| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:RET-NCOA4 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: RET-NCOA4 | FusionPDB ID: 73564 | FusionGDB2.0 ID: 73564 | Hgene | Tgene | Gene symbol | RET | NCOA4 | Gene ID | 5979 | 8031 |
| Gene name | ret proto-oncogene | nuclear receptor coactivator 4 | |
| Synonyms | CDHF12|CDHR16|HSCR1|MEN2A|MEN2B|MTC1|PTC|RET-ELE1 | ARA70|ELE1|PTC3|RFG | |
| Cytomap | 10q11.21 | 10q11.22 | |
| Type of gene | protein-coding | protein-coding | |
| Description | proto-oncogene tyrosine-protein kinase receptor RetRET receptor tyrosine kinasecadherin family member 12cadherin-related family member 16proto-oncogene c-Retrearranged during transfectionret proto-oncogene (multiple endocrine neoplasia and medullary | nuclear receptor coactivator 470 kDa AR-activator70 kDa androgen receptor coactivatorNCoA-4RET-activating gene ELE1androgen receptor-associated protein of 70 kDaret fused | |
| Modification date | 20200322 | 20200313 | |
| UniProtAcc | RTL1 | Q13772 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000340058, ENST00000355710, | ENST00000344348, ENST00000374082, ENST00000374087, ENST00000414907, ENST00000430396, ENST00000443446, ENST00000498586, ENST00000438493, ENST00000452682, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 9 X 6 X 4=216 | 10 X 11 X 3=330 |
| # samples | 9 | 12 | |
| ** MAII score | log2(9/216*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(12/330*10)=-1.4594316186373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: RET [Title/Abstract] AND NCOA4 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | NCOA4(51582939)-RET(43612032), # samples:12 RET(43610184)-NCOA4(51584616), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | NCOA4-RET seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. NCOA4-RET seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. RET-NCOA4 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. RET-NCOA4 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. RET-NCOA4 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. RET-NCOA4 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NCOA4-RET seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. NCOA4-RET seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. NCOA4-RET seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. NCOA4-RET seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. NCOA4-RET seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. NCOA4-RET seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | RET | GO:0030155 | regulation of cell adhesion | 21357690 |
| Hgene | RET | GO:0030335 | positive regulation of cell migration | 20702524 |
| Hgene | RET | GO:0033619 | membrane protein proteolysis | 21357690 |
| Hgene | RET | GO:0033630 | positive regulation of cell adhesion mediated by integrin | 20702524 |
| Hgene | RET | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway | 28953886 |
| Hgene | RET | GO:0043410 | positive regulation of MAPK cascade | 28846099 |
| Tgene | NCOA4 | GO:0006622 | protein targeting to lysosome | 25327288 |
 Fusion gene breakpoints across RET (5'-gene) Fusion gene breakpoints across RET (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
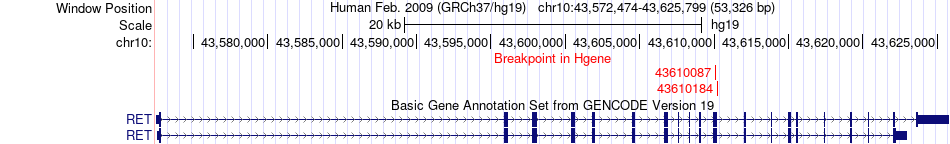 |
 Fusion gene breakpoints across NCOA4 (3'-gene) Fusion gene breakpoints across NCOA4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
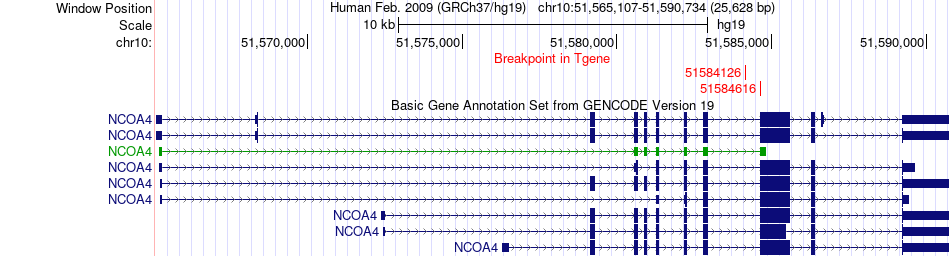 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | THCA | TCGA-CE-A482-01A | RET | chr10 | 43610184 | - | NCOA4 | chr10 | 51584616 | + |
| ChimerDB4 | THCA | TCGA-CE-A482-01A | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51584616 | - |
| ChimerDB4 | THCA | TCGA-CE-A482-01A | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51584616 | + |
| ChimerKB3 | . | . | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51580879 | + |
| ChimerKB3 | . | . | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51582795 | + |
| ChimerKB3 | . | . | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51584615 | + |
| ChiTaRS5.0 | N/A | S83049 | RET | chr10 | 43610087 | + | NCOA4 | chr10 | 51584126 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000355710 | RET | chr10 | 43610184 | + | ENST00000438493 | NCOA4 | chr10 | 51580879 | + | 5432 | 2368 | 232 | 3930 | 1232 |
| ENST00000355710 | RET | chr10 | 43610184 | + | ENST00000452682 | NCOA4 | chr10 | 51580879 | + | 5521 | 2368 | 232 | 3990 | 1252 |
| ENST00000340058 | RET | chr10 | 43610184 | + | ENST00000438493 | NCOA4 | chr10 | 51580879 | + | 5380 | 2316 | 180 | 3878 | 1232 |
| ENST00000340058 | RET | chr10 | 43610184 | + | ENST00000452682 | NCOA4 | chr10 | 51580879 | + | 5469 | 2316 | 180 | 3938 | 1252 |
| ENST00000355710 | RET | chr10 | 43610184 | + | ENST00000438493 | NCOA4 | chr10 | 51582795 | + | 5144 | 2368 | 232 | 3642 | 1136 |
| ENST00000355710 | RET | chr10 | 43610184 | + | ENST00000452682 | NCOA4 | chr10 | 51582795 | + | 5233 | 2368 | 232 | 3702 | 1156 |
| ENST00000340058 | RET | chr10 | 43610184 | + | ENST00000438493 | NCOA4 | chr10 | 51582795 | + | 5092 | 2316 | 180 | 3590 | 1136 |
| ENST00000340058 | RET | chr10 | 43610184 | + | ENST00000452682 | NCOA4 | chr10 | 51582795 | + | 5181 | 2316 | 180 | 3650 | 1156 |
| ENST00000355710 | RET | chr10 | 43610184 | + | ENST00000438493 | NCOA4 | chr10 | 51584615 | + | 5000 | 2368 | 232 | 3498 | 1088 |
| ENST00000355710 | RET | chr10 | 43610184 | + | ENST00000452682 | NCOA4 | chr10 | 51584615 | + | 5089 | 2368 | 232 | 3558 | 1108 |
