| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ATIC-GIGYF2 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ATIC-GIGYF2 | FusionPDB ID: 7630 | FusionGDB2.0 ID: 7630 | Hgene | Tgene | Gene symbol | ATIC | GIGYF2 | Gene ID | 471 | 26058 |
| Gene name | 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase | GRB10 interacting GYF protein 2 | |
| Synonyms | AICAR|AICARFT|HEL-S-70p|IMPCHASE|PURH | GYF2|PARK11|PERQ2|PERQ3|TNRC15 | |
| Cytomap | 2q35 | 2q37.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | bifunctional purine biosynthesis protein PURH5-aminoimidazole-4-carboxamide-1-beta-D-ribonucleotide transformylase/inosinicaseAICAR formyltransferase/IMP cyclohydrolase bifunctional enzymeAICARFT/IMPCHASEepididymis secretory sperm binding protein Li 7 | GRB10-interacting GYF protein 2PERQ amino acid rich, with GYF domain 3PERQ amino acid-rich with GYF domain-containing protein 2Parkinson disease (autosomal recessive, early onset) 11trinucleotide repeat-containing gene 15 protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | P31939 | Q6Y7W6 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000236959, ENST00000435675, ENST00000540518, | ENST00000452341, ENST00000482666, ENST00000373563, ENST00000373566, ENST00000409196, ENST00000409451, ENST00000409480, ENST00000409547, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 9 X 9 X 4=324 | 17 X 18 X 6=1836 |
| # samples | 9 | 19 | |
| ** MAII score | log2(9/324*10)=-1.84799690655495 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(19/1836*10)=-3.27249473508286 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ATIC [Title/Abstract] AND GIGYF2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ATIC(216184454)-GIGYF2(233612325), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ATIC-GIGYF2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ATIC-GIGYF2 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ATIC-GIGYF2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ATIC-GIGYF2 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | GIGYF2 | GO:0016441 | posttranscriptional gene silencing | 27157137 |
| Tgene | GIGYF2 | GO:0061157 | mRNA destabilization | 27157137 |
 Fusion gene breakpoints across ATIC (5'-gene) Fusion gene breakpoints across ATIC (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
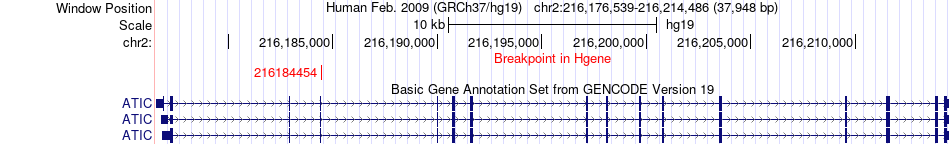 |
 Fusion gene breakpoints across GIGYF2 (3'-gene) Fusion gene breakpoints across GIGYF2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
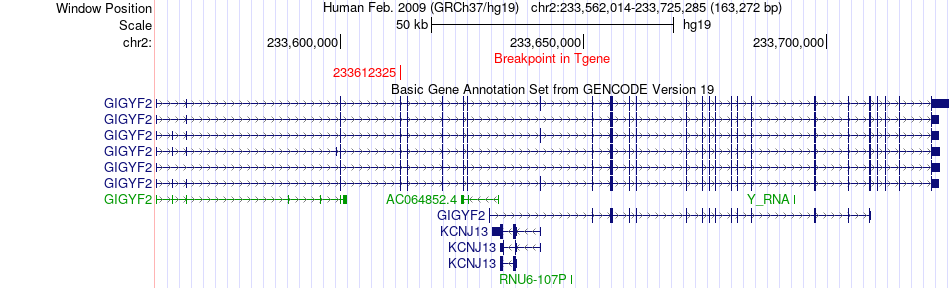 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-D8-A143-01A | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000236959 | ATIC | chr2 | 216184454 | + | ENST00000373566 | GIGYF2 | chr2 | 233612325 | + | 8256 | 616 | 266 | 4540 | 1424 |
| ENST00000236959 | ATIC | chr2 | 216184454 | + | ENST00000373563 | GIGYF2 | chr2 | 233612325 | + | 6227 | 616 | 266 | 4474 | 1402 |
| ENST00000236959 | ATIC | chr2 | 216184454 | + | ENST00000409480 | GIGYF2 | chr2 | 233612325 | + | 6289 | 616 | 266 | 4540 | 1424 |
| ENST00000236959 | ATIC | chr2 | 216184454 | + | ENST00000409547 | GIGYF2 | chr2 | 233612325 | + | 6242 | 616 | 266 | 4474 | 1402 |
| ENST00000236959 | ATIC | chr2 | 216184454 | + | ENST00000409196 | GIGYF2 | chr2 | 233612325 | + | 6224 | 616 | 266 | 4456 | 1396 |
| ENST00000236959 | ATIC | chr2 | 216184454 | + | ENST00000409451 | GIGYF2 | chr2 | 233612325 | + | 6286 | 616 | 266 | 4537 | 1423 |
| ENST00000540518 | ATIC | chr2 | 216184454 | + | ENST00000373566 | GIGYF2 | chr2 | 233612325 | + | 8230 | 590 | 279 | 4514 | 1411 |
| ENST00000540518 | ATIC | chr2 | 216184454 | + | ENST00000373563 | GIGYF2 | chr2 | 233612325 | + | 6201 | 590 | 279 | 4448 | 1389 |
| ENST00000540518 | ATIC | chr2 | 216184454 | + | ENST00000409480 | GIGYF2 | chr2 | 233612325 | + | 6263 | 590 | 279 | 4514 | 1411 |
| ENST00000540518 | ATIC | chr2 | 216184454 | + | ENST00000409547 | GIGYF2 | chr2 | 233612325 | + | 6216 | 590 | 279 | 4448 | 1389 |
| ENST00000540518 | ATIC | chr2 | 216184454 | + | ENST00000409196 | GIGYF2 | chr2 | 233612325 | + | 6198 | 590 | 279 | 4430 | 1383 |
| ENST00000540518 | ATIC | chr2 | 216184454 | + | ENST00000409451 | GIGYF2 | chr2 | 233612325 | + | 6260 | 590 | 279 | 4511 | 1410 |
| ENST00000435675 | ATIC | chr2 | 216184454 | + | ENST00000373566 | GIGYF2 | chr2 | 233612325 | + | 8318 | 678 | 391 | 4602 | 1403 |
| ENST00000435675 | ATIC | chr2 | 216184454 | + | ENST00000373563 | GIGYF2 | chr2 | 233612325 | + | 6289 | 678 | 391 | 4536 | 1381 |
| ENST00000435675 | ATIC | chr2 | 216184454 | + | ENST00000409480 | GIGYF2 | chr2 | 233612325 | + | 6351 | 678 | 391 | 4602 | 1403 |
| ENST00000435675 | ATIC | chr2 | 216184454 | + | ENST00000409547 | GIGYF2 | chr2 | 233612325 | + | 6304 | 678 | 391 | 4536 | 1381 |
| ENST00000435675 | ATIC | chr2 | 216184454 | + | ENST00000409196 | GIGYF2 | chr2 | 233612325 | + | 6286 | 678 | 391 | 4518 | 1375 |
| ENST00000435675 | ATIC | chr2 | 216184454 | + | ENST00000409451 | GIGYF2 | chr2 | 233612325 | + | 6348 | 678 | 391 | 4599 | 1402 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000236959 | ENST00000373566 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.001053311 | 0.9989467 |
