| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:RERE-CYP4A11 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: RERE-CYP4A11 | FusionPDB ID: 73492 | FusionGDB2.0 ID: 73492 | Hgene | Tgene | Gene symbol | RERE | CYP4A11 | Gene ID | 473 | 1579 |
| Gene name | arginine-glutamic acid dipeptide repeats | cytochrome P450 family 4 subfamily A member 11 | |
| Synonyms | ARG|ARP|ATN1L|DNB1|NEDBEH | CP4Y|CYP4A2|CYP4AII|CYPIVA11 | |
| Cytomap | 1p36.23 | 1p33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | arginine-glutamic acid dipeptide repeats proteinarginine-glutamic acid dipeptide (RE) repeatsatrophin 2atrophin-1 like proteinatrophin-1 related protein | cytochrome P450 4A1120-HETE synthase20-hydroxyeicosatetraenoic acid synthaseP450HL-omegaalkane-1 monooxygenasecytochrome P-450HK-omegacytochrome P450, family 4, subfamily A, polypeptide 11cytochrome P450, subfamily IVA, polypeptide 11cytochrome P4 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q02928 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000337907, ENST00000377464, ENST00000400907, ENST00000400908, ENST00000460659, ENST00000476556, | ENST00000457840, ENST00000496519, ENST00000310638, ENST00000371904, ENST00000371905, ENST00000462347, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 27 X 28 X 11=8316 | 1 X 1 X 1=1 |
| # samples | 43 | 1 | |
| ** MAII score | log2(43/8316*10)=-4.27348119326891 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(1/1*10)=3.32192809488736 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: RERE [Title/Abstract] AND CYP4A11 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | RERE(8525985)-CYP4A11(47403809), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | RERE-CYP4A11 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. RERE-CYP4A11 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. RERE-CYP4A11 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. RERE-CYP4A11 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. RERE-CYP4A11 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CYP4A11 | GO:0001676 | long-chain fatty acid metabolic process | 18433732 |
| Tgene | CYP4A11 | GO:0006691 | leukotriene metabolic process | 9799565 |
| Tgene | CYP4A11 | GO:0019369 | arachidonic acid metabolic process | 10660572 |
| Tgene | CYP4A11 | GO:0019373 | epoxygenase P450 pathway | 9618440 |
| Tgene | CYP4A11 | GO:0055114 | oxidation-reduction process | 9618440|9799565|18433732 |
 Fusion gene breakpoints across RERE (5'-gene) Fusion gene breakpoints across RERE (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
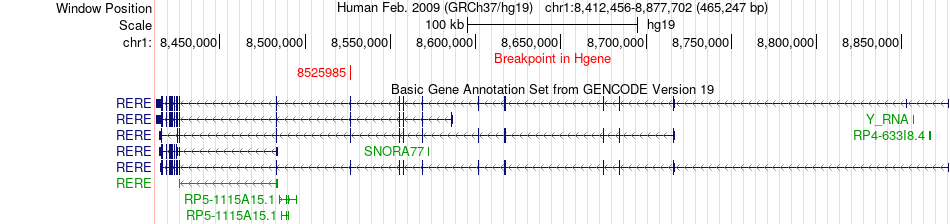 |
 Fusion gene breakpoints across CYP4A11 (3'-gene) Fusion gene breakpoints across CYP4A11 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