| ENST00000340058 | RET | chr10 | 43610184 | + | ENST00000438493 | NCOA4 | chr10 | 51584615 | + | 4948 | 2316 | 180 | 3446 | 1088 |
| ENST00000340058 | RET | chr10 | 43610184 | + | ENST00000452682 | NCOA4 | chr10 | 51584615 | + | 5037 | 2316 | 180 | 3506 | 1108 |
| ENST00000355710 | RET | chr10 | 43610184 | + | ENST00000438493 | NCOA4 | chr10 | 51584616 | - | 5000 | 2368 | 232 | 3498 | 1088 |
| ENST00000355710 | RET | chr10 | 43610184 | + | ENST00000452682 | NCOA4 | chr10 | 51584616 | - | 5089 | 2368 | 232 | 3558 | 1108 |
| ENST00000340058 | RET | chr10 | 43610184 | + | ENST00000438493 | NCOA4 | chr10 | 51584616 | - | 4948 | 2316 | 180 | 3446 | 1088 |
| ENST00000340058 | RET | chr10 | 43610184 | + | ENST00000452682 | NCOA4 | chr10 | 51584616 | - | 5037 | 2316 | 180 | 3506 | 1108 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000355710 | ENST00000438493 | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51584616 | - | 0.001067746 | 0.9989323 |
| ENST00000355710 | ENST00000452682 | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51584616 | - | 0.00056626 | 0.9994337 |
| ENST00000340058 | ENST00000438493 | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51584616 | - | 0.000985496 | 0.9990145 |
| ENST00000340058 | ENST00000452682 | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51584616 | - | 0.000527557 | 0.99947244 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >73564_73564_1_RET-NCOA4_RET_chr10_43610184_ENST00000340058_NCOA4_chr10_51580879_ENST00000438493_length(amino acids)=1232AA_BP=0 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILLLGQFNCL THQLECTQNKDLANQVSVCLERLGSLTLKPEDSTVLLFEADTITLRQTITTFGSLKTIQIPEHLMAHASSANIGPFLEKRGCISMPEQKS ASGIVAVPFSEWLLGSKPASGYQAPYIPSTDPQDWLTQKQTLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFS IEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDH LEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLC PRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQL -------------------------------------------------------------- >73564_73564_2_RET-NCOA4_RET_chr10_43610184_ENST00000340058_NCOA4_chr10_51580879_ENST00000452682_length(amino acids)=1252AA_BP=0 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILLLGQFNCL THQLECTQNKDLANQVSVCLERLGSLTLKPEDSTVLLFEADTITLRQTITTFGSLKTIQIPEHLMAHASSANIGPFLEKRGCISMPEQKS ASGIVAVPFSEWLLGSKPASGYQAPYIPSTDPQDWLTQKQTLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFS IEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDH LEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLC PRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQL -------------------------------------------------------------- >73564_73564_3_RET-NCOA4_RET_chr10_43610184_ENST00000340058_NCOA4_chr10_51582795_ENST00000438493_length(amino acids)=1136AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILKSASGIVA VPFSEWLLGSKPASGYQAPYIPSTDPQDWLTQKQTLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKV GDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKP LSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVI EQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDK -------------------------------------------------------------- >73564_73564_4_RET-NCOA4_RET_chr10_43610184_ENST00000340058_NCOA4_chr10_51582795_ENST00000452682_length(amino acids)=1156AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILKSASGIVA VPFSEWLLGSKPASGYQAPYIPSTDPQDWLTQKQTLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKV GDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKP LSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVI EQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDK -------------------------------------------------------------- >73564_73564_5_RET-NCOA4_RET_chr10_43610184_ENST00000340058_NCOA4_chr10_51584615_ENST00000438493_length(amino acids)=1088AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILTSSRACNF FNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYN VNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEA LYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTED RAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEHNFPPDHYGLPAVCDLFACMQLKVDKEKW -------------------------------------------------------------- >73564_73564_6_RET-NCOA4_RET_chr10_43610184_ENST00000340058_NCOA4_chr10_51584615_ENST00000452682_length(amino acids)=1108AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILTSSRACNF FNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYN VNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEA LYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTED RAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEHNFPPDHYGLPAVCDLFACMQLKVDKEKW -------------------------------------------------------------- >73564_73564_7_RET-NCOA4_RET_chr10_43610184_ENST00000340058_NCOA4_chr10_51584616_ENST00000438493_length(amino acids)=1088AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILTSSRACNF FNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYN VNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEA LYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTED RAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEHNFPPDHYGLPAVCDLFACMQLKVDKEKW -------------------------------------------------------------- >73564_73564_8_RET-NCOA4_RET_chr10_43610184_ENST00000340058_NCOA4_chr10_51584616_ENST00000452682_length(amino acids)=1108AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILTSSRACNF FNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYN VNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEA LYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTED RAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEHNFPPDHYGLPAVCDLFACMQLKVDKEKW -------------------------------------------------------------- >73564_73564_9_RET-NCOA4_RET_chr10_43610184_ENST00000355710_NCOA4_chr10_51580879_ENST00000438493_length(amino acids)=1232AA_BP=0 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILLLGQFNCL THQLECTQNKDLANQVSVCLERLGSLTLKPEDSTVLLFEADTITLRQTITTFGSLKTIQIPEHLMAHASSANIGPFLEKRGCISMPEQKS ASGIVAVPFSEWLLGSKPASGYQAPYIPSTDPQDWLTQKQTLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFS IEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDH LEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLC PRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQL -------------------------------------------------------------- >73564_73564_10_RET-NCOA4_RET_chr10_43610184_ENST00000355710_NCOA4_chr10_51580879_ENST00000452682_length(amino acids)=1252AA_BP=0 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILLLGQFNCL THQLECTQNKDLANQVSVCLERLGSLTLKPEDSTVLLFEADTITLRQTITTFGSLKTIQIPEHLMAHASSANIGPFLEKRGCISMPEQKS ASGIVAVPFSEWLLGSKPASGYQAPYIPSTDPQDWLTQKQTLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFS IEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDH LEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLC PRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQL -------------------------------------------------------------- >73564_73564_11_RET-NCOA4_RET_chr10_43610184_ENST00000355710_NCOA4_chr10_51582795_ENST00000438493_length(amino acids)=1136AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILKSASGIVA VPFSEWLLGSKPASGYQAPYIPSTDPQDWLTQKQTLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKV GDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKP LSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVI EQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDK -------------------------------------------------------------- >73564_73564_12_RET-NCOA4_RET_chr10_43610184_ENST00000355710_NCOA4_chr10_51582795_ENST00000452682_length(amino acids)=1156AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILKSASGIVA VPFSEWLLGSKPASGYQAPYIPSTDPQDWLTQKQTLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKV GDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKP LSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVI EQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDK -------------------------------------------------------------- >73564_73564_13_RET-NCOA4_RET_chr10_43610184_ENST00000355710_NCOA4_chr10_51584615_ENST00000438493_length(amino acids)=1088AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILTSSRACNF FNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYN VNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEA LYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTED RAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEHNFPPDHYGLPAVCDLFACMQLKVDKEKW -------------------------------------------------------------- >73564_73564_14_RET-NCOA4_RET_chr10_43610184_ENST00000355710_NCOA4_chr10_51584615_ENST00000452682_length(amino acids)=1108AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILTSSRACNF FNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYN VNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEA LYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTED RAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEHNFPPDHYGLPAVCDLFACMQLKVDKEKW -------------------------------------------------------------- >73564_73564_15_RET-NCOA4_RET_chr10_43610184_ENST00000355710_NCOA4_chr10_51584616_ENST00000438493_length(amino acids)=1088AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILTSSRACNF FNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYN VNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEA LYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTED RAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEHNFPPDHYGLPAVCDLFACMQLKVDKEKW -------------------------------------------------------------- >73564_73564_16_RET-NCOA4_RET_chr10_43610184_ENST00000355710_NCOA4_chr10_51584616_ENST00000452682_length(amino acids)=1108AA_BP=712 MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQE DTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENR PPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAP TFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNR NLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGR CEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLC DELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILTSSRACNF FNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLFQSYN VNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEA LYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTED RAGKQKFKSPMNTSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEHNFPPDHYGLPAVCDLFACMQLKVDKEKW -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:51582939/chr10:43612032) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| RET | NCOA4 |