| ENST00000236959 | ENST00000373563 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003477236 | 0.9965228 |
| ENST00000236959 | ENST00000409480 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003458201 | 0.9965418 |
| ENST00000236959 | ENST00000409547 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003478834 | 0.99652123 |
| ENST00000236959 | ENST00000409196 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003392623 | 0.9966074 |
| ENST00000236959 | ENST00000409451 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.004141981 | 0.995858 |
| ENST00000540518 | ENST00000373566 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.001224655 | 0.9987753 |
| ENST00000540518 | ENST00000373563 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003968561 | 0.99603146 |
| ENST00000540518 | ENST00000409480 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003946721 | 0.9960532 |
| ENST00000540518 | ENST00000409547 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003970339 | 0.99602973 |
| ENST00000540518 | ENST00000409196 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003871049 | 0.9961289 |
| ENST00000540518 | ENST00000409451 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.004694712 | 0.9953053 |
| ENST00000435675 | ENST00000373566 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.001130312 | 0.9988697 |
| ENST00000435675 | ENST00000373563 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003567767 | 0.9964322 |
| ENST00000435675 | ENST00000409480 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.00355906 | 0.99644095 |
| ENST00000435675 | ENST00000409547 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003570284 | 0.99642974 |
| ENST00000435675 | ENST00000409196 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.003486554 | 0.9965134 |
| ENST00000435675 | ENST00000409451 | ATIC | chr2 | 216184454 | + | GIGYF2 | chr2 | 233612325 | + | 0.004228185 | 0.9957718 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >7630_7630_1_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000236959_GIGYF2_chr2_233612325_ENST00000373563_length(amino acids)=1402AA_BP=116 MRTWCRRCCLPLALNPVPAAMAPGQLALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHP AVHAGILARNIPEDNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLP PLALVPFTEEEQRNFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDR RFEKPGRKDVGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREHMERRRRFEF DFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAKE PDKTNKKEGEKTDRVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQPL SQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSIP LMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQELT ALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEGG SVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRRQ REQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKEL EVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQQ QQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIWS NADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPTF VSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTNQ -------------------------------------------------------------- >7630_7630_2_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000236959_GIGYF2_chr2_233612325_ENST00000373566_length(amino acids)=1424AA_BP=116 MRTWCRRCCLPLALNPVPAAMAPGQLALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHP AVHAGILARNIPEDNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLP PLALVPFTEEEQRNFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDR RFEKPGRKDVGKKNGYYCMYSPVLLLGQPLCQGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPH SPDGPRSAGWREHMERRRRFEFDFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDF RPVDEGEECSDSEGSHNEEAKEPDKTNKKEGEKTDRVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRME NSLPAKVPSRGDEMVADVQQPLSQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDS ALDDERLASKLQEHRAKGVSIPLMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPG PAPPPHMGELDQERLTRQQELTALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSV PDTGSIWELQPTASQPTVWEGGSVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEE ERKRREEEELARRKQEEALRRQREQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENR LRMEEEAARLRHEEEERKRKELEVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEE RERQLREEQRRQQRELMKALQQQQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSV WGSINTGPPNQWASDLVSSIWSNADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGF TQWCEQMLHALNTANNLDVPTFVSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQP -------------------------------------------------------------- >7630_7630_3_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000236959_GIGYF2_chr2_233612325_ENST00000409196_length(amino acids)=1396AA_BP=116 MRTWCRRCCLPLALNPVPAAMAPGQLALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHP AVHAGILARNIPEDNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLP PLALVPFTEEEQRNFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDR RFEKPGRKDVGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPGWREHMERRRRFEFDFRDRD DERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAKEPDKTNK KEGEKTDRVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQPLSQIPSD TASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSIPLMHEAM QKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQELTALYQMQ HLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEGGSVWDLP LDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRRQREQEIA LRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKELEVQRQK ELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQQQQQQQQ QKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIWSNADTKN SNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPTFVSFLKE VESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTNQSNNQQS -------------------------------------------------------------- >7630_7630_4_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000236959_GIGYF2_chr2_233612325_ENST00000409451_length(amino acids)=1423AA_BP=116 MRTWCRRCCLPLALNPVPAAMAPGQLALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHP AVHAGILARNIPEDNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLP PLALVPFTEEEQRNFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDR RFEKPGRKDVGKKNGYYCMYSPVLLLGQPLCQGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPH SPDGPRSAGWREHMERRRRFEFDFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDF RPVDEGEECSDSEGSHNEEAKEPDKTNKKEGEKTDRVGVASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMEN SLPAKVPSRGDEMVADVQQPLSQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSA LDDERLASKLQEHRAKGVSIPLMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGP APPPHMGELDQERLTRQQELTALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVP DTGSIWELQPTASQPTVWEGGSVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEE RKRREEEELARRKQEEALRRQREQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRL RMEEEAARLRHEEEERKRKELEVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEER ERQLREEQRRQQRELMKALQQQQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVW GSINTGPPNQWASDLVSSIWSNADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFT QWCEQMLHALNTANNLDVPTFVSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQ -------------------------------------------------------------- >7630_7630_5_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000236959_GIGYF2_chr2_233612325_ENST00000409480_length(amino acids)=1424AA_BP=116 MRTWCRRCCLPLALNPVPAAMAPGQLALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHP AVHAGILARNIPEDNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLP PLALVPFTEEEQRNFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDR RFEKPGRKDVGKKNGYYCMYSPVLLLGQPLCQGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPH SPDGPRSAGWREHMERRRRFEFDFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDF RPVDEGEECSDSEGSHNEEAKEPDKTNKKEGEKTDRVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRME NSLPAKVPSRGDEMVADVQQPLSQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDS ALDDERLASKLQEHRAKGVSIPLMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPG PAPPPHMGELDQERLTRQQELTALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSV PDTGSIWELQPTASQPTVWEGGSVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEE ERKRREEEELARRKQEEALRRQREQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENR LRMEEEAARLRHEEEERKRKELEVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEE RERQLREEQRRQQRELMKALQQQQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSV WGSINTGPPNQWASDLVSSIWSNADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGF TQWCEQMLHALNTANNLDVPTFVSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQP -------------------------------------------------------------- >7630_7630_6_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000236959_GIGYF2_chr2_233612325_ENST00000409547_length(amino acids)=1402AA_BP=116 MRTWCRRCCLPLALNPVPAAMAPGQLALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHP AVHAGILARNIPEDNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLP PLALVPFTEEEQRNFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDR RFEKPGRKDVGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREHMERRRRFEF DFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAKE PDKTNKKEGEKTDRVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQPL SQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSIP LMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQELT ALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEGG SVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRRQ REQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKEL EVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQQ QQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIWS NADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPTF VSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTNQ -------------------------------------------------------------- >7630_7630_7_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000435675_GIGYF2_chr2_233612325_ENST00000373563_length(amino acids)=1381AA_BP=95 MSSLSALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPEDNADMARL DFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQRNFSMSVNS AAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGRPNFEEGGPT SVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREHMERRRRFEFDFRDRDDERGYRRVRSGSGSI DDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAKEPDKTNKKEGEKTDRVGVEASE ETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQPLSQIPSDTASPLLILPPPVPNP SPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSIPLMHEAMQKWYYKDPQGEIQGP FNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQELTALYQMQHLQYQQFLIQQQYAQ VLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEGGSVWDLPLDTTTPGPALEQLQQ LEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRRQREQEIALRRQREEEERQQQEE ALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKELEVQRQKELMRQRQQQQEALRR LQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQQQQQQQQQKLSGWGNVSKPSGT TKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIWSNADTKNSNMGFWDDAVKEVGP RNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPTFVSFLKEVESPYEVHDYIRAYL GDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTNQSNNQQSNFEAVQSGKKKKKQK -------------------------------------------------------------- >7630_7630_8_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000435675_GIGYF2_chr2_233612325_ENST00000373566_length(amino acids)=1403AA_BP=95 MSSLSALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPEDNADMARL DFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQRNFSMSVNS AAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGKKNGYYCMYS PVLLLGQPLCQGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREHMERRRRFE FDFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAK EPDKTNKKEGEKTDRVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQP LSQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSI PLMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQEL TALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEG GSVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRR QREQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKE LEVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQ QQQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIW SNADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPT FVSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTN -------------------------------------------------------------- >7630_7630_9_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000435675_GIGYF2_chr2_233612325_ENST00000409196_length(amino acids)=1375AA_BP=95 MSSLSALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPEDNADMARL DFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQRNFSMSVNS AAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGRPNFEEGGPT SVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPGWREHMERRRRFEFDFRDRDDERGYRRVRSGSGSIDDDRDS LPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAKEPDKTNKKEGEKTDRVGVEASEETPQTS SSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQPLSQIPSDTASPLLILPPPVPNPSPTLRP VETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSIPLMHEAMQKWYYKDPQGEIQGPFNNQEM AEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQELTALYQMQHLQYQQFLIQQQYAQVLAQQQ KAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEGGSVWDLPLDTTTPGPALEQLQQLEKAKA AKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRRQREQEIALRRQREEEERQQQEEALRRLE ERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKELEVQRQKELMRQRQQQQEALRRLQQQQQ QQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQQQQQQQQQKLSGWGNVSKPSGTTKSLLE IQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIWSNADTKNSNMGFWDDAVKEVGPRNSTNK NKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPTFVSFLKEVESPYEVHDYIRAYLGDTSEA KEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTNQSNNQQSNFEAVQSGKKKKKQKMVRADP -------------------------------------------------------------- >7630_7630_10_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000435675_GIGYF2_chr2_233612325_ENST00000409451_length(amino acids)=1402AA_BP=95 MSSLSALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPEDNADMARL DFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQRNFSMSVNS AAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGKKNGYYCMYS PVLLLGQPLCQGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREHMERRRRFE FDFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAK EPDKTNKKEGEKTDRVGVASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQPL SQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSIP LMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQELT ALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEGG SVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRRQ REQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKEL EVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQQ QQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIWS NADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPTF VSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTNQ -------------------------------------------------------------- >7630_7630_11_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000435675_GIGYF2_chr2_233612325_ENST00000409480_length(amino acids)=1403AA_BP=95 MSSLSALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPEDNADMARL DFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQRNFSMSVNS AAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGKKNGYYCMYS PVLLLGQPLCQGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREHMERRRRFE FDFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAK EPDKTNKKEGEKTDRVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQP LSQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSI PLMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQEL TALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEG GSVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRR QREQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKE LEVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQ QQQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIW SNADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPT FVSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTN -------------------------------------------------------------- >7630_7630_12_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000435675_GIGYF2_chr2_233612325_ENST00000409547_length(amino acids)=1381AA_BP=95 MSSLSALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPEDNADMARL DFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQRNFSMSVNS AAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGRPNFEEGGPT SVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREHMERRRRFEFDFRDRDDERGYRRVRSGSGSI DDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAKEPDKTNKKEGEKTDRVGVEASE ETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQPLSQIPSDTASPLLILPPPVPNP SPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSIPLMHEAMQKWYYKDPQGEIQGP FNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQELTALYQMQHLQYQQFLIQQQYAQ VLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEGGSVWDLPLDTTTPGPALEQLQQ LEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRRQREQEIALRRQREEEERQQQEE ALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKELEVQRQKELMRQRQQQQEALRR LQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQQQQQQQQQKLSGWGNVSKPSGT TKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIWSNADTKNSNMGFWDDAVKEVGP RNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPTFVSFLKEVESPYEVHDYIRAYL GDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTNQSNNQQSNFEAVQSGKKKKKQK -------------------------------------------------------------- >7630_7630_13_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000540518_GIGYF2_chr2_233612325_ENST00000373563_length(amino acids)=1389AA_BP=103 MGRPGPGLRTQGPALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPE DNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQR NFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGRP NFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREHMERRRRFEFDFRDRDDERGYRR VRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAKEPDKTNKKEGEKTD RVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQPLSQIPSDTASPLLI LPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSIPLMHEAMQKWYYKD PQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQELTALYQMQHLQYQQF LIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEGGSVWDLPLDTTTPG PALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRRQREQEIALRRQREE EERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKELEVQRQKELMRQRQ QQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQQQQQQQQQKLSGWG NVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIWSNADTKNSNMGFWD DAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPTFVSFLKEVESPYEV HDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTNQSNNQQSNFEAVQS -------------------------------------------------------------- >7630_7630_14_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000540518_GIGYF2_chr2_233612325_ENST00000373566_length(amino acids)=1411AA_BP=103 MGRPGPGLRTQGPALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPE DNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQR NFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGKK NGYYCMYSPVLLLGQPLCQGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREH MERRRRFEFDFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSE GSHNEEAKEPDKTNKKEGEKTDRVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDE MVADVQQPLSQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQE HRAKGVSIPLMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQE RLTRQQELTALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTA SQPTVWEGGSVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARR KQEEALRRQREQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHE EEERKRKELEVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQ RELMKALQQQQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWA SDLVSSIWSNADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNT ANNLDVPTFVSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHST -------------------------------------------------------------- >7630_7630_15_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000540518_GIGYF2_chr2_233612325_ENST00000409196_length(amino acids)=1383AA_BP=103 MGRPGPGLRTQGPALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPE DNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQR NFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGRP NFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPGWREHMERRRRFEFDFRDRDDERGYRRVRSGSG SIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAKEPDKTNKKEGEKTDRVGVEA SEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQPLSQIPSDTASPLLILPPPVP NPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSIPLMHEAMQKWYYKDPQGEIQ GPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQELTALYQMQHLQYQQFLIQQQY AQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEGGSVWDLPLDTTTPGPALEQL QQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRRQREQEIALRRQREEEERQQQ EEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKELEVQRQKELMRQRQQQQEAL RRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQQQQQQQQQKLSGWGNVSKPS GTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIWSNADTKNSNMGFWDDAVKEV GPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPTFVSFLKEVESPYEVHDYIRA YLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTNQSNNQQSNFEAVQSGKKKKK -------------------------------------------------------------- >7630_7630_16_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000540518_GIGYF2_chr2_233612325_ENST00000409451_length(amino acids)=1410AA_BP=103 MGRPGPGLRTQGPALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPE DNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQR NFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGKK NGYYCMYSPVLLLGQPLCQGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREH MERRRRFEFDFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSE GSHNEEAKEPDKTNKKEGEKTDRVGVASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEM VADVQQPLSQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEH RAKGVSIPLMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQER LTRQQELTALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTAS QPTVWEGGSVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRK QEEALRRQREQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEE EERKRKELEVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQR ELMKALQQQQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWAS DLVSSIWSNADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTA NNLDVPTFVSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTL -------------------------------------------------------------- >7630_7630_17_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000540518_GIGYF2_chr2_233612325_ENST00000409480_length(amino acids)=1411AA_BP=103 MGRPGPGLRTQGPALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPE DNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQR NFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGKK NGYYCMYSPVLLLGQPLCQGRPNFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREH MERRRRFEFDFRDRDDERGYRRVRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSE GSHNEEAKEPDKTNKKEGEKTDRVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDE MVADVQQPLSQIPSDTASPLLILPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQE HRAKGVSIPLMHEAMQKWYYKDPQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQE RLTRQQELTALYQMQHLQYQQFLIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTA SQPTVWEGGSVWDLPLDTTTPGPALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARR KQEEALRRQREQEIALRRQREEEERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHE EEERKRKELEVQRQKELMRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQ RELMKALQQQQQQQQQKLSGWGNVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWA SDLVSSIWSNADTKNSNMGFWDDAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNT ANNLDVPTFVSFLKEVESPYEVHDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHST -------------------------------------------------------------- >7630_7630_18_ATIC-GIGYF2_ATIC_chr2_216184454_ENST00000540518_GIGYF2_chr2_233612325_ENST00000409547_length(amino acids)=1389AA_BP=103 MGRPGPGLRTQGPALFSVSDKTGLVEFARNLTALGLNLVASGGTAKALRDAGLAVRDVSELTGFPEMLGGRVKTLHPAVHAGILARNIPE DNADMARLDFNLIRLRALSSGGSITSPPLSPALPKYKLADYRYGREEMLALFLKDNKIPSDLLDKEFLPILQEEPLPPLALVPFTEEEQR NFSMSVNSAAVLRLTGRGGGGTVVGAPRGRSSSRGRGRGRGECGFYQRSFDEVEGVFGRGGGREMHRSQSWEERGDRRFEKPGRKDVGRP NFEEGGPTSVGRKHEFIRSESENWRIFREEQNGEDEDGGWRLAGSRRDGERWRPHSPDGPRSAGWREHMERRRRFEFDFRDRDDERGYRR VRSGSGSIDDDRDSLPEWCLEDAEEEMGTFDSSGAFLSLKKVQKEPIPEEQEMDFRPVDEGEECSDSEGSHNEEAKEPDKTNKKEGEKTD RVGVEASEETPQTSSSSARPGTPSDHQSQEASQFERKDEPKTEQTEKAEEETRMENSLPAKVPSRGDEMVADVQQPLSQIPSDTASPLLI LPPPVPNPSPTLRPVETPVVGAPGMGSVSTEPDDEEGLKHLEQQAEKMVAYLQDSALDDERLASKLQEHRAKGVSIPLMHEAMQKWYYKD PQGEIQGPFNNQEMAEWFQAGYFTMSLLVKRACDESFQPLGDIMKMWGRVPFSPGPAPPPHMGELDQERLTRQQELTALYQMQHLQYQQF LIQQQYAQVLAQQQKAALSSQQQQQLALLLQQFQTLKMRISDQNIIPSVTRSVSVPDTGSIWELQPTASQPTVWEGGSVWDLPLDTTTPG PALEQLQQLEKAKAAKLEQERREAEMRAKREEEERKRQEELRRQQEEILRRQQEEERKRREEEELARRKQEEALRRQREQEIALRRQREE EERQQQEEALRRLEERRREEEERRKQEELLRKQEEEAAKWAREEEEAQRRLEENRLRMEEEAARLRHEEEERKRKELEVQRQKELMRQRQ QQQEALRRLQQQQQQQQLAQMKLPSSSTWGQQSNTTACQSQATLSLAEIQKLEEERERQLREEQRRQQRELMKALQQQQQQQQQKLSGWG NVSKPSGTTKSLLEIQQEEARQMQKQQQQQQQHQQPNRARNNTHSNLHTSIGNSVWGSINTGPPNQWASDLVSSIWSNADTKNSNMGFWD DAVKEVGPRNSTNKNKNNASLSKSVGVSNRQNKKVEEEEKLLKLFQGVNKAQDGFTQWCEQMLHALNTANNLDVPTFVSFLKEVESPYEV HDYIRAYLGDTSEAKEFAKQFLERRAKQKANQQRQQQQLPQQQQQQPPQQPPQQPQQQDSVWGMNHSTLHSVFQTNQSNNQQSNFEAVQS -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:216184454/chr2:233612325) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ATIC | GIGYF2 |