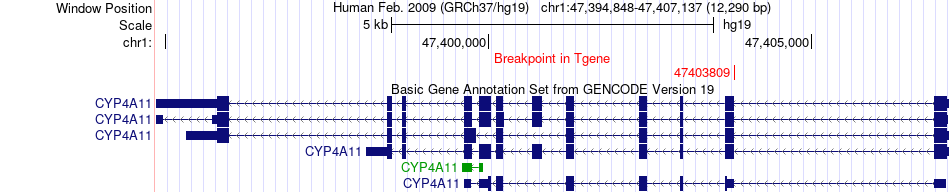 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-D8-A1XZ-01A | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000337907 | RERE | chr1 | 8525985 | - | ENST00000310638 | CYP4A11 | chr1 | 47403809 | - | 4141 | 1838 | 533 | 3202 | 889 |
| ENST00000337907 | RERE | chr1 | 8525985 | - | ENST00000371904 | CYP4A11 | chr1 | 47403809 | - | 3380 | 1838 | 533 | 3205 | 890 |
| ENST00000337907 | RERE | chr1 | 8525985 | - | ENST00000462347 | CYP4A11 | chr1 | 47403809 | - | 3387 | 1838 | 533 | 2908 | 791 |
| ENST00000337907 | RERE | chr1 | 8525985 | - | ENST00000371905 | CYP4A11 | chr1 | 47403809 | - | 3337 | 1838 | 533 | 3010 | 825 |
| ENST00000377464 | RERE | chr1 | 8525985 | - | ENST00000310638 | CYP4A11 | chr1 | 47403809 | - | 2857 | 554 | 155 | 1918 | 587 |
| ENST00000377464 | RERE | chr1 | 8525985 | - | ENST00000371904 | CYP4A11 | chr1 | 47403809 | - | 2096 | 554 | 155 | 1921 | 588 |
| ENST00000377464 | RERE | chr1 | 8525985 | - | ENST00000462347 | CYP4A11 | chr1 | 47403809 | - | 2103 | 554 | 155 | 1624 | 489 |
| ENST00000377464 | RERE | chr1 | 8525985 | - | ENST00000371905 | CYP4A11 | chr1 | 47403809 | - | 2053 | 554 | 155 | 1726 | 523 |
| ENST00000400907 | RERE | chr1 | 8525985 | - | ENST00000310638 | CYP4A11 | chr1 | 47403809 | - | 3580 | 1277 | 74 | 2641 | 855 |
| ENST00000400907 | RERE | chr1 | 8525985 | - | ENST00000371904 | CYP4A11 | chr1 | 47403809 | - | 2819 | 1277 | 74 | 2644 | 856 |
| ENST00000400907 | RERE | chr1 | 8525985 | - | ENST00000462347 | CYP4A11 | chr1 | 47403809 | - | 2826 | 1277 | 74 | 2347 | 757 |
| ENST00000400907 | RERE | chr1 | 8525985 | - | ENST00000371905 | CYP4A11 | chr1 | 47403809 | - | 2776 | 1277 | 74 | 2449 | 791 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000337907 | ENST00000310638 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.001417378 | 0.9985826 |
| ENST00000337907 | ENST00000371904 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.002346561 | 0.9976534 |
| ENST00000337907 | ENST00000462347 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.003002701 | 0.99699736 |
| ENST00000337907 | ENST00000371905 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.001988069 | 0.99801195 |
| ENST00000377464 | ENST00000310638 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.010778805 | 0.9892212 |
| ENST00000377464 | ENST00000371904 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.012364891 | 0.9876351 |
| ENST00000377464 | ENST00000462347 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.033593964 | 0.966406 |
| ENST00000377464 | ENST00000371905 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.011682247 | 0.9883178 |
| ENST00000400907 | ENST00000310638 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.001610611 | 0.99838936 |
| ENST00000400907 | ENST00000371904 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.00280792 | 0.9971921 |
| ENST00000400907 | ENST00000462347 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.00355919 | 0.9964408 |