| 1358 | FUNCTION: Enhances the androgen receptor transcriptional activity in prostate cancer cells. Ligand-independent coactivator of the peroxisome proliferator-activated receptor (PPAR) gamma. {ECO:0000269|PubMed:10347167}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000340058 | + | 11 | 19 | 168_272 | 712.0 | 1073.0 | Domain | Cadherin |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000355710 | + | 11 | 20 | 168_272 | 712.0 | 1115.0 | Domain | Cadherin |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000340058 | + | 11 | 19 | 29_635 | 712.0 | 1073.0 | Topological domain | Extracellular |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000355710 | + | 11 | 20 | 29_635 | 712.0 | 1115.0 | Topological domain | Extracellular |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000340058 | + | 11 | 19 | 636_657 | 712.0 | 1073.0 | Transmembrane | Helical |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000355710 | + | 11 | 20 | 636_657 | 712.0 | 1115.0 | Transmembrane | Helical |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000340058 | + | 11 | 19 | 724_1016 | 712.0 | 1073.0 | Domain | Protein kinase |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000355710 | + | 11 | 20 | 724_1016 | 712.0 | 1115.0 | Domain | Protein kinase |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000340058 | + | 11 | 19 | 730_738 | 712.0 | 1073.0 | Nucleotide binding | ATP |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000355710 | + | 11 | 20 | 730_738 | 712.0 | 1115.0 | Nucleotide binding | ATP |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000340058 | + | 11 | 19 | 805_807 | 712.0 | 1073.0 | Region | Note=Inhibitors binding |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000355710 | + | 11 | 20 | 805_807 | 712.0 | 1115.0 | Region | Note=Inhibitors binding |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000340058 | + | 11 | 19 | 658_1114 | 712.0 | 1073.0 | Topological domain | Cytoplasmic |
| Hgene | RET | chr10:43610184 | chr10:51584616 | ENST00000355710 | + | 11 | 20 | 658_1114 | 712.0 | 1115.0 | Topological domain | Cytoplasmic |
Top |
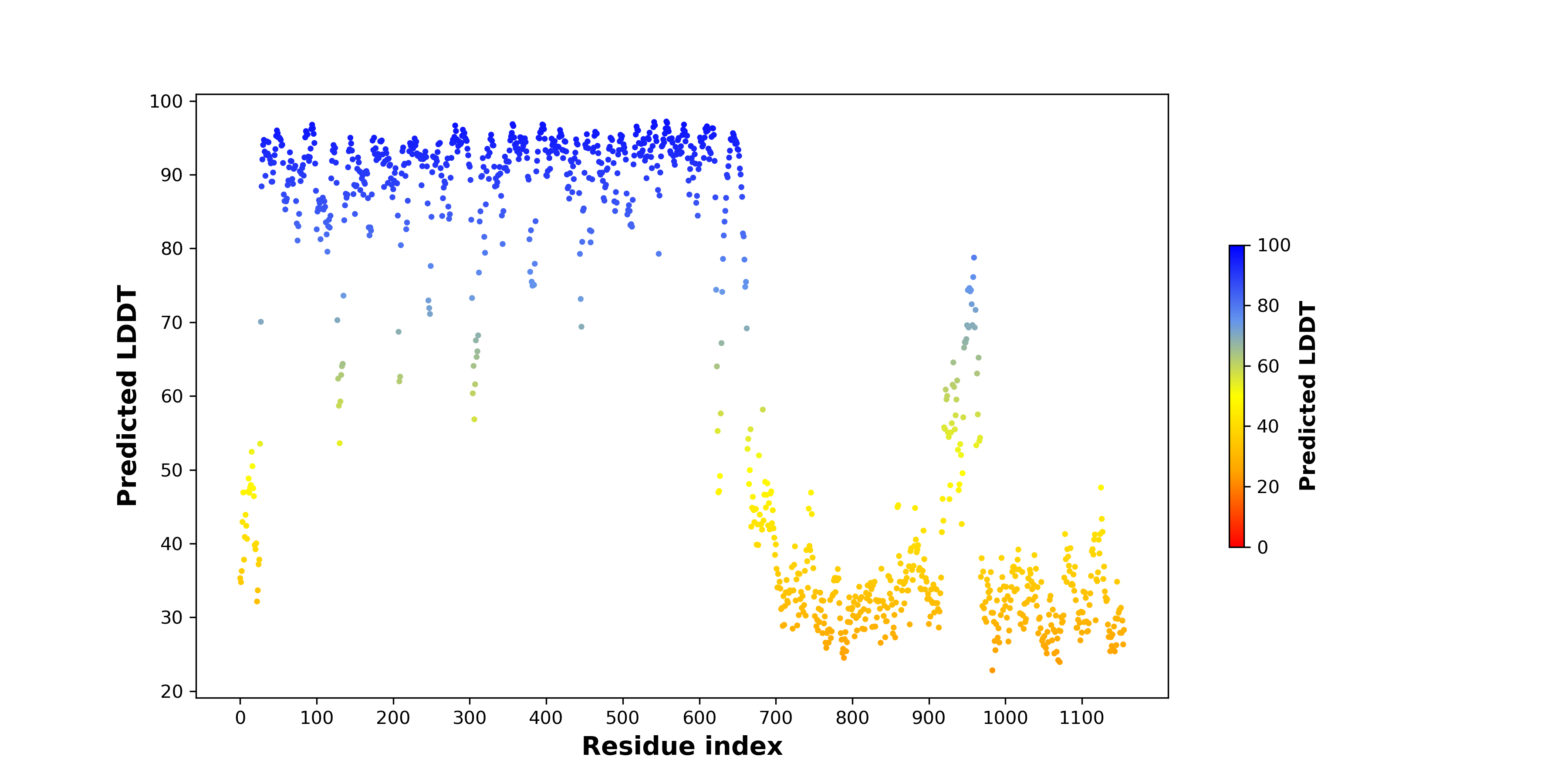
Fusion Protein Structures |
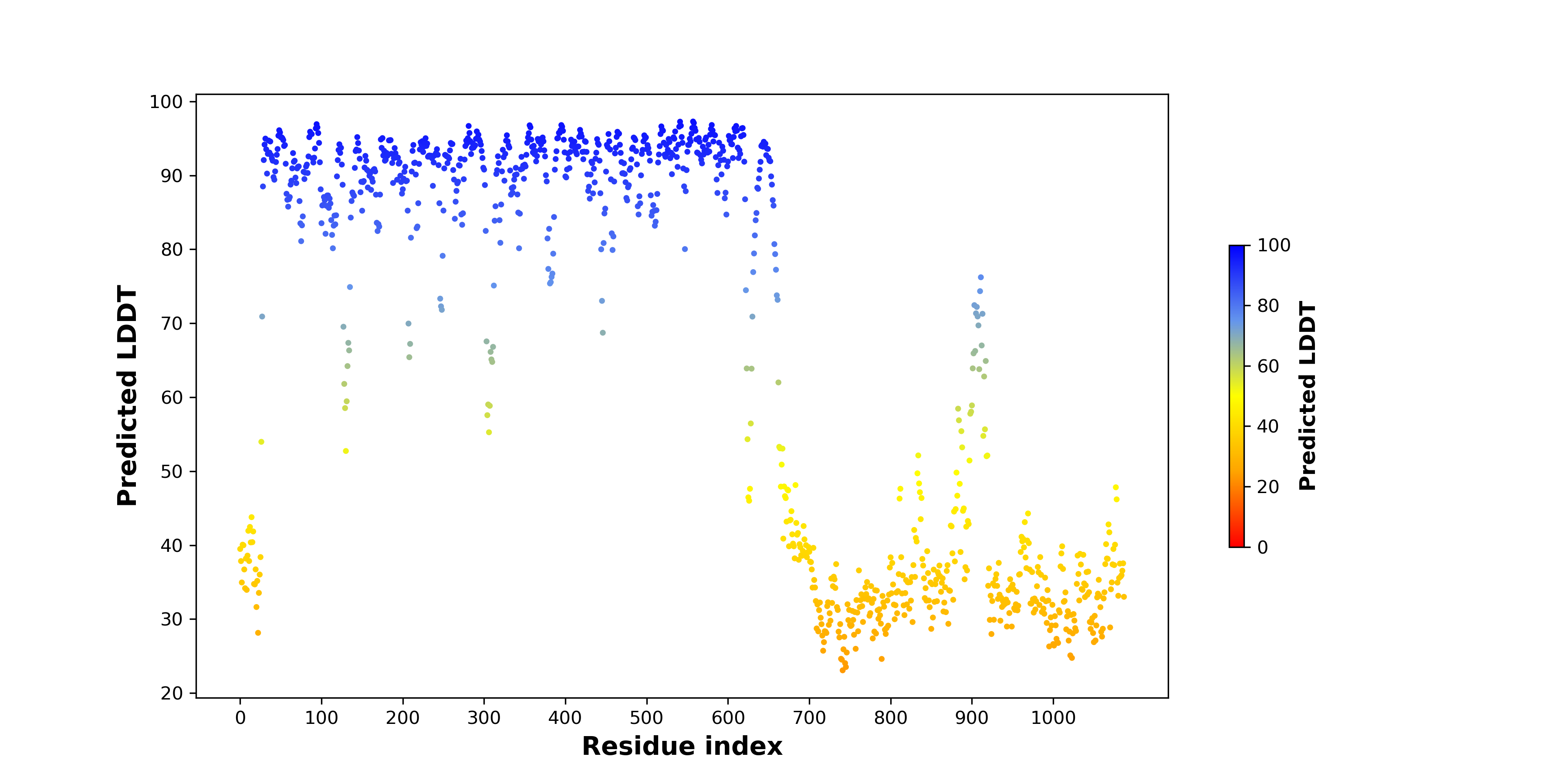
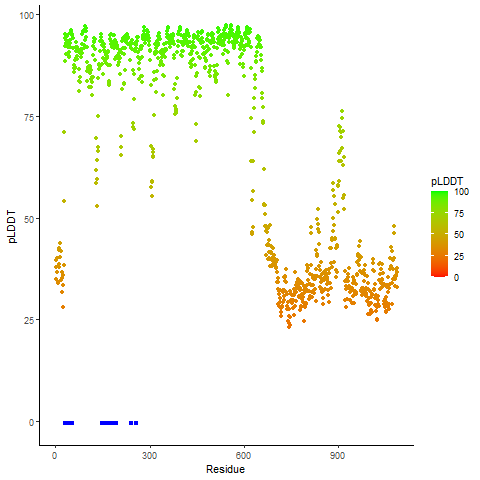
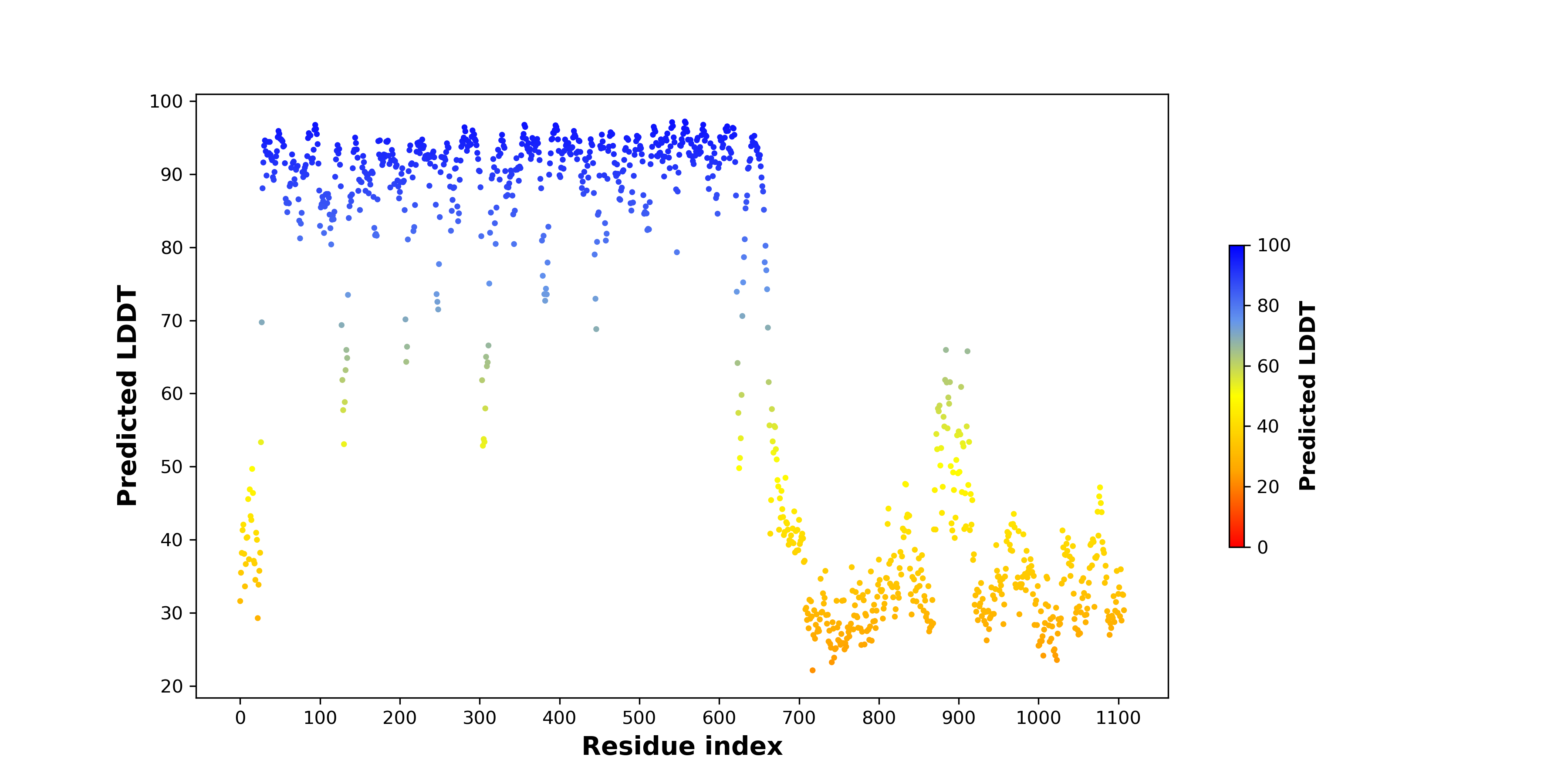
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (796) >>>796.pdbFusion protein BP residue: 712 CIF file (796) >>>796.cif | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51584615 | + | MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPL LYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQEDTGLLYLNRS LDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFF NTSFPACSSLKPRELCFPETRPSFRIRENRPPGTFHQFRLLPVQFLCPNI SVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAR EEVVMVPFPVTVYDEDDSAPTFPAGVDTASAVVEFKRKEDTVVATLRVFD ADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRAT VHDYRLVLNRNLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPV SLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQL LVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGRCEWRQGDGKG ITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPR GIKAGYGTCNCFPEEEKCFCEPEDIQDPLCDELCRTVIAAAVLFSFIVSV LLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSM ENQVSVDAFKILTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHS TTSSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSE KFKLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAK KPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEA LYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKA MTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSP MNTSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQE | 1088 |
| 3D view using mol* of 796 (AA BP:712) | ||||||||||
 | ||||||||||
| PDB file (804) >>>804.pdbFusion protein BP residue: 712 CIF file (804) >>>804.cif | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51584615 | + | MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPL LYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQEDTGLLYLNRS LDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFF NTSFPACSSLKPRELCFPETRPSFRIRENRPPGTFHQFRLLPVQFLCPNI SVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAR EEVVMVPFPVTVYDEDDSAPTFPAGVDTASAVVEFKRKEDTVVATLRVFD ADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRAT VHDYRLVLNRNLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPV SLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQL LVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGRCEWRQGDGKG ITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPR GIKAGYGTCNCFPEEEKCFCEPEDIQDPLCDELCRTVIAAAVLFSFIVSV LLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSM ENQVSVDAFKILTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHS TTSSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSE KFKLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAK KPLSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEA LYKWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKA MTPSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSP MNTSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQE EHNFPPDHYGLPAVCDLFACMQLKVDKEKWLYRTPLQAYFKMNFQDVTVG | 1108 |
| 3D view using mol* of 804 (AA BP:712) | ||||||||||
 | ||||||||||
| PDB file (826) >>>826.pdbFusion protein BP residue: 712 CIF file (826) >>>826.cif | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51582795 | + | MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPL LYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQEDTGLLYLNRS LDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFF NTSFPACSSLKPRELCFPETRPSFRIRENRPPGTFHQFRLLPVQFLCPNI SVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAR EEVVMVPFPVTVYDEDDSAPTFPAGVDTASAVVEFKRKEDTVVATLRVFD ADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRAT VHDYRLVLNRNLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPV SLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQL LVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGRCEWRQGDGKG ITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPR GIKAGYGTCNCFPEEEKCFCEPEDIQDPLCDELCRTVIAAAVLFSFIVSV LLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSM ENQVSVDAFKILKSASGIVAVPFSEWLLGSKPASGYQAPYIPSTDPQDWL TQKQTLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTT SSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKF KLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKP LSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALY KWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMT PSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMN TSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEH | 1136 |