| FUNCTION: Bifunctional enzyme that catalyzes the last two steps of purine biosynthesis (PubMed:11948179, PubMed:14756554). Acts as a transformylase that incorporates a formyl group to the AMP analog AICAR (5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4-carboxamide) to produce the intermediate formyl-AICAR (FAICAR) (PubMed:9378707, PubMed:11948179, PubMed:10985775). Can use both 10-formyldihydrofolate and 10-formyltetrahydrofolate as the formyl donor in this reaction (PubMed:10985775). Also catalyzes the cyclization of FAICAR to IMP (PubMed:11948179, PubMed:14756554). Is able to convert thio-AICAR to 6-mercaptopurine ribonucleotide, an inhibitor of purine biosynthesis used in the treatment of human leukemias (PubMed:10985775). Promotes insulin receptor/INSR autophosphorylation and is involved in INSR internalization (PubMed:25687571). {ECO:0000269|PubMed:10985775, ECO:0000269|PubMed:11948179, ECO:0000269|PubMed:14756554, ECO:0000269|PubMed:25687571, ECO:0000269|PubMed:9378707}. | FUNCTION: Key component of the 4EHP-GYF2 complex, a multiprotein complex that acts as a repressor of translation initiation (PubMed:22751931, PubMed:31439631). In the 4EHP-GYF2 complex, acts as a factor that bridges EIF4E2 to ZFP36/TTP, linking translation repression with mRNA decay (PubMed:31439631). Also recruits and bridges the association of the 4EHP complex with the decapping effector protein DDX6, which is required for the ZFP36/TTP-mediated down-regulation of AU-rich mRNA (PubMed:31439631). May act cooperatively with GRB10 to regulate tyrosine kinase receptor signaling, including IGF1 and insulin receptors (PubMed:12771153). {ECO:0000269|PubMed:12771153, ECO:0000269|PubMed:22751931, ECO:0000269|PubMed:31439631}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000236959 | + | 4 | 16 | 12_14 | 96.66666666666667 | 593.0 | Nucleotide binding | IMP |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000236959 | + | 4 | 16 | 34_37 | 96.66666666666667 | 593.0 | Nucleotide binding | IMP |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000236959 | + | 4 | 16 | 64_67 | 96.66666666666667 | 593.0 | Nucleotide binding | IMP |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000435675 | + | 3 | 15 | 12_14 | 95.66666666666667 | 592.0 | Nucleotide binding | IMP |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000435675 | + | 3 | 15 | 34_37 | 95.66666666666667 | 592.0 | Nucleotide binding | IMP |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000435675 | + | 3 | 15 | 64_67 | 95.66666666666667 | 592.0 | Nucleotide binding | IMP |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000373563 | 2 | 29 | 106_111 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000373563 | 2 | 29 | 118_272 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Arg-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000373563 | 2 | 29 | 1198_1252 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Gln-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000373563 | 2 | 29 | 436_473 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Pro-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000373563 | 2 | 29 | 607_1025 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Gln-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000373563 | 2 | 29 | 738_888 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Glu-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409451 | 3 | 31 | 106_111 | 13.666666666666666 | 1321.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409451 | 3 | 31 | 118_272 | 13.666666666666666 | 1321.0 | Compositional bias | Note=Arg-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409451 | 3 | 31 | 1198_1252 | 13.666666666666666 | 1321.0 | Compositional bias | Note=Gln-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409451 | 3 | 31 | 436_473 | 13.666666666666666 | 1321.0 | Compositional bias | Note=Pro-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409451 | 3 | 31 | 607_1025 | 13.666666666666666 | 1321.0 | Compositional bias | Note=Gln-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409451 | 3 | 31 | 738_888 | 13.666666666666666 | 1321.0 | Compositional bias | Note=Glu-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409547 | 4 | 31 | 106_111 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409547 | 4 | 31 | 118_272 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Arg-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409547 | 4 | 31 | 1198_1252 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Gln-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409547 | 4 | 31 | 436_473 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Pro-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409547 | 4 | 31 | 607_1025 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Gln-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409547 | 4 | 31 | 738_888 | 13.666666666666666 | 1300.0 | Compositional bias | Note=Glu-rich | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000373563 | 2 | 29 | 533_581 | 13.666666666666666 | 1300.0 | Domain | GYF | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409451 | 3 | 31 | 533_581 | 13.666666666666666 | 1321.0 | Domain | GYF | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409547 | 4 | 31 | 533_581 | 13.666666666666666 | 1300.0 | Domain | GYF | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000373563 | 2 | 29 | 280_310 | 13.666666666666666 | 1300.0 | Motif | DDX6 binding motif | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000373563 | 2 | 29 | 40_50 | 13.666666666666666 | 1300.0 | Motif | 4EHP-binding motif | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409451 | 3 | 31 | 280_310 | 13.666666666666666 | 1321.0 | Motif | DDX6 binding motif | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409451 | 3 | 31 | 40_50 | 13.666666666666666 | 1321.0 | Motif | 4EHP-binding motif | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409547 | 4 | 31 | 280_310 | 13.666666666666666 | 1300.0 | Motif | DDX6 binding motif | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409547 | 4 | 31 | 40_50 | 13.666666666666666 | 1300.0 | Motif | 4EHP-binding motif | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000373563 | 2 | 29 | 547_563 | 13.666666666666666 | 1300.0 | Region | Required for GRB10-binding | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409451 | 3 | 31 | 547_563 | 13.666666666666666 | 1321.0 | Region | Required for GRB10-binding | |