| ENST00000400907 | ENST00000371905 | RERE | chr1 | 8525985 | - | CYP4A11 | chr1 | 47403809 | - | 0.002334131 | 0.9976659 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >73492_73492_1_RERE-CYP4A11_RERE_chr1_8525985_ENST00000337907_CYP4A11_chr1_47403809_ENST00000310638_length(amino acids)=889AA_BP=824 MELSEKFRRLKPRLTVYLSRRPSRERTEEGTWRIMTADKDKDKDKEKDRDRDRDREREKRDKARESENSRPRRSCTLEGGAKNYAESDHS EDEDNDNNSATAEESTKKNKKKPPKKKSRYERTDTGEITSYITEDDVVYRPGDCVYIESRRPNTPYFICSIQDFKLVHNSQACCRSPTPA LCDPPACSLPVASQPPQHLSEAGRGPVGSKRDHLLMNVKWYYRQSEVPDSVYQHLVQDRHNENDSGRELVITDPVIKNRELFISDYVDTY HAAALRGKCNISHFSDIFAAREFKARVDSFFYILGYNPETRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCD LLMYLRAARSMAAFAGMCDGGSTEDGCVAASRDDTTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVE TFPSACPHWLWGGKVRVQLYDPDYMKVILGRSDPKSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVML DKWEELLGQDSPLEVFQHVSLMTLDTIMKCAFSHQGSIQVDRNSQSYIQAISDLNNLVFSRVRNAFHQNDTIYSLTSAGRWTHRACQLAH QHTDQVIQLRKAQLQKEGELEKIKRKRHLDFLDILLLAKMENGSILSDKDLRAEVDTFMFEGHDTTASGISWILYALATHPKHQERCREE IHSLLGDGASITWNHLDQMPYTTMCIKEALRLYPPVPGIGRELSTPVTFPDGRSLPKGIMVLLSIYGLHHNPKVWPNPEVFDPFRFAPGS -------------------------------------------------------------- >73492_73492_2_RERE-CYP4A11_RERE_chr1_8525985_ENST00000337907_CYP4A11_chr1_47403809_ENST00000371904_length(amino acids)=890AA_BP=825 MELSEKFRRLKPRLTVYLSRRPSRERTEEGTWRIMTADKDKDKDKEKDRDRDRDREREKRDKARESENSRPRRSCTLEGGAKNYAESDHS EDEDNDNNSATAEESTKKNKKKPPKKKSRYERTDTGEITSYITEDDVVYRPGDCVYIESRRPNTPYFICSIQDFKLVHNSQACCRSPTPA LCDPPACSLPVASQPPQHLSEAGRGPVGSKRDHLLMNVKWYYRQSEVPDSVYQHLVQDRHNENDSGRELVITDPVIKNRELFISDYVDTY HAAALRGKCNISHFSDIFAAREFKARVDSFFYILGYNPETRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCD LLMYLRAARSMAAFAGMCDGGSTEDGCVAASRDDTTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVE TFPSACPHWLWGGKVRVQLYDPDYMKVILGRSDPKSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVML DKWEELLGQDSPLEVFQHVSLMTLDTIMKCAFSHQGSIQVDRNSQSYIQAISDLNNLVFSRVRNAFHQNDTIYSLTSAGRWTHRACQLAH QHTGDQVIQLRKAQLQKEGELEKIKRKRHLDFLDILLLAKMENGSILSDKDLRAEVDTFMFEGHDTTASGISWILYALATHPKHQERCRE EIHSLLGDGASITWNHLDQMPYTTMCIKEALRLYPPVPGIGRELSTPVTFPDGRSLPKGIMVLLSIYGLHHNPKVWPNPEVFDPFRFAPG -------------------------------------------------------------- >73492_73492_3_RERE-CYP4A11_RERE_chr1_8525985_ENST00000337907_CYP4A11_chr1_47403809_ENST00000371905_length(amino acids)=825AA_BP=435 MELSEKFRRLKPRLTVYLSRRPSRERTEEGTWRIMTADKDKDKDKEKDRDRDRDREREKRDKARESENSRPRRSCTLEGGAKNYAESDHS EDEDNDNNSATAEESTKKNKKKPPKKKSRYERTDTGEITSYITEDDVVYRPGDCVYIESRRPNTPYFICSIQDFKLVHNSQACCRSPTPA LCDPPACSLPVASQPPQHLSEAGRGPVGSKRDHLLMNVKWYYRQSEVPDSVYQHLVQDRHNENDSGRELVITDPVIKNRELFISDYVDTY HAAALRGKCNISHFSDIFAAREFKARVDSFFYILGYNPETRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCD LLMYLRAARSMAAFAGMCDGGSTEDGCVAASRDDTTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVE TFPSACPHWLWGGKVRVQLYDPDYMKVILGRSDPKSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVML DKWEELLGQDSPLEVFQHVSLMTLDTIMKCAFSHQGSIQVDRNSQSYIQAISDLNNLVFSRVRNAFHQNDTIYSLTSAGRWTHRACQLAH QHTDQVIQLRKAQLQKEGELEKIKRKRHLDFLDILLLAKMENGSILSDKDLRAEVDTFMFEGHDTTASGISWILYALATHPKHQERCREE IHSLLGDGASITWNHLDQMPYTTMCIKEALRLYPPVPGIGRELSTPVTFPDGRSLPKGIMVLLSIYGLHHNPKVWPNPEVFDPFRFAPGS -------------------------------------------------------------- >73492_73492_4_RERE-CYP4A11_RERE_chr1_8525985_ENST00000337907_CYP4A11_chr1_47403809_ENST00000462347_length(amino acids)=791AA_BP=726 MELSEKFRRLKPRLTVYLSRRPSRERTEEGTWRIMTADKDKDKDKEKDRDRDRDREREKRDKARESENSRPRRSCTLEGGAKNYAESDHS EDEDNDNNSATAEESTKKNKKKPPKKKSRYERTDTGEITSYITEDDVVYRPGDCVYIESRRPNTPYFICSIQDFKLVHNSQACCRSPTPA LCDPPACSLPVASQPPQHLSEAGRGPVGSKRDHLLMNVKWYYRQSEVPDSVYQHLVQDRHNENDSGRELVITDPVIKNRELFISDYVDTY HAAALRGKCNISHFSDIFAAREFKARVDSFFYILGYNPETRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCD LLMYLRAARSMAAFAGMCDGGSTEDGCVAASRDDTTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVE TFPSACPHWLWGGKVRVQLYDPDYMKVILGRSDPKSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVML DKWEELLGQDSPLEVFQHVSLMTLDTIMKCAFSHQGSIQVDRPSDPTEEGSTTEGGGAGEDQEEEAFGFSGYPPLGQRAPGLPRPCWCSG WNCFRNHLDQMPYTTMCIKEALRLYPPVPGIGRELSTPVTFPDGRSLPKGIMVLLSIYGLHHNPKVWPNPEVFDPFRFAPGSAQHSHAFL -------------------------------------------------------------- >73492_73492_5_RERE-CYP4A11_RERE_chr1_8525985_ENST00000377464_CYP4A11_chr1_47403809_ENST00000310638_length(amino acids)=587AA_BP=522 MDDPFSPCRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCDLLMYLRAARSMAAFAGMCDGGSTEDGCVAASR DDTTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVETFPSACPHWLWGGKVRVQLYDPDYMKVILGRS DPKSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVMLDKWEELLGQDSPLEVFQHVSLMTLDTIMKCAF SHQGSIQVDRNSQSYIQAISDLNNLVFSRVRNAFHQNDTIYSLTSAGRWTHRACQLAHQHTDQVIQLRKAQLQKEGELEKIKRKRHLDFL DILLLAKMENGSILSDKDLRAEVDTFMFEGHDTTASGISWILYALATHPKHQERCREEIHSLLGDGASITWNHLDQMPYTTMCIKEALRL YPPVPGIGRELSTPVTFPDGRSLPKGIMVLLSIYGLHHNPKVWPNPEVFDPFRFAPGSAQHSHAFLPFSGGSRNCIGKQFAMNELKVATA -------------------------------------------------------------- >73492_73492_6_RERE-CYP4A11_RERE_chr1_8525985_ENST00000377464_CYP4A11_chr1_47403809_ENST00000371904_length(amino acids)=588AA_BP=523 MDDPFSPCRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCDLLMYLRAARSMAAFAGMCDGGSTEDGCVAASR DDTTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVETFPSACPHWLWGGKVRVQLYDPDYMKVILGRS DPKSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVMLDKWEELLGQDSPLEVFQHVSLMTLDTIMKCAF SHQGSIQVDRNSQSYIQAISDLNNLVFSRVRNAFHQNDTIYSLTSAGRWTHRACQLAHQHTGDQVIQLRKAQLQKEGELEKIKRKRHLDF LDILLLAKMENGSILSDKDLRAEVDTFMFEGHDTTASGISWILYALATHPKHQERCREEIHSLLGDGASITWNHLDQMPYTTMCIKEALR LYPPVPGIGRELSTPVTFPDGRSLPKGIMVLLSIYGLHHNPKVWPNPEVFDPFRFAPGSAQHSHAFLPFSGGSRNCIGKQFAMNELKVAT -------------------------------------------------------------- >73492_73492_7_RERE-CYP4A11_RERE_chr1_8525985_ENST00000377464_CYP4A11_chr1_47403809_ENST00000371905_length(amino