| 3D view using mol* of 826 (AA BP:712) | ||||||||||
 | ||||||||||
| PDB file (834) >>>834.pdbFusion protein BP residue: 712 CIF file (834) >>>834.cif | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51582795 | + | MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPL LYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQEDTGLLYLNRS LDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFF NTSFPACSSLKPRELCFPETRPSFRIRENRPPGTFHQFRLLPVQFLCPNI SVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAR EEVVMVPFPVTVYDEDDSAPTFPAGVDTASAVVEFKRKEDTVVATLRVFD ADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRAT VHDYRLVLNRNLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPV SLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQL LVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGRCEWRQGDGKG ITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPR GIKAGYGTCNCFPEEEKCFCEPEDIQDPLCDELCRTVIAAAVLFSFIVSV LLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSM ENQVSVDAFKILKSASGIVAVPFSEWLLGSKPASGYQAPYIPSTDPQDWL TQKQTLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTT SSFSIEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKF KLLFQSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKP LSTPSMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALY KWLLKKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMT PSRIADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMN TSWCSFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEH NFPPDHYGLPAVCDLFACMQLKVDKEKWLYRTPLQAYFKMNFQDVTVGNF | 1156 |
| 3D view using mol* of 834 (AA BP:712) | ||||||||||
 | ||||||||||
| PDB file (864) >>>864.pdbFusion protein BP residue: 0 CIF file (864) >>>864.cif | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51580879 | + | MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPL LYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQEDTGLLYLNRS LDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFF NTSFPACSSLKPRELCFPETRPSFRIRENRPPGTFHQFRLLPVQFLCPNI SVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAR EEVVMVPFPVTVYDEDDSAPTFPAGVDTASAVVEFKRKEDTVVATLRVFD ADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRAT VHDYRLVLNRNLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPV SLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQL LVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGRCEWRQGDGKG ITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPR GIKAGYGTCNCFPEEEKCFCEPEDIQDPLCDELCRTVIAAAVLFSFIVSV LLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSM ENQVSVDAFKILLLGQFNCLTHQLECTQNKDLANQVSVCLERLGSLTLKP EDSTVLLFEADTITLRQTITTFGSLKTIQIPEHLMAHASSANIGPFLEKR GCISMPEQKSASGIVAVPFSEWLLGSKPASGYQAPYIPSTDPQDWLTQKQ TLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFS IEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLF QSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKPLSTP SMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALYKWLL KKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMTPSRI ADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMNTSWC SFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEHNFPP | 1232 |
| 3D view using mol* of 864 (AA BP:0) | ||||||||||
 | ||||||||||
| PDB file (869)CIF file (869) >>>869.cif | RET | chr10 | 43610184 | + | NCOA4 | chr10 | 51580879 | + | MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPL LYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQEDTGLLYLNRS LDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFF NTSFPACSSLKPRELCFPETRPSFRIRENRPPGTFHQFRLLPVQFLCPNI SVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAR EEVVMVPFPVTVYDEDDSAPTFPAGVDTASAVVEFKRKEDTVVATLRVFD ADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRAT VHDYRLVLNRNLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPV SLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCS TLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQL LVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGRCEWRQGDGKG ITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPR GIKAGYGTCNCFPEEEKCFCEPEDIQDPLCDELCRTVIAAAVLFSFIVSV LLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSM ENQVSVDAFKILLLGQFNCLTHQLECTQNKDLANQVSVCLERLGSLTLKP EDSTVLLFEADTITLRQTITTFGSLKTIQIPEHLMAHASSANIGPFLEKR GCISMPEQKSASGIVAVPFSEWLLGSKPASGYQAPYIPSTDPQDWLTQKQ TLENSQTSSRACNFFNNVGGNLKGLENWLLKSEKSSYQKCNSHSTTSSFS IEMEKVGDQELPDQDEMDLSDWLVTPQESHKLRKPENGSRETSEKFKLLF QSYNVNDWLVKTDSCTNCQGNQPKGVEIENLGNLKCLNDHLEAKKPLSTP SMVTEDWLVQNHQDPCKVEEVCRANEPCTSFAECVCDENCEKEALYKWLL KKEGKDKNGMPVEPKPEPEKHKDSLNMWLCPRKEVIEQTKAPKAMTPSRI ADSFQVIKNSPLSEWLIRPPYKEGSPKEVPGTEDRAGKQKFKSPMNTSWC SFNTADWVLPGKKMGNLSQLSSGEDKWLLRKKAQEVLLNSPLQEEHNFPP DHYGLPAVCDLFACMQLKVDKEKWLYRTPLQAYFKMNFQDVTVGNFQIPC | 1252 |
| 3D view using mol* of 869 (AA BP:) | ||||||||||
 | ||||||||||
Top |
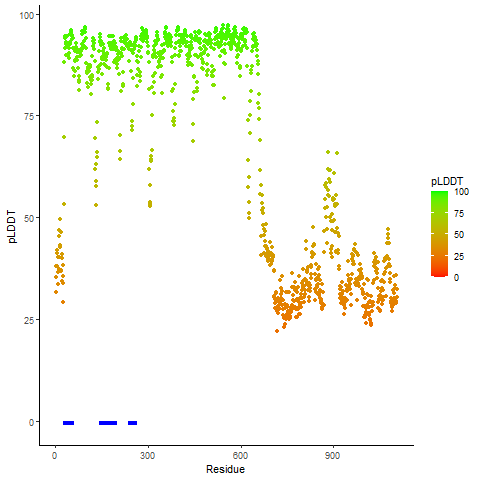
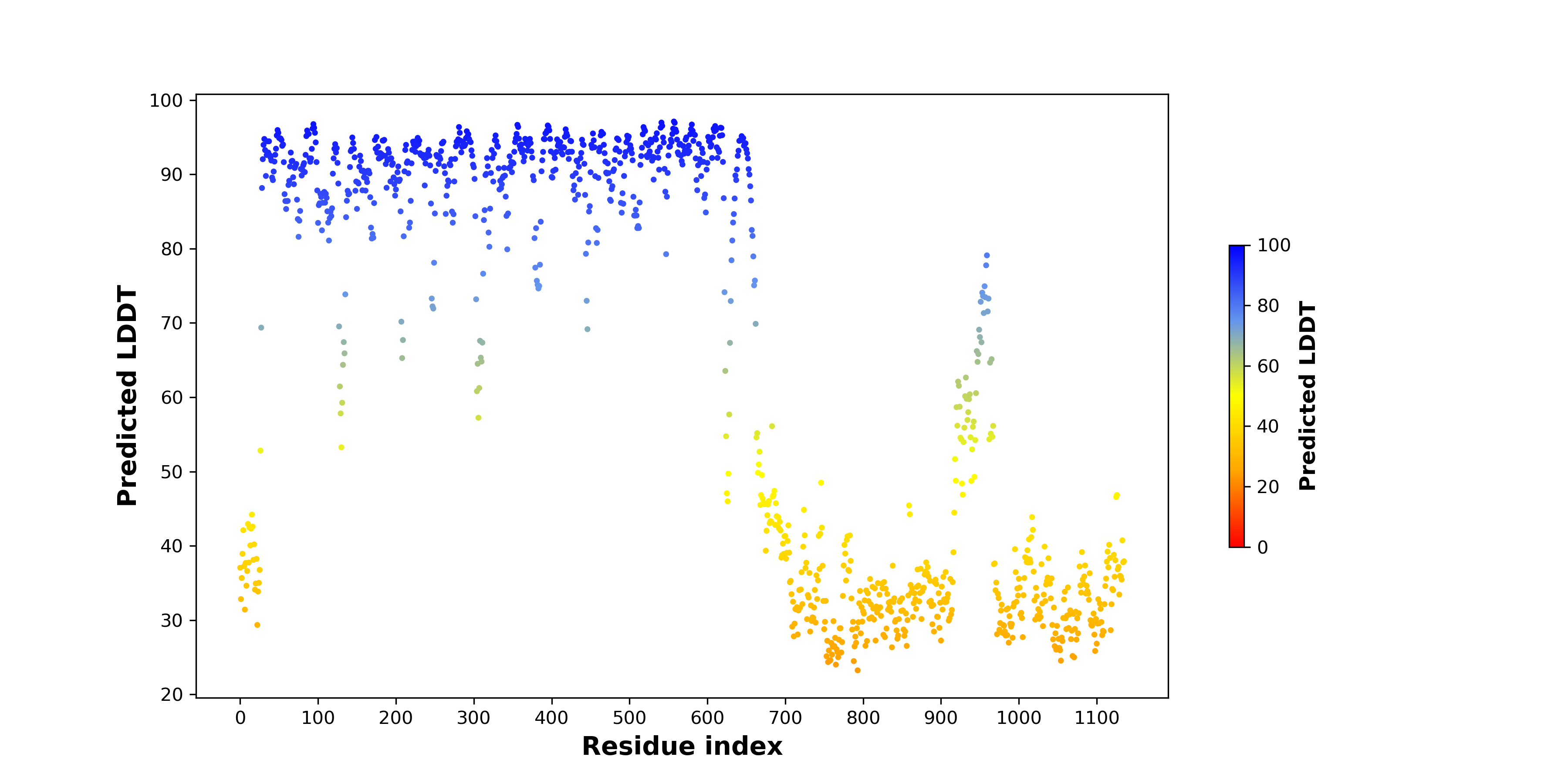
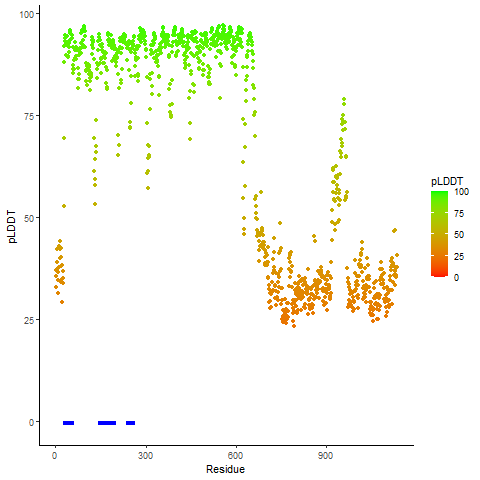
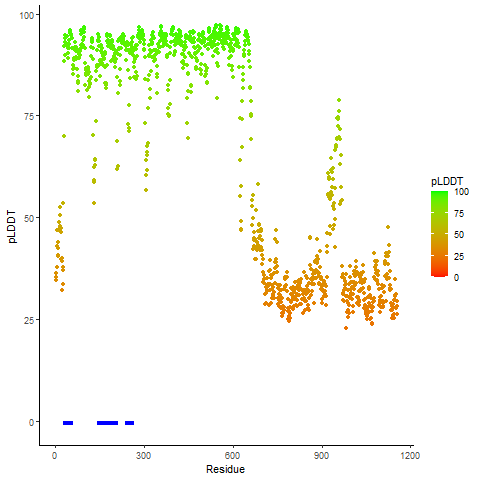
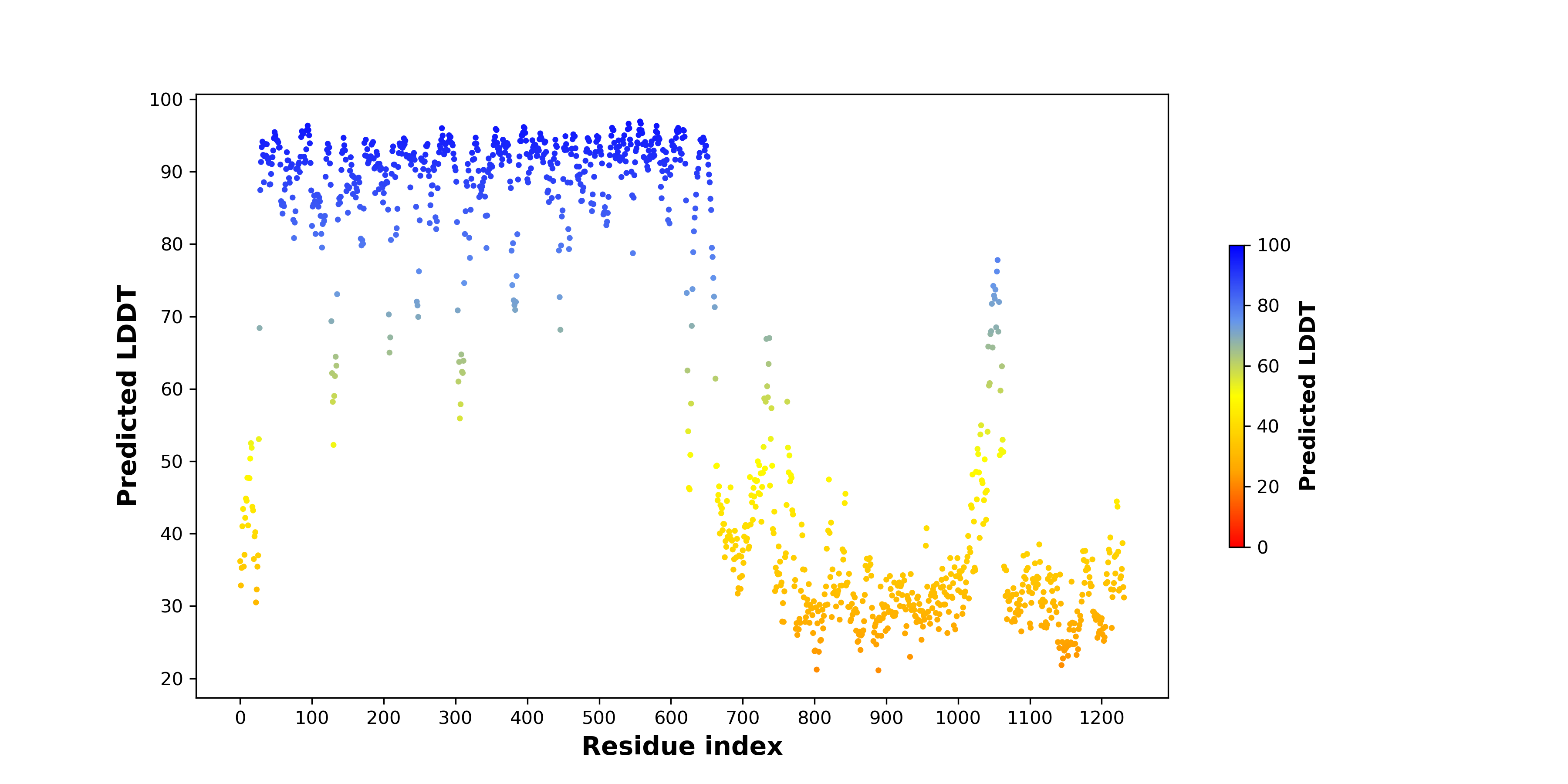
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
RET_pLDDT.png |
NCOA4_pLDDT.png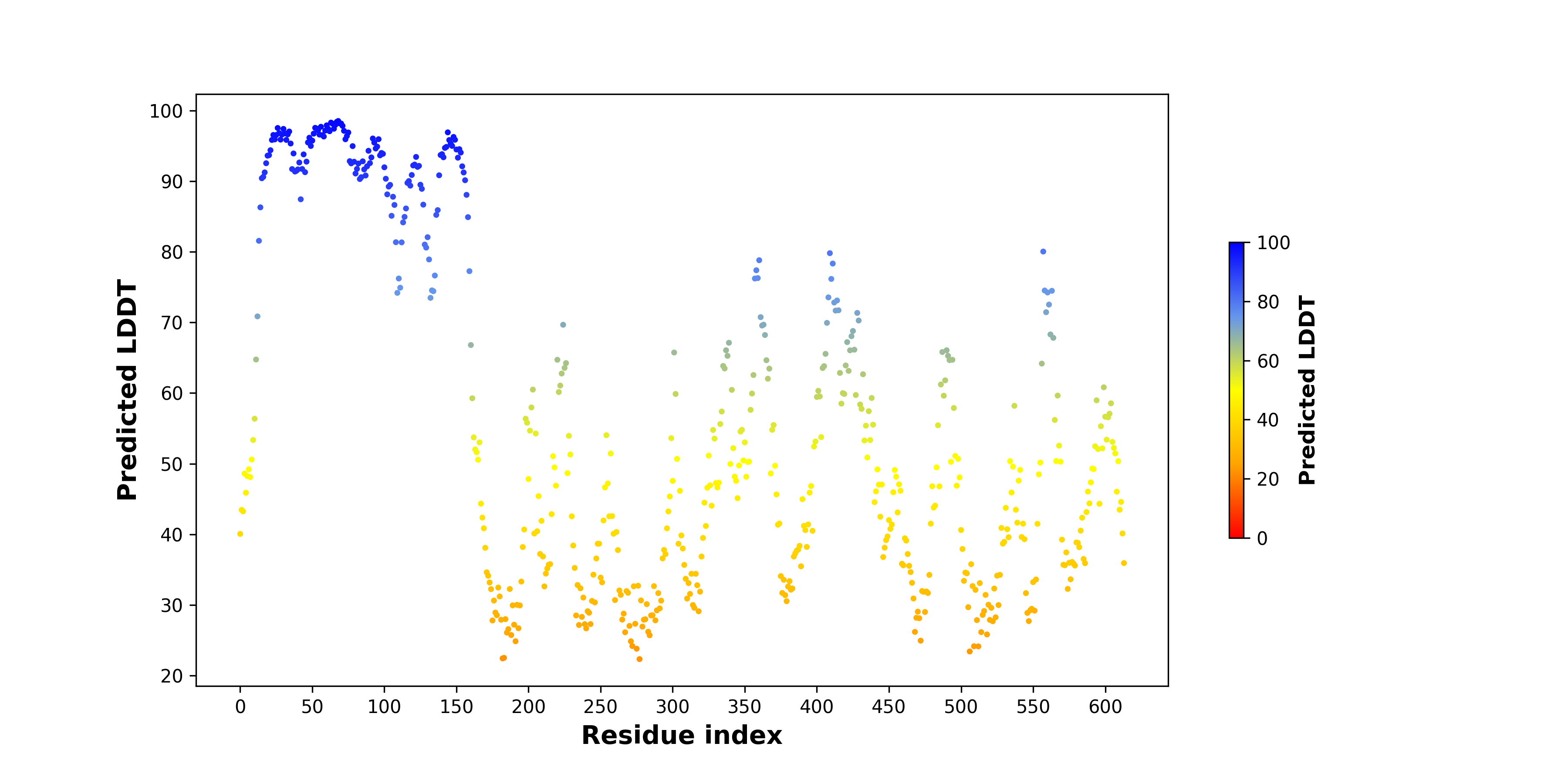 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| RET_NCOA4_796.png |
