSANRENFSPERFLPLNIRNSPFTRGRREDYVGG QSHRSRNIGSNYPEKLSARDGHNQKDNTKSKEKESENAPGDGKGNKHKKHRKRRKGEESEGFLNPELLETSRKSREPTGVEENKTDSLFV LPSRDDATPVRDEPMDAESITFKSVSEKDKRERDKPKAKGDKTKRKNDGSAVSKKENIVKPAKGPQEKVDGERERSPRSEPPIKKAKEET PKTDNTKSSSSSQKDEKITGTPRKAHSKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKAKKPNEKNKPLDNKGEKRKRKTEEKGV DKDFESSSMKISKLEVTEIVKPSPKRKMEPDTEKMDRTPEKDKISLSAPAKKIKLNRETGKKIGSTENISNTKEPSEKLESTSSKVKQEK VKGKVRRKVTGTEGSSSTLVDYTSTSSTGGSPVRKSEEKTDTKRTVIKTMEEYNNDNTAPAEDVIIMIQVPQSKWDKDDFESEEEDVKST QPISSVGKPASVIKNVSTKPSNIVKYPEKESEPSEKIQKFTKDVSHEIIQHEVKSSKNSASSEKGKTKDRDYSVLEKENPEKRKNSTQPE KESNLDRLNEQGNFKSLSQSSKEARTSDKHDSTRASSNKDFTPNRDKKTDYDTREYSSSKRRDEKNELTRRKDSPSRNKDSASGQKNKPR EERDLPKKGTGDSKKSNSSPSRDRKPHDHKATYDTKRPNEETKSVDKNPCKDREKHVLEARNNKESSGNKLLYILNPPETQVEKEQITGQ IDKSTVKPKPQLSHSSRLSSDLTRETDEAAFEPDYNESDSESNVSVKEEESSGNISKDLKDKIVEKAKESLDTAAVVQVGISRNQSHSSP SVSPSRSHSPSGSQTRSHSSSASSAESQDSKKKKKKKEKKKHKKHKKHKKHKKHAGTEVELEKSQKHKHKKKKSKKNKDKEKEKEKDDQK -------------------------------------------------------------- >73043_73043_8_RBMS3-RBBP6_RBMS3_chr3_29323247_ENST00000456853_RBBP6_chr16_24564834_ENST00000319715_length(amino acids)=1716AA_BP=1647 MGKRLDQPQMYPQYTYYYPHYLQTKIDDSSASISLAQLTKTANLAEANASEEDKIKAMMSQSGHEYDPINYMKKPLGPPPPSYTCFRCGK PGHYIKNCPTNGDKNFESGPRIKKSTGIPRSFMMEVKDPNMKGAMLTNTGKYAIPTIDAEAYAIGKKEKPPFLPEEPSSSSEEDDPIPDE LLCLICKDIMTDAVVIPCCGNSYCDECIRTALLESDEHTCPTCHQNDVSPDALIANKFLRQAVNNFKNETGYTKRLRKQLPPPPPPIPPP RPLIQRNLQPLMRSPISRQQDPLMIPVTSSSTHPAPSISSLTSNQSSLAPPVSGNPSSAPAPVPDITATVSISVHSEKSDGPFRDSDNKI LPAAALASEHSKGTSSIAITALMEEKGYQVPVLGTPSLLGQSLLHGQLIPTTGPVRINTARPGGGRPGWEHSNKLGYLVSPPQQIRRGER SCYRSINRGRHHSERSQRTQGPSLPATPVFVPVPPPPLYPPPPHTLPLPPGVPPPQFSPQFPPGQPPPAGYSVPPPGFPPAPANLSTPWV SSGVQTAHSNTIPTTQAPPLSREEFYREQRRLKEEEKKKSKLDEFTNDFAKELMEYKKIQKERRRSFSRSKSPYSGSSYSRSSYTYSKSR SGSTRSRSYSRSFSRSHSRSYSRSPPYPRRGRGKSRNYRSRSRSHGYHRSRSRSPPYRRYHSRSRSPQAFRGQSPNKRNVPQGETEREYF NRYREVPPPYDMKAYYGRSVDFRDPFEKERYREWERKYREWYEKYYKGYAAGAQPRPSANRENFSPERFLPLNIRNSPFTRGRREDYVGG QSHRSRNIGSNYPEKLSARDGHNQKDNTKSKEKESENAPGDGKGNKHKKHRKRRKGEESEGFLNPELLETSRKSREPTGVEENKTDSLFV LPSRDDATPVRDEPMDAESITFKSVSEKDKRERDKPKAKGDKTKRKNDGSAVSKKENIVKPAKGPQEKVDGERERSPRSEPPIKKAKEET PKTDNTKSSSSSQKDEKITGTPRKAHSKSAKEHQETKPVKEEKVKKDYSKDVKSEKLTTKEEKAKKPNEKNKPLDNKGEKRKRKTEEKGV DKDFESSSMKISKLEVTEIVKPSPKRKMEPDTEKMDRTPEKDKISLSAPAKKIKLNRETGKKIGSTENISNTKEPSEKLESTSSKVKQEK VKGKVRRKVTGTEGSSSTLVDYTSTSSTGGSPVRKSEEKTDTKRTVIKTMEEYNNDNTAPAEDVIIMIQVPQSKWDKDDFESEEEDVKST QPISSVGKPASVIKNVSTKPSNIVKYPEKESEPSEKIQKFTKDVSHEIIQHEVKSSKNSASSEKGKTKDRDYSVLEKENPEKRKNSTQPE KESNLDRLNEQGNFKSLSQSSKEARTSDKHDSTRASSNKDFTPNRDKKTDYDTREYSSSKRRDEKNELTRRKDSPSRNKDSASGQKNKPR EERDLPKKGTGDSKKSNSSPSRDRKPHDHKATYDTKRPNEETKSVDKNPCKDREKHVLEARNNKESSGNKLLYILNPPETQVEKEQITGQ IDKSTVKPKPQLSHSSRLSSDLTRETDEAAFEPDYNESDSESNVSVKEEESSGNISKDLKDKIVEKAKESLDTAAVVQVGISRNQSHSSP SVSPSRSHSPSGSQTRSHSSSASSAESQDSKKKKKKKEKKKHKKHKKHKKHKKHAGTEVELEKSQKHKHKKKKSKKNKDKEKEKEKDDQK -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:29323247/chr16:24564834) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000452655 | 0 | 3 | 4_76 | 0 | 119.0 | Domain | DWNN | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000319715 | 2 | 18 | 560_565 | 101.0 | 1793.0 | Motif | PPxPxY | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000348022 | 2 | 17 | 560_565 | 101.0 | 1759.0 | Motif | PPxPxY | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000381039 | 2 | 11 | 560_565 | 101.0 | 953.0 | Motif | PPxPxY | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000452655 | 0 | 3 | 560_565 | 0 | 119.0 | Motif | PPxPxY | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000319715 | 2 | 18 | 159_176 | 101.0 | 1793.0 | Zinc finger | CCHC-type | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000319715 | 2 | 18 | 259_300 | 101.0 | 1793.0 | Zinc finger | RING-type%3B degenerate | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000348022 | 2 | 17 | 159_176 | 101.0 | 1759.0 | Zinc finger | CCHC-type | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000348022 | 2 | 17 | 259_300 | 101.0 | 1759.0 | Zinc finger | RING-type%3B degenerate | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000381039 | 2 | 11 | 159_176 | 101.0 | 953.0 | Zinc finger | CCHC-type | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000381039 | 2 | 11 | 259_300 | 101.0 | 953.0 | Zinc finger | RING-type%3B degenerate | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000452655 | 0 | 3 | 159_176 | 0 | 119.0 | Zinc finger | CCHC-type | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000452655 | 0 | 3 | 259_300 | 0 | 119.0 | Zinc finger | RING-type%3B degenerate |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RBMS3 | chr3:29323247 | chr16:24564834 | ENST00000273139 | + | 1 | 13 | 140_225 | 25.0 | 421.0 | Domain | RRM 2 |
| Hgene | RBMS3 | chr3:29323247 | chr16:24564834 | ENST00000273139 | + | 1 | 13 | 61_134 | 25.0 | 421.0 | Domain | RRM 1 |
| Hgene | RBMS3 | chr3:29323247 | chr16:24564834 | ENST00000383766 | + | 1 | 14 | 140_225 | 25.0 | 420.0 | Domain | RRM 2 |
| Hgene | RBMS3 | chr3:29323247 | chr16:24564834 | ENST00000383766 | + | 1 | 14 | 61_134 | 25.0 | 420.0 | Domain | RRM 1 |
| Hgene | RBMS3 | chr3:29323247 | chr16:24564834 | ENST00000383767 | + | 1 | 15 | 140_225 | 25.0 | 438.0 | Domain | RRM 2 |
| Hgene | RBMS3 | chr3:29323247 | chr16:24564834 | ENST00000383767 | + | 1 | 15 | 61_134 | 25.0 | 438.0 | Domain | RRM 1 |
| Hgene | RBMS3 | chr3:29323247 | chr16:24564834 | ENST00000452462 | + | 1 | 14 | 140_225 | 25.0 | 422.0 | Domain | RRM 2 |
| Hgene | RBMS3 | chr3:29323247 | chr16:24564834 | ENST00000452462 | + | 1 | 14 | 61_134 | 25.0 | 422.0 | Domain | RRM 1 |
| Hgene | RBMS3 | chr3:29323247 | chr16:24564834 | ENST00000456853 | + | 1 | 14 | 140_225 | 25.0 | 434.0 | Domain | RRM 2 |
| Hgene | RBMS3 | chr3:29323247 | chr16:24564834 | ENST00000456853 | + | 1 | 14 | 61_134 | 25.0 | 434.0 | Domain | RRM 1 |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000319715 | 2 | 18 | 4_76 | 101.0 | 1793.0 | Domain | DWNN | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000348022 | 2 | 17 | 4_76 | 101.0 | 1759.0 | Domain | DWNN | |
| Tgene | RBBP6 | chr3:29323247 | chr16:24564834 | ENST00000381039 | 2 | 11 | 4_76 | 101.0 | 953.0 | Domain | DWNN |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| RBMS3 | |
| RBBP6 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to RBMS3-RBBP6 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to RBMS3-RBBP6 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