acids)=523AA_BP=133 MDDPFSPCRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCDLLMYLRAARSMAAFAGMCDGGSTEDGCVAASR DDTTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVETFPSACPHWLWGGKVRVQLYDPDYMKVILGRS DPKSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVMLDKWEELLGQDSPLEVFQHVSLMTLDTIMKCAF SHQGSIQVDRNSQSYIQAISDLNNLVFSRVRNAFHQNDTIYSLTSAGRWTHRACQLAHQHTDQVIQLRKAQLQKEGELEKIKRKRHLDFL DILLLAKMENGSILSDKDLRAEVDTFMFEGHDTTASGISWILYALATHPKHQERCREEIHSLLGDGASITWNHLDQMPYTTMCIKEALRL -------------------------------------------------------------- >73492_73492_8_RERE-CYP4A11_RERE_chr1_8525985_ENST00000377464_CYP4A11_chr1_47403809_ENST00000462347_length(amino acids)=489AA_BP=424 MDDPFSPCRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCDLLMYLRAARSMAAFAGMCDGGSTEDGCVAASR DDTTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVETFPSACPHWLWGGKVRVQLYDPDYMKVILGRS DPKSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVMLDKWEELLGQDSPLEVFQHVSLMTLDTIMKCAF SHQGSIQVDRPSDPTEEGSTTEGGGAGEDQEEEAFGFSGYPPLGQRAPGLPRPCWCSGWNCFRNHLDQMPYTTMCIKEALRLYPPVPGIG RELSTPVTFPDGRSLPKGIMVLLSIYGLHHNPKVWPNPEVFDPFRFAPGSAQHSHAFLPFSGGSRNCIGKQFAMNELKVATALTLLRFEL -------------------------------------------------------------- >73492_73492_9_RERE-CYP4A11_RERE_chr1_8525985_ENST00000400907_CYP4A11_chr1_47403809_ENST00000310638_length(amino acids)=855AA_BP=790 MTADKDKDKDKEKDRDRDRDREREKRDKARESENSRPRRSCTLEGGAKNYAESDHSEDEDNDNNSATAEESTKKNKKKPPKKKSRYERTD TGEITSYITEDDVVYRPGDCVYIESRRPNTPYFICSIQDFKLVHNSQACCRSPTPALCDPPACSLPVASQPPQHLSEAGRGPVGSKRDHL LMNVKWYYRQSEVPDSVYQHLVQDRHNENDSGRELVITDPVIKNRELFISDYVDTYHAAALRGKCNISHFSDIFAAREFKARVDSFFYIL GYNPETRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCDLLMYLRAARSMAAFAGMCDGGSTEDGCVAASRDD TTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVETFPSACPHWLWGGKVRVQLYDPDYMKVILGRSDP KSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVMLDKWEELLGQDSPLEVFQHVSLMTLDTIMKCAFSH QGSIQVDRNSQSYIQAISDLNNLVFSRVRNAFHQNDTIYSLTSAGRWTHRACQLAHQHTDQVIQLRKAQLQKEGELEKIKRKRHLDFLDI LLLAKMENGSILSDKDLRAEVDTFMFEGHDTTASGISWILYALATHPKHQERCREEIHSLLGDGASITWNHLDQMPYTTMCIKEALRLYP PVPGIGRELSTPVTFPDGRSLPKGIMVLLSIYGLHHNPKVWPNPEVFDPFRFAPGSAQHSHAFLPFSGGSRNCIGKQFAMNELKVATALT -------------------------------------------------------------- >73492_73492_10_RERE-CYP4A11_RERE_chr1_8525985_ENST00000400907_CYP4A11_chr1_47403809_ENST00000371904_length(amino acids)=856AA_BP=791 MTADKDKDKDKEKDRDRDRDREREKRDKARESENSRPRRSCTLEGGAKNYAESDHSEDEDNDNNSATAEESTKKNKKKPPKKKSRYERTD TGEITSYITEDDVVYRPGDCVYIESRRPNTPYFICSIQDFKLVHNSQACCRSPTPALCDPPACSLPVASQPPQHLSEAGRGPVGSKRDHL LMNVKWYYRQSEVPDSVYQHLVQDRHNENDSGRELVITDPVIKNRELFISDYVDTYHAAALRGKCNISHFSDIFAAREFKARVDSFFYIL GYNPETRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCDLLMYLRAARSMAAFAGMCDGGSTEDGCVAASRDD TTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVETFPSACPHWLWGGKVRVQLYDPDYMKVILGRSDP KSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVMLDKWEELLGQDSPLEVFQHVSLMTLDTIMKCAFSH