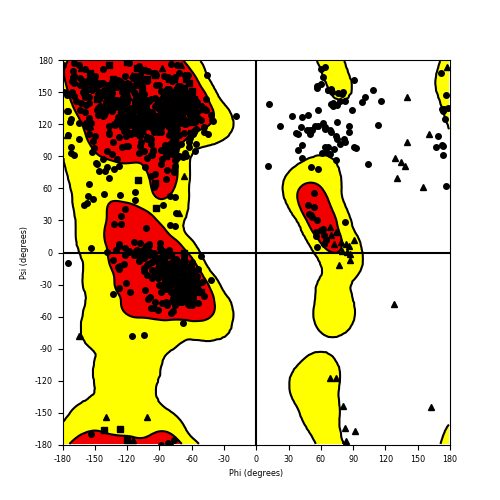 |
| RET_NCOA4_804.png |
 |
| RET_NCOA4_826.png |
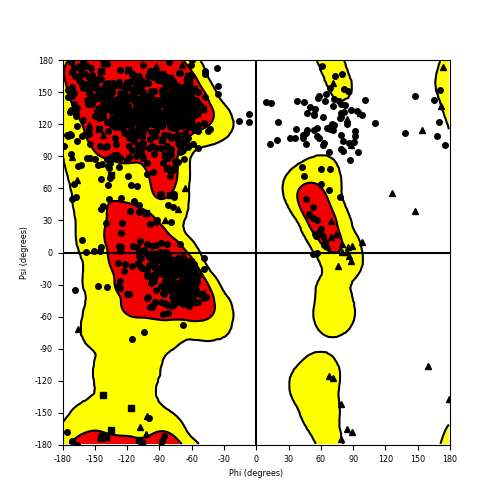 |
| RET_NCOA4_834.png |
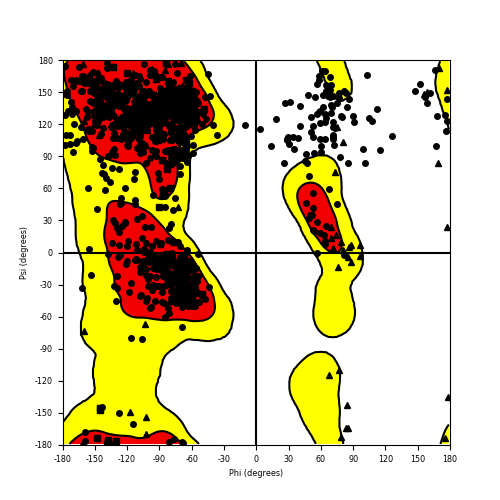 |
| RET_NCOA4_864.png |
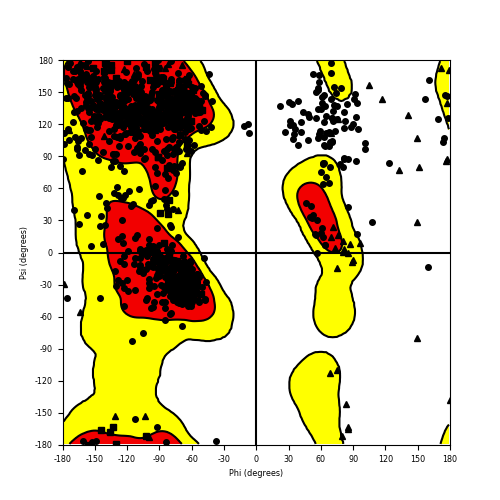 |
Top |
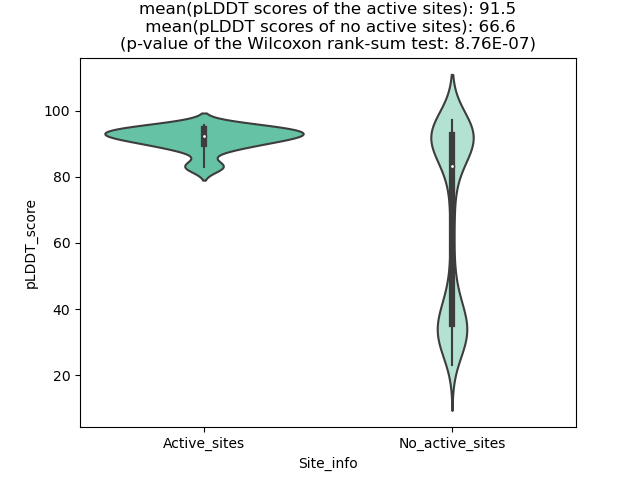
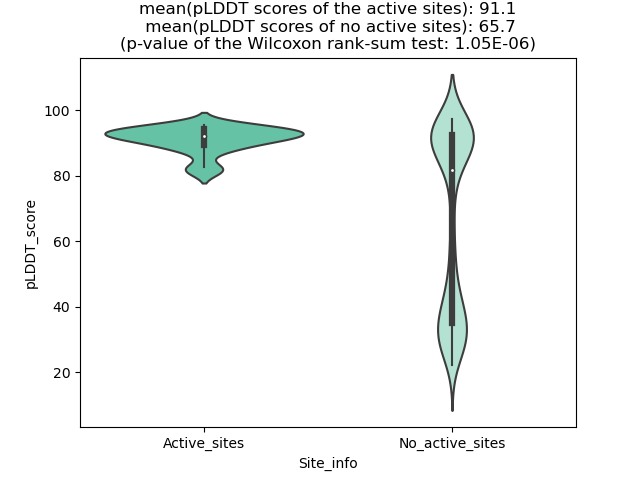
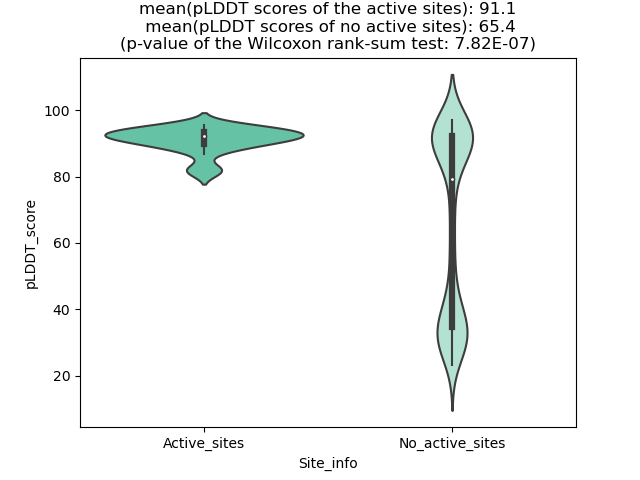
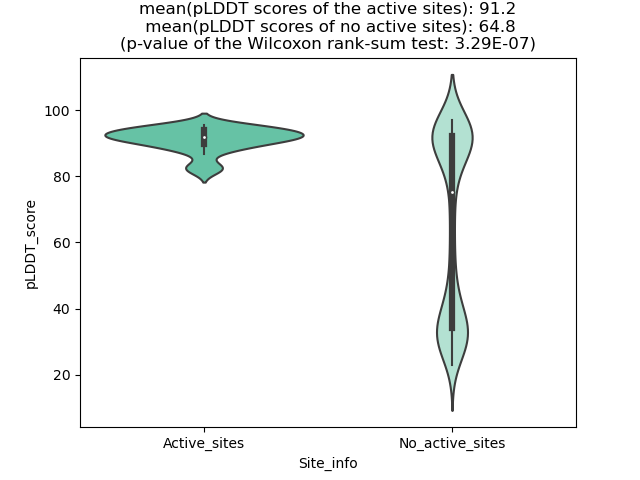
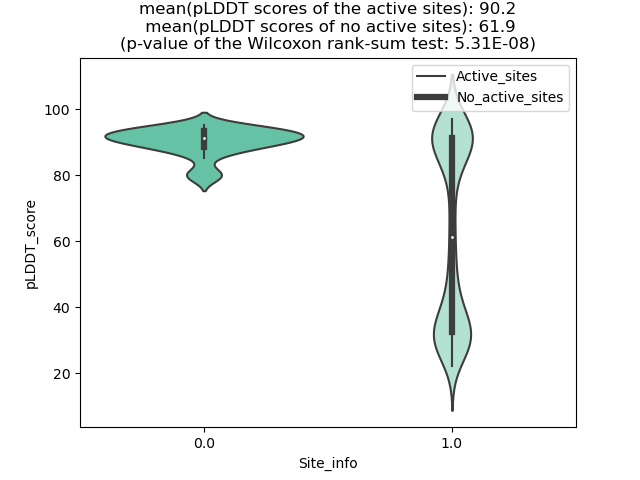
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 796 | 1.073 | 261 | 0.949 | 420.518 | 0.415 | 0.808 | 1.061 | 0.493 | 1.459 | 0.338 | 0.764 | Chain A: 32,33,36,37,38,39,41,50,51,52,54,148,150, 154,160,163,164,166,167,168,169,170,171,172,174,18 5,186,187,188,189,190,191,192,193,241,256,258 |
| 804 | 1.0708 | 253 | 0.9911 | 428.407 | 0.4276 | 0.8039 | 1.0629 | 0.674 | 1.3227 | 0.5096 | 0.7687 | Chain A: 32,33,36,37,38,39,41,50,51,52,54,148,150, 154,160,163,164,167,168,169,170,171,172,174,185,18 6,187,188,189,190,191,192,193,241,256,258 |
| 826 | 1.0836 | 248 | 0.9696 | 435.953 | 0.401 | 0.8229 | 1.0863 | 0.6137 | 1.4238 | 0.431 | 0.6878 | Chain A: 33,36,37,38,39,41,44,50,51,52,54,148,150, 154,155,160,163,164,166,167,168,169,170,171,172,17 4,185,186,187,188,189,190,191,192,193,241,256,258 |
| 834 | 1.0949 | 238 | 0.9283 | 363.237 | 0.385 | 0.84 | 1.1478 | 0.4423 | 1.5819 | 0.2796 | 0.6841 | Chain A: 32,33,36,37,38,39,41,50,51,52,54,148,150, 154,160,163,164,166,167,168,169,170,171,172,174,18 5,186,187,188,189,190,191,192,193,204,241,256,257, 258 |
| 864 | 1.058 | 271 | 0.97 | 419.489 | 0.394 | 0.785 | 1.061 | 0.633 | 1.353 | 0.468 | 0.616 | Chain A: 32,33,36,37,38,39,41,44,50,51,52,54,148,1 50,154,160,163,164,166,167,168,169,170,171,172,174 ,185,186,187,188,189,190,191,192,193,204,241,254,2 56,257,258 |
| 869 | 1.108 | 236 | 0.932 | 397.194 | 0.355 | 0.859 | 1.184 | 0.528 | 1.605 | 0.329 | 0.71 | Chain A: 32,33,36,37,38,39,41,44,50,51,52,54,148,1 50,154,160,163,164,166,167,168,169,170,171,172,174 ,185,186,187,188,189,190,191,192,193,204,256,257,2 58 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
| 796 | ZINC000002568036 | DB01219 | Dantrolene | -6.46879 | -7.74989 |
| 796 | ZINC000252679615 | DB01219 | Dantrolene | -6.46879 | -7.74989 |
| 796 | ZINC000003875368 | DB00698 | Nitrofurantoin | -6.44203 | -7.72313 |
| 796 | ZINC000000004319 | DB00598 | Labetalol | -7.48731 | -7.51491 |
| 796 | ZINC000000034157 | DB06707 | Levonordefrin | -7.40451 | -7.42201 |
| 796 | ZINC000052971887 | DB06243 | Eflornithine | -6.84476 | -7.12526 |
| 796 | ZINC000000896546 | DB01099 | Flucytosine | -7.04448 | -7.04498 |
| 796 | ZINC000000004321 | DB00909 | Zonisamide | -7.01525 | -7.01665 |
| 796 | ZINC000000001735 | DB00350 | Minoxidil | -6.91019 | -6.91019 |
| 796 | ZINC000001530600 | DB00369 | Cidofovir | -6.32425 | -6.87755 |
| 796 | ZINC000006036847 | DB00182 | Amphetamine | -6.74 | -6.7408 |
| 796 | ZINC000003801919 | DB00630 | Alendronic acid | -6.50568 | -6.70478 |
| 796 | ZINC000000897258 | DB00983 | Formoterol | -6.67858 | -6.69078 |
| 796 | ZINC000100032379 | DB02631 | -6.48414 | -6.65934 | |
| 796 | ZINC000000000882 | DB00173 | Adenine | -6.61164 | -6.63234 |
| 796 | ZINC000000897256 | DB00983 | Formoterol | -6.5687 | -6.5809 |
| 796 | ZINC000000001644 | DB06795 | Mafenide | -6.54747 | -6.57707 |
| 796 | ZINC000003812862 | DB00282 | Pamidronic acid | -6.45839 | -6.56889 |
| 796 | ZINC000006021033 | DB00182 | Amphetamine | -6.48317 | -6.48397 |
| 796 | ZINC000001482184 | DB00853 | Temozolomide | -6.36783 | -6.36943 |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
| ZINC000000034157 | DB06707 | Levonordefrin | Small molecule | C[C@H](N)[C@H](O)C1=CC(O)=C(O)C=C1 | Approved |
| ZINC000000896546 | DB01099 | Flucytosine | Small molecule | NC1=C(F)C=NC(=O)N1 | Approved|Investigational |
| ZINC000000004321 | DB00909 | Zonisamide | Small molecule | NS(=O)(=O)CC1=NOC2=CC=CC=C12 | Approved|Investigational |
| ZINC000000001735 | DB00350 | Minoxidil | Small molecule | NC1=CC(=NC(N)=[N+]1[O-])N1CCCCC1 | Approved|Investigational |
| ZINC000001530600 | DB00369 | Cidofovir | Small molecule | NC1=NC(=O)N(C[C@@H](CO)OCP(O)(O)=O)C=C1 | Approved |
| ZINC000006036847 | DB00182 | Amphetamine | Small molecule | CC(N)CC1=CC=CC=C1 | Approved|Illicit|Investigational |
| ZINC000003801919 | DB00630 | Alendronic acid | Small molecule | NCCCC(O)(P(O)(O)=O)P(O)(O)=O | Approved |
| ZINC000000897258 | DB00983 | Formoterol | Small molecule | COC1=CC=C(CC(C)NCC(O)C2=CC(NC=O)=C(O)C=C2)C=C1 | Approved|Investigational |
| ZINC000000000882 | DB00173 | Adenine | Small molecule | NC1=C2NC=NC2=NC=N1 | Approved|Nutraceutical |
| ZINC000000001644 | DB06795 | Mafenide | Small molecule | NCC1=CC=C(C=C1)S(N)(=O)=O | Approved|Vet_approved |
| ZINC000003812862 | DB00282 | Pamidronic acid | Small molecule | NCCC(O)(P(O)(O)=O)P(O)(O)=O | Approved |
| ZINC000001482184 | DB00853 | Temozolomide | Small molecule | CN1N=NC2=C(N=CN2C1=O)C(N)=O | Approved|Investigational |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
| ZINC000002568036 | 314.257 | 3.867 | 548.758 | 54.323 | 243.185 | 251.25 | 0 | 924.46 | 1 | 5 | 9.205 | 3 | 67.058 | 0 | 0 |
| ZINC000002568036 | 314.257 | 4.186 | 547.3 | 54.729 | 241.127 | 251.444 | 0 | 922.097 | 1 | 5 | 9.191 | 3 | 67.408 | 0 | 0 |
| ZINC000252679615 | 314.257 | 3.867 | 548.758 | 54.323 | 243.185 | 251.25 | 0 | 924.46 | 1 | 5 | 9.205 | 3 | 67.058 | 0 | 0 |
| ZINC000252679615 | 314.257 | 2.557 | 527.996 | 64.383 | 238.233 | 225.38 | 0 | 905.494 | 1 | 5 | 9.686 | 3 | 67.506 | 0 | 0 |