| Tgene | GIGYF2 | chr2:216184454 | chr2:233612325 | ENST00000409547 | 4 | 31 | 547_563 | 13.666666666666666 | 1300.0 | Region | Required for GRB10-binding |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000236959 | + | 4 | 16 | 2_146 | 96.66666666666667 | 593.0 | Domain | MGS-like |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000435675 | + | 3 | 15 | 2_146 | 95.66666666666667 | 592.0 | Domain | MGS-like |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000236959 | + | 4 | 16 | 101_102 | 96.66666666666667 | 593.0 | Nucleotide binding | IMP |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000236959 | + | 4 | 16 | 125_126 | 96.66666666666667 | 593.0 | Nucleotide binding | IMP |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000435675 | + | 3 | 15 | 101_102 | 95.66666666666667 | 592.0 | Nucleotide binding | IMP |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000435675 | + | 3 | 15 | 125_126 | 95.66666666666667 | 592.0 | Nucleotide binding | IMP |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000236959 | + | 4 | 16 | 199_592 | 96.66666666666667 | 593.0 | Region | AICAR formyltransferase |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000236959 | + | 4 | 16 | 207_208 | 96.66666666666667 | 593.0 | Region | AICAR binding |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000236959 | + | 4 | 16 | 2_198 | 96.66666666666667 | 593.0 | Region | IMP cyclohydrolase |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000236959 | + | 4 | 16 | 565_566 | 96.66666666666667 | 593.0 | Region | 10-formyltetrahydrofolate binding |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000435675 | + | 3 | 15 | 199_592 | 95.66666666666667 | 592.0 | Region | AICAR formyltransferase |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000435675 | + | 3 | 15 | 207_208 | 95.66666666666667 | 592.0 | Region | AICAR binding |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000435675 | + | 3 | 15 | 2_198 | 95.66666666666667 | 592.0 | Region | IMP cyclohydrolase |
| Hgene | ATIC | chr2:216184454 | chr2:233612325 | ENST00000435675 | + | 3 | 15 | 565_566 | 95.66666666666667 | 592.0 | Region | 10-formyltetrahydrofolate binding |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| ATIC | PSTPIP1, IKBKE, ELF3, JAK1, RADIL, STOM, HNRNPDL, SNRPA, NDUFAF1, CMBL, NME2, EGFR, SIRT7, ISG15, CDK2, CLNS1A, TYMS, OXCT1, RBBP6, CRK, TARDBP, SUMO2, TXN, ASB6, AHCY, CAPN2, CTH, FH, GMDS, MCTS1, STK26, MYO1E, OGFOD1, PAPSS1, PTMA, TATDN1, UBA1, IPO5, ISOC1, MSN, NAE1, NSFL1C, PAFAH1B1, PROSC, YWHAE, CUL7, INSR, ALDH16A1, ACOT13, ADSL, C6orf211, CTBP1, DERA, DNM1L, GLO1, HSPB1, KCNAB1, KCNAB2, PAICS, PGM1, PLIN3, SHMT1, SPR, EEF2, ELAC2, GART, RTN4, TRNT1, NTRK1, U2AF2, CDH1, MTNR1A, SERPINB5, CTNNB1, UBE2M, RNF123, AGR2, MYC, PRNP, BCAT1, DIABLO, ECH1, YWHAG, CAPNS1, LDHA, BIRC3, LMBR1L, TEX101, PINK1, LINC01554, CAV1, PRKCE, SH3GL3, SHOC2, EMC2, CIT, PRC1, IFI16, PYHIN1, FASN, SREBF2, NDN, BRD4, DDX58, OGT, ATG7, NAA40, KAT6A, CTSL, IFITM1, IFITM3, CLEC4D, CLEC4E, CCNF, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ATIC |  |
| GIGYF2 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to ATIC-GIGYF2 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ATIC-GIGYF2 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | ATIC | C0001787 | Osteoporosis, Age-Related | 1 | CTD_human |
| Hgene | ATIC | C0003873 | Rheumatoid Arthritis | 1 | CTD_human |
| Hgene | ATIC | C0013221 | Drug toxicity | 1 | CTD_human |
| Hgene | ATIC | C0029456 | Osteoporosis | 1 | CTD_human |
| Hgene | ATIC | C0029459 | Osteoporosis, Senile | 1 | CTD_human |
| Hgene | ATIC | C0041755 | Adverse reaction to drug | 1 | CTD_human |
| Hgene | ATIC | C0155003 | Blindness, Transient | 1 | CTD_human |
| Hgene | ATIC | C0221473 | Blindness, Hysterical | 1 | CTD_human |
| Hgene | ATIC | C0271215 | Blindness, Legal | 1 | CTD_human |
| Hgene | ATIC | C0339730 | Blindness, Acquired | 1 | CTD_human |
| Hgene | ATIC | C0376288 | Amaurosis | 1 | CTD_human |
| Hgene | ATIC | C0456909 | Blindness | 1 | CTD_human |
| Hgene | ATIC | C0750958 | Blindness, Monocular | 1 | CTD_human |
| Hgene | ATIC | C0751406 | Post-Traumatic Osteoporosis | 1 | CTD_human |
| Hgene | ATIC | C1837530 | AICAR Transformylase Inosine Monophosphate Cyclohydrolase Deficiency | 1 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | ATIC | C1879328 | Blindness both eyes NOS (disorder) | 1 | CTD_human |
| Hgene | ATIC | C3714756 | Intellectual Disability | 1 | GENOMICS_ENGLAND |