QGSIQVDRNSQSYIQAISDLNNLVFSRVRNAFHQNDTIYSLTSAGRWTHRACQLAHQHTGDQVIQLRKAQLQKEGELEKIKRKRHLDFLD ILLLAKMENGSILSDKDLRAEVDTFMFEGHDTTASGISWILYALATHPKHQERCREEIHSLLGDGASITWNHLDQMPYTTMCIKEALRLY PPVPGIGRELSTPVTFPDGRSLPKGIMVLLSIYGLHHNPKVWPNPEVFDPFRFAPGSAQHSHAFLPFSGGSRNCIGKQFAMNELKVATAL -------------------------------------------------------------- >73492_73492_11_RERE-CYP4A11_RERE_chr1_8525985_ENST00000400907_CYP4A11_chr1_47403809_ENST00000371905_length(amino acids)=791AA_BP=401 MTADKDKDKDKEKDRDRDRDREREKRDKARESENSRPRRSCTLEGGAKNYAESDHSEDEDNDNNSATAEESTKKNKKKPPKKKSRYERTD TGEITSYITEDDVVYRPGDCVYIESRRPNTPYFICSIQDFKLVHNSQACCRSPTPALCDPPACSLPVASQPPQHLSEAGRGPVGSKRDHL LMNVKWYYRQSEVPDSVYQHLVQDRHNENDSGRELVITDPVIKNRELFISDYVDTYHAAALRGKCNISHFSDIFAAREFKARVDSFFYIL GYNPETRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCDLLMYLRAARSMAAFAGMCDGGSTEDGCVAASRDD TTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVETFPSACPHWLWGGKVRVQLYDPDYMKVILGRSDP KSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVMLDKWEELLGQDSPLEVFQHVSLMTLDTIMKCAFSH QGSIQVDRNSQSYIQAISDLNNLVFSRVRNAFHQNDTIYSLTSAGRWTHRACQLAHQHTDQVIQLRKAQLQKEGELEKIKRKRHLDFLDI LLLAKMENGSILSDKDLRAEVDTFMFEGHDTTASGISWILYALATHPKHQERCREEIHSLLGDGASITWNHLDQMPYTTMCIKEALRLYP -------------------------------------------------------------- >73492_73492_12_RERE-CYP4A11_RERE_chr1_8525985_ENST00000400907_CYP4A11_chr1_47403809_ENST00000462347_length(amino acids)=757AA_BP=692 MTADKDKDKDKEKDRDRDRDREREKRDKARESENSRPRRSCTLEGGAKNYAESDHSEDEDNDNNSATAEESTKKNKKKPPKKKSRYERTD TGEITSYITEDDVVYRPGDCVYIESRRPNTPYFICSIQDFKLVHNSQACCRSPTPALCDPPACSLPVASQPPQHLSEAGRGPVGSKRDHL LMNVKWYYRQSEVPDSVYQHLVQDRHNENDSGRELVITDPVIKNRELFISDYVDTYHAAALRGKCNISHFSDIFAAREFKARVDSFFYIL GYNPETRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCDLLMYLRAARSMAAFAGMCDGGSTEDGCVAASRDD TTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVLQQDQELQRIQKWVETFPSACPHWLWGGKVRVQLYDPDYMKVILGRSDP KSHGSYRFLAPWIGYGLLLLNGQTWFQHRRMLTPAFHYDILKPYVGLMADSVRVMLDKWEELLGQDSPLEVFQHVSLMTLDTIMKCAFSH QGSIQVDRPSDPTEEGSTTEGGGAGEDQEEEAFGFSGYPPLGQRAPGLPRPCWCSGWNCFRNHLDQMPYTTMCIKEALRLYPPVPGIGRE LSTPVTFPDGRSLPKGIMVLLSIYGLHHNPKVWPNPEVFDPFRFAPGSAQHSHAFLPFSGGSRNCIGKQFAMNELKVATALTLLRFELLP -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:8525985/chr1:47403809) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | CYP4A11 |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | FUNCTION: A cytochrome P450 monooxygenase involved in the metabolism of fatty acids and their oxygenated derivatives (oxylipins) (PubMed:7679927, PubMed:1739747, PubMed:8914854, PubMed:10553002, PubMed:10660572, PubMed:15611369). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase) (PubMed:7679927, PubMed:1739747, PubMed:8914854, PubMed:10553002, PubMed:10660572, PubMed:15611369). Catalyzes predominantly the oxidation of the terminal carbon (omega-oxidation) of saturated and unsaturated fatty acids, the catalytic efficiency decreasing in the following order: dodecanoic > tetradecanoic > (9Z)-octadecenoic > (9Z,12Z)-octadecadienoic > hexadecanoic acid (PubMed:10553002, PubMed:10660572). Acts as a major omega-hydroxylase for dodecanoic (lauric) acid in liver (PubMed:7679927, PubMed:1739747, PubMed:8914854, PubMed:15611369). Participates in omega-hydroxylation of (5Z,8Z,11Z,14Z)-eicosatetraenoic acid (arachidonate) to 20-hydroxyeicosatetraenoic acid (20-HETE), a signaling molecule acting both as vasoconstrictive and natriuretic with overall effect on arterial blood pressure (PubMed:10620324, PubMed:10660572, PubMed:15611369). Can also catalyze the oxidation of the penultimate carbon (omega-1 oxidation) of fatty acids with lower efficiency (PubMed:7679927). May contribute to the degradation of saturated very long-chain fatty acids (VLCFAs) such as docosanoic acid, by catalyzing successive omega-oxidations to the corresponding dicarboxylic acid, thereby initiating chain shortening (PubMed:18182499). Omega-hydroxylates (9R,10S)-epoxy-octadecanoate stereoisomer (PubMed:15145985). Plays a minor role in omega-oxidation of long-chain 3-hydroxy fatty acids (PubMed:18065749). Has little activity toward prostaglandins A1 and E1 (PubMed:7679927). {ECO:0000269|PubMed:10553002, ECO:0000269|PubMed:10620324, ECO:0000269|PubMed:10660572, ECO:0000269|PubMed:15145985, ECO:0000269|PubMed:15611369, ECO:0000269|PubMed:1739747, ECO:0000269|PubMed:18065749, ECO:0000269|PubMed:18182499, ECO:0000269|PubMed:7679927, ECO:0000269|PubMed:8914854}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000337907 | - | 12 | 24 | 103_283 | 401.0 | 1567.0 | Domain | BAH |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000337907 | - | 12 | 24 | 284_387 | 401.0 | 1567.0 | Domain | ELM2 |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000400908 | - | 11 | 23 | 103_283 | 401.0 | 1567.0 | Domain | BAH |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000400908 | - | 11 | 23 | 284_387 | 401.0 | 1567.0 | Domain | ELM2 |
| Tgene | CYP4A11 | chr1:8525985 | chr1:47403809 | ENST00000310638 | 0 | 12 | 131_134 | 65.0 | 520.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | CYP4A11 | chr1:8525985 | chr1:47403809 | ENST00000371905 | 0 | 11 | 131_134 | 65.0 | 456.0 | Compositional bias | Note=Poly-Leu |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000337907 | - | 12 | 24 | 1156_1211 | 401.0 | 1567.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000400908 | - | 11 | 23 | 1156_1211 | 401.0 | 1567.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000476556 | - | 1 | 13 | 1156_1211 | 0 | 1013.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000337907 | - | 12 | 24 | 1179_1204 | 401.0 | 1567.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000337907 | - | 12 | 24 | 1300_1322 | 401.0 | 1567.