| ZINC000252679615 | 314.257 | 4.186 | 547.3 | 54.729 | 241.127 | 251.444 | 0 | 922.097 | 1 | 5 | 9.191 | 3 | 67.408 | 0 | 0 |
| ZINC000252679615 | 314.257 | 2.845 | 525.884 | 65.15 | 235.939 | 224.795 | 0 | 902.055 | 1 | 5 | 9.671 | 3 | 67.875 | 0 | 0 |
| ZINC000003875368 | 238.159 | 4.578 | 425.423 | 58.928 | 250.52 | 115.975 | 0 | 683.161 | 1 | 5 | 9.961 | 2 | 53.722 | 0 | 0 |
| ZINC000003875368 | 238.159 | 4.877 | 423.946 | 59.354 | 248.434 | 116.158 | 0 | 680.796 | 1 | 5 | 9.943 | 2 | 54.077 | 0 | 0 |
| ZINC000000004319 | 328.41 | 5.655 | 664.549 | 169.051 | 199.646 | 295.852 | 0 | 1140.615 | 4 | 5.45 | 9.067 | 2 | 70.589 | 0 | 0 |
| ZINC000000004319 | 328.41 | 3.937 | 652.909 | 170.739 | 198.631 | 283.539 | 0 | 1134.882 | 4 | 5.45 | 9.364 | 2 | 70.662 | 0 | 0 |
| ZINC000000034157 | 183.207 | 1.584 | 399.847 | 106.772 | 186.848 | 106.227 | 0 | 646.199 | 5 | 4.2 | 8.948 | 2 | 52.109 | 0 | 0 |
| ZINC000000034157 | 183.207 | 2.284 | 398.836 | 105.841 | 186.106 | 106.889 | 0 | 643.154 | 5 | 4.2 | 8.781 | 2 | 52.19 | 0 | 0 |
| ZINC000052971887 | 182.17 | 4.815 | 375.577 | 120.033 | 183.002 | 0 | 72.541 | 593.19 | 5 | 4 | 9.616 | 1 | 19.752 | 0 | 1 |
| ZINC000052971887 | 182.17 | 4.577 | 376.305 | 117.956 | 183.727 | 0 | 74.623 | 594.335 | 5 | 4 | 9.723 | 1 | 19.678 | 0 | 1 |
| ZINC000000896546 | 129.094 | 5.933 | 283.803 | 0 | 174.821 | 65.196 | 43.787 | 412.204 | 3 | 4 | 9.417 | 2 | 59.957 | 0 | 0 |
| ZINC000000004321 | 212.223 | 6.978 | 398.517 | 40.32 | 185.554 | 172.644 | 0 | 642.453 | 2 | 6 | 9.667 | 3 | 65.959 | 0 | 0 |
| ZINC000000001735 | 209.25 | 5.141 | 437.436 | 217.593 | 169.974 | 49.869 | 0 | 711.864 | 4 | 3.5 | 7.733 | 3 | 76.873 | 0 | 0 |
| ZINC000001530600 | 279.189 | 10.973 | 485.753 | 106.478 | 301.213 | 75.118 | 2.945 | 819.936 | 5 | 12.4 | 8.977 | 1 | 18.286 | 0 | 1 |
| ZINC000001530600 | 279.189 | 13.189 | 481.116 | 107.174 | 307.995 | 61.465 | 4.483 | 817.63 | 5 | 12.4 | 8.916 | 1 | 16.855 | 0 | 1 |
| ZINC000006036847 | 135.208 | 1.199 | 369.378 | 132.77 | 47.27 | 189.339 | 0 | 584.758 | 2 | 1 | 9.281 | 3 | 89.947 | 0 | 0 |
| ZINC000003801919 | 249.097 | 6.598 | 414.717 | 86.372 | 321.667 | 0 | 6.679 | 680 | 3 | 7.75 | 9.26 | 1 | 0 | 1 | 1 |
| ZINC000003801919 | 249.097 | 13.583 | 415.33 | 88.33 | 320.807 | 0 | 6.194 | 681.18 | 3 | 7.75 | 9.122 | 1 | 0 | 1 | 1 |
| ZINC000003801919 | 249.097 | 7.425 | 414.525 | 87.312 | 321.455 | 0 | 5.758 | 680.611 | 3 | 7.75 | 9.244 | 1 | 0 | 1 | 1 |
| ZINC000000897258 | 344.41 | 7.083 | 674.489 | 258.736 | 187.926 | 227.826 | 0 | 1168.666 | 4 | 7.2 | 8.855 | 2 | 65.222 | 0 | 0 |
| ZINC000000897258 | 344.41 | 4.982 | 586.764 | 251.201 | 138.136 | 197.426 | 0 | 1091.391 | 4 | 7.2 | 8.677 | 3 | 73.278 | 0 | 0 |
| ZINC000000000882 | 135.128 | 2.734 | 303.153 | 0 | 170.481 | 132.672 | 0 | 451.892 | 3 | 4 | 8.538 | 3 | 68.38 | 0 | 0 |
| ZINC000000000882 | 135.128 | 9.438 | 304.278 | 0 | 171.466 | 132.812 | 0 | 454.287 | 3 | 4 | 8.639 | 3 | 68.214 | 0 | 0 |
| ZINC000000897256 | 344.41 | 5.264 | 632.493 | 247.284 | 157.494 | 227.715 | 0 | 1135.048 | 4 | 7.2 | 8.825 | 3 | 70.837 | 0 | 0 |
| ZINC000000897256 | 344.41 | 6.478 | 644.939 | 257.318 | 179.642 | 207.979 | 0 | 1149.057 | 4 | 7.2 | 8.565 | 2 | 66.251 | 0 | 0 |
| ZINC000000001644 | 186.228 | 7.234 | 386.689 | 48.38 | 203.852 | 132.644 | 1.813 | 608.208 | 4 | 5.5 | 9.936 | 2 | 46.461 | 0 | 0 |
| ZINC000000001644 | 186.228 | 8.543 | 384.513 | 48.79 | 201.796 | 132.114 | 1.813 | 604.747 | 4 | 5.5 | 9.902 | 2 | 46.8 | 0 | 0 |
| ZINC000003812862 | 235.07 | 9.08 | 382.849 | 49.638 | 327.049 | 0 | 6.162 | 620.98 | 3 | 7.75 | 9.11 | 1 | 0 | 1 | 1 |
| ZINC000003812862 | 235.07 | 11.837 | 382.227 | 54.693 | 321.348 | 0 | 6.185 | 620.451 | 3 | 7.75 | 9.075 | 1 | 0 | 1 | 1 |
| ZINC000003812862 | 235.07 | 4.081 | 379.433 | 56.179 | 318.684 | 0 | 4.57 | 620.406 | 3 | 7.75 | 9.224 | 1 | 0 | 1 | 1 |
| ZINC000006021033 | 135.208 | 1.199 | 369.378 | 132.77 | 47.27 | 189.339 | 0 | 584.758 | 2 | 1 | 9.281 | 3 | 89.947 | 0 | 0 |
| ZINC000001482184 | 194.152 | 3.411 | 379.128 | 87.887 | 235.051 | 56.19 | 0 | 597.216 | 2 | 7 | 9.649 | 2 | 51.425 | 0 | 0 |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
| ZINC000002568036 | O=C1CN(/N=C/c2ccc(-c3ccc([N+](=O)[O-])cc3)o2)C(=O)N1 | 0.204070823 | 0.461176527 |
| ZINC000252679615 | O=C1CN(N=Cc2ccc(-c3ccc([N+](=O)[O-])cc3)o2)C(O)=N1 | 0.149132109 | 0.500589886 |
| ZINC000003875368 | O=C1CN(/N=C/c2ccc([N+](=O)[O-])o2)C(=O)N1 | 0.144023256 | 0.52186584 |
| ZINC000000004319 | C[C@H](CCc1ccccc1)NC[C@H](O)c1ccc(O)c(C(N)=O)c1 | 0.163888793 | 0.349082082 |
| ZINC000000034157 | C[C@H](N)[C@H](O)c1ccc(O)c(O)c1 | 0.135079192 | 0.302260037 |
| ZINC000052971887 | NCCC[C@](N)(C(=O)O)C(F)F | 0.086413284 | 0.375086254 |
| ZINC000000896546 | Nc1nc(=O)[nH]cc1F | 0.116019695 | 0.475808288 |
| ZINC000000004321 | NS(=O)(=O)Cc1noc2ccccc12 | 0.254569254 | 0.46155884 |
| ZINC000000001735 | Nc1cc(N2CCCCC2)nc(N)[n+]1[O-] | 0.129740946 | 0.395964399 |
| ZINC000001530600 | Nc1ccn(C[C@@H](CO)OCP(=O)(O)O)c(=O)n1 | 0.080149721 | 0.307248667 |
| ZINC000006036847 | C[C@@H](N)Cc1ccccc1 | 0.432320606 | 0.4471075 |
| ZINC000003801919 | NCCCC(O)(P(=O)(O)O)P(=O)(O)O | 0.111910059 | 0.475661392 |
| ZINC000000897258 | COc1ccc(C[C@@H](C)NC[C@@H](O)c2ccc(O)c(NC=O)c2)cc1 | 0.133863078 | 0.246482573 |
| ZINC000100032379 | N=C1CCCN1Cc1[nH]c(=O)[nH]c(=O)c1Cl | 0.113188199 | 0.388278234 |
| ZINC000000000882 | Nc1ncnc2[nH]cnc12 | 0.174637229 | 0.465480638 |
| ZINC000000897256 | COc1ccc(C[C@H](C)NC[C@H](O)c2ccc(O)c(NC=O)c2)cc1 | 0.133863078 | 0.246482573 |