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000337907 | - | 12 | 24 | 1425_1445 | 401.0 | 1567.0 | Compositional bias | Note=His-rich |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000337907 | - | 12 | 24 | 1451_1510 | 401.0 | 1567.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000337907 | - | 12 | 24 | 738_1118 | 401.0 | 1567.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000400908 | - | 11 | 23 | 1179_1204 | 401.0 | 1567.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000400908 | - | 11 | 23 | 1300_1322 | 401.0 | 1567.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000400908 | - | 11 | 23 | 1425_1445 | 401.0 | 1567.0 | Compositional bias | Note=His-rich |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000400908 | - | 11 | 23 | 1451_1510 | 401.0 | 1567.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000400908 | - | 11 | 23 | 738_1118 | 401.0 | 1567.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000476556 | - | 1 | 13 | 1179_1204 | 0 | 1013.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000476556 | - | 1 | 13 | 1300_1322 | 0 | 1013.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000476556 | - | 1 | 13 | 1425_1445 | 0 | 1013.0 | Compositional bias | Note=His-rich |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000476556 | - | 1 | 13 | 1451_1510 | 0 | 1013.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000476556 | - | 1 | 13 | 738_1118 | 0 | 1013.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000337907 | - | 12 | 24 | 391_443 | 401.0 | 1567.0 | Domain | SANT |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000400908 | - | 11 | 23 | 391_443 | 401.0 | 1567.0 | Domain | SANT |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000476556 | - | 1 | 13 | 103_283 | 0 | 1013.0 | Domain | BAH |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000476556 | - | 1 | 13 | 284_387 | 0 | 1013.0 | Domain | ELM2 |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000476556 | - | 1 | 13 | 391_443 | 0 | 1013.0 | Domain | SANT |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000337907 | - | 12 | 24 | 507_532 | 401.0 | 1567.0 | Zinc finger | Note=GATA-type |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000400908 | - | 11 | 23 | 507_532 | 401.0 | 1567.0 | Zinc finger | Note=GATA-type |
| Hgene | RERE | chr1:8525985 | chr1:47403809 | ENST00000476556 | - | 1 | 13 | 507_532 | 0 | 1013.0 | Zinc finger | Note=GATA-type |
| Tgene | CYP4A11 | chr1:8525985 | chr1:47403809 | ENST00000310638 | 0 | 12 | 24_31 | 65.0 | 520.0 | Compositional bias | Note=Poly-Leu | |
| Tgene | CYP4A11 | chr1:8525985 | chr1:47403809 | ENST00000371905 | 0 | 11 | 24_31 | 65.0 | 456.0 | Compositional bias | Note=Poly-Leu |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| RERE | |
| CYP4A11 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to RERE-CYP4A11 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to RERE-CYP4A11 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