| ZINC000000001644 | NCc1ccc(S(N)(=O)=O)cc1 | 0.497774604 | 0.389970963 |
| ZINC000003812862 | NCCC(O)(P(=O)(O)O)P(=O)(O)O | 0.095344218 | 0.466020225 |
| ZINC000006021033 | C[C@H](N)Cc1ccccc1 | 0.432320606 | 0.4471075 |
| ZINC000001482184 | Cn1nnc2c(C(N)=O)ncn2c1=O | 0.160886409 | 0.261932701 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| NCOA4 | AR, PPARG, ESR1, Nr3c1, GTF2B, KAT2B, NR3C1, VDR, AKT1, ATXN2, RXRA, HERC2, XPO1, GRIA2, KDR, KIFC3, PSMC1, XRCC3, HUWE1, PSMD14, TFG, KIF21A, USP43, Bmpr1a, Kif1b, Cdc23, Wdr61, Psmd6, FTH1, FTL, GMCL1, NEURL4, TAX1BP1, EGLN3, MAPK6, MID2, ORF3a, nsp13, nsp16, RNF10, PRELID2, PIPSL, BTF3, MAP1LC3B, NBR1, CALCOCO2, OPTN, SQSTM1, |
| RET | STAT3, DOK6, DOK5, SRC, SHC1, PTPRF, DOK2, DOK4, DOK1, GRB2, CRK, PLCG1, PDLIM7, GRB7, FRS2, NRTN, GFRA1, GRB10, IKBKG, MCRS1, CBL, CBLC, RET, HSP90AA1, NOTCH2NL, NOTCH3, AIP, EGFR, NTRK1, ZBTB48, HNRNPD, SORT1, SYNCRIP, SGTB, BAG6, HIST1H3A, KYNU, TXNL4A, ZCCHC8, ICE2, MAPK3, PTK2, MAPK1, PTPRR, DUSP26, Shc1, PIK3R1, FBXO7, NEDD4, SHANK2, LRRK1, PTK6, CTNNB1, SHC4, ERBB2, ADAM10, SPINT2, PCDH7, LRIG2, APP, GDNF, IGF1R, CLU, EPCAM, CDH2, TGFBR1, PTK7, FAT1, DHCR24, SMN1, PCDH19, NCSTN, CERS2, CHP1, FANCD2, ITM2C, SEL1L, PCDHGB5, CKB, NRP1, PLXNB2, NCK2, CTNND1, TULP3, PTPN1, NF2, CRKL, CAMLG, NUMB, BASP1, ROR2, NOTCH2, PTPN13, ANK3, RAB3GAP1, PEA15, LLGL1, GPRIN3, RAPH1, MARK2, CYFIP1, ARC, GPRIN1, ANKLE2, VRK2, MIB1, PHACTR4, UNC5B, TMEM57, SYAP1, FAF2, ERBB2IP, FAM129B, KIAA1715, PEAK1, VANGL2, BAIAP2, EPN1, NUMBL, PRKAR2A, GAB1, PTPRA, DDX39A, TIMP1, EPHB4, MANEA, SRY, RABAC1, LEMD3, TMEM214, PDZD8, GPR50, SMIM5, RNF149, SDF2L1, BMPR1A, PLEKHH3, ACVR2A, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| RET |  |
| NCOA4 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to RET-NCOA4 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to RET-NCOA4 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| RET | NCOA4 | Thyroid Gland Papillary Carcinoma | MyCancerGenome | |
| RET | NCOA4 | Adenocarcinoma Of Unknown Primary | MyCancerGenome | |
| RET | NCOA4 | Cholangiocarcinoma | MyCancerGenome | |
| RET | NCOA4 | Colon Adenocarcinoma | MyCancerGenome | |
| RET | NCOA4 | Colorectal Signet Ring Cell Carcinoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | RET | C1833921 | Familial medullary thyroid carcinoma | 23 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | RET | C3888239 | HIRSCHSPRUNG DISEASE, SUSCEPTIBILITY TO, 1 | 16 | GENOMICS_ENGLAND;UNIPROT |
| Hgene | RET | C0025268 | Multiple Endocrine Neoplasia Type 2a | 15 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | RET | C1708353 | Hereditary Paraganglioma-Pheochromocytoma Syndrome | 12 | CLINGEN |
| Hgene | RET | C0025269 | Multiple Endocrine Neoplasia Type 2b | 10 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | RET | C0238463 | Papillary thyroid carcinoma | 3 | CTD_human;ORPHANET |
| Hgene | RET | C1275808 | Congenital central hypoventilation | 3 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | RET | C1859049 | CCHS WITH HIRSCHSPRUNG DISEASE | 3 | CTD_human;ORPHANET |
| Hgene | RET | C0009402 | Colorectal Carcinoma | 2 | CTD_human;UNIPROT |
| Hgene | RET | C0009404 | Colorectal Neoplasms | 2 | CTD_human |
| Hgene | RET | C0019569 | Hirschsprung Disease | 2 | CTD_human |
| Hgene | RET | C0027662 | Multiple Endocrine Neoplasia | 2 | CTD_human;GENOMICS_ENGLAND |
| Hgene | RET | C0085758 | Aganglionosis, Colonic | 2 | CTD_human |
| Hgene | RET | C0266294 | Unilateral agenesis of kidney | 2 | ORPHANET |
| Hgene | RET | C1257840 | Aganglionosis, Rectosigmoid Colon | 2 | CTD_human |
| Hgene | RET | C3661523 | Congenital Intestinal Aganglionosis | 2 | CTD_human |
| Hgene | RET | C0006413 | Burkitt Lymphoma | 1 | CTD_human |
| Hgene | RET | C0031511 | Pheochromocytoma | 1 | CGI;CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Hgene | RET | C0038220 | Status Epilepticus | 1 | CTD_human |
| Hgene | RET | C0040136 | Thyroid Neoplasm | 1 | CGI;CTD_human |
| Hgene | RET | C0151468 | Thyroid Gland Follicular Adenoma | 1 | CTD_human |
| Hgene | RET | C0206693 | Medullary carcinoma | 1 | CTD_human |
| Hgene | RET | C0238462 | Medullary carcinoma of thyroid | 1 | CGI;CTD_human |
| Hgene | RET | C0270823 | Petit mal status | 1 | CTD_human |
| Hgene | RET | C0311335 | Grand Mal Status Epilepticus | 1 | CTD_human |
| Hgene | RET | C0343640 | African Burkitt's lymphoma | 1 | CTD_human |
| Hgene | RET | C0393734 | Complex Partial Status Epilepticus | 1 | CTD_human |
| Hgene | RET | C0549473 | Thyroid carcinoma | 1 | CGI;CTD_human;UNIPROT |
| Hgene | RET | C0740340 | Amyloidosis, Familial | 1 | CTD_human |
| Hgene | RET | C0751522 | Status Epilepticus, Subclinical | 1 | CTD_human |
| Hgene | RET | C0751523 | Non-Convulsive Status Epilepticus | 1 | CTD_human |
| Hgene | RET | C0751524 | Simple Partial Status Epilepticus | 1 | CTD_human |
| Hgene | RET | C1257877 | Pheochromocytoma, Extra-Adrenal | 1 | CTD_human |
| Hgene | RET | C1609433 | Congenital absence of kidneys syndrome | 1 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Hgene | RET | C3501843 | Nonmedullary Thyroid Carcinoma | 1 | CTD_human |
| Hgene | RET | C3501844 | Familial Nonmedullary Thyroid Cancer | 1 | CTD_human |
| Hgene | RET | C4721444 | Burkitt Leukemia | 1 | CTD_human |
| Tgene | NCOA4 | C0238463 | Papillary thyroid carcinoma | 2 | ORPHANET |
| Tgene | NCOA4 | C0040136 | Thyroid Neoplasm | 1 | CTD_human |
| Tgene | NCOA4 | C0151468 | Thyroid Gland Follicular Adenoma | 1 | CTD_human |
| Tgene | NCOA4 | C0549473 | Thyroid carcinoma | 1 | CTD_human |