| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:ATP1B1-NRG1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: ATP1B1-NRG1 | FusionPDB ID: 7804 | FusionGDB2.0 ID: 7804 | Hgene | Tgene | Gene symbol | ATP1B1 | NRG1 | Gene ID | 481 | 3084 |
| Gene name | ATPase Na+/K+ transporting subunit beta 1 | neuregulin 1 | |
| Synonyms | ATP1B | ARIA|GGF|GGF2|HGL|HRG|HRG1|HRGA|MST131|MSTP131|NDF|NRG1-IT2|SMDF | |
| Cytomap | 1q24.2 | 8p12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | sodium/potassium-transporting ATPase subunit beta-1ATPase, Na+/K+ transporting, beta 1 polypeptideBeta 1-subunit of Na(+),K(+)-ATPaseNa, K-ATPase beta-1 polypeptideadenosinetriphosphatasesodium pump subunit beta-1sodium-potassium ATPase subunit beta | pro-neuregulin-1, membrane-bound isoformacetylcholine receptor-inducing activityglial growth factor 2heregulin, alpha (45kD, ERBB2 p185-activator)neu differentiation factorpro-NRG1sensory and motor neuron derived factor | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | P05026 | Q02297 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000367813, ENST00000367815, ENST00000367816, ENST00000499679, | ENST00000520502, ENST00000523681, ENST00000539990, ENST00000287842, ENST00000287845, ENST00000338921, ENST00000341377, ENST00000356819, ENST00000405005, ENST00000519301, ENST00000520407, ENST00000521670, ENST00000523079, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 11 X 12 X 7=924 | 25 X 17 X 14=5950 |
| # samples | 16 | 27 | |
| ** MAII score | log2(16/924*10)=-2.5298209465287 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(27/5950*10)=-4.46185835603184 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: ATP1B1 [Title/Abstract] AND NRG1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | ATP1B1(169080736)-NRG1(32453346), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | ATP1B1-NRG1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ATP1B1-NRG1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. ATP1B1-NRG1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. ATP1B1-NRG1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ATP1B1 | GO:0006883 | cellular sodium ion homeostasis | 10636900|19542013 |
| Hgene | ATP1B1 | GO:0030007 | cellular potassium ion homeostasis | 10636900|19542013 |
| Hgene | ATP1B1 | GO:0032781 | positive regulation of ATPase activity | 10636900 |
| Hgene | ATP1B1 | GO:0036376 | sodium ion export across plasma membrane | 10636900|19542013 |
| Hgene | ATP1B1 | GO:0046034 | ATP metabolic process | 23954377 |
| Hgene | ATP1B1 | GO:0050821 | protein stabilization | 10636900 |
| Hgene | ATP1B1 | GO:0072659 | protein localization to plasma membrane | 18522992 |
| Hgene | ATP1B1 | GO:0086009 | membrane repolarization | 19542013 |
| Hgene | ATP1B1 | GO:1901018 | positive regulation of potassium ion transmembrane transporter activity | 10636900 |
| Hgene | ATP1B1 | GO:1903278 | positive regulation of sodium ion export across plasma membrane | 10636900 |
| Hgene | ATP1B1 | GO:1903288 | positive regulation of potassium ion import | 10636900 |
| Hgene | ATP1B1 | GO:1990573 | potassium ion import across plasma membrane | 10636900|19542013 |
| Tgene | NRG1 | GO:0003222 | ventricular trabecula myocardium morphogenesis | 17336907 |
| Tgene | NRG1 | GO:0031334 | positive regulation of protein complex assembly | 10559227 |
| Tgene | NRG1 | GO:0038127 | ERBB signaling pathway | 11389077 |
| Tgene | NRG1 | GO:0038129 | ERBB3 signaling pathway | 27353365 |
| Tgene | NRG1 | GO:0045892 | negative regulation of transcription, DNA-templated | 15073182 |
| Tgene | NRG1 | GO:0051048 | negative regulation of secretion | 10559227 |
| Tgene | NRG1 | GO:0060379 | cardiac muscle cell myoblast differentiation | 17336907 |
| Tgene | NRG1 | GO:0060956 | endocardial cell differentiation | 17336907 |
 Fusion gene breakpoints across ATP1B1 (5'-gene) Fusion gene breakpoints across ATP1B1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
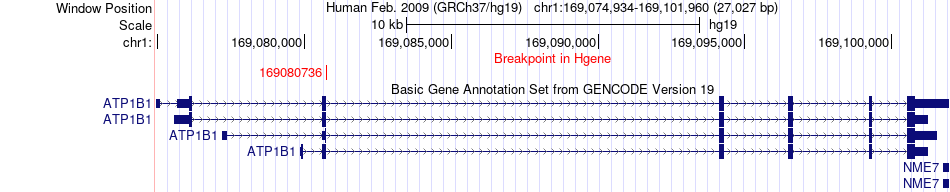 |
 Fusion gene breakpoints across NRG1 (3'-gene) Fusion gene breakpoints across NRG1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
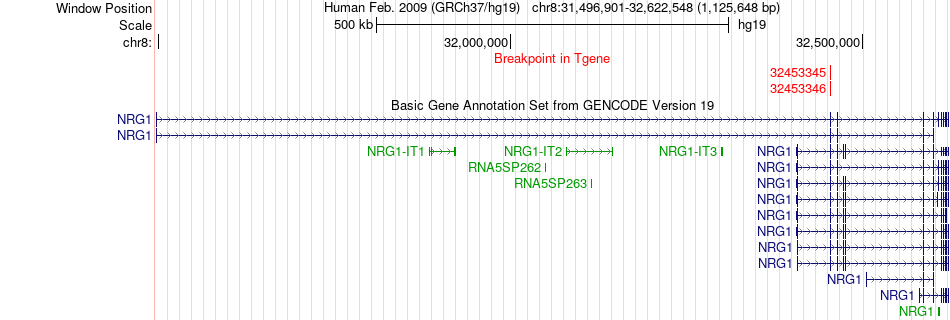 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PAAD | TCGA-3A-A9I5-01A | ATP1B1 | chr1 | 169080736 | - | NRG1 | chr8 | 32453346 | + |
| ChimerDB4 | PAAD | TCGA-3A-A9I5-01A | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + |
| ChimerDB4 | PAAD | TCGA-3A-A9I5 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + |
| ChimerDB4 | PAAD | TCGA-3A-A9I5 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + |
| ChimerKB4 | . | . | ATP1B1 | chr1 | 169080607 | + | NRG1 | chr8 | 169080607 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000519301 | NRG1 | chr8 | 32453346 | + | 2494 | 755 | 529 | 2490 | 653 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000520407 | NRG1 | chr8 | 32453346 | + | 1735 | 755 | 529 | 1278 | 249 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000523079 | NRG1 | chr8 | 32453346 | + | 2497 | 755 | 529 | 1917 | 462 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000338921 | NRG1 | chr8 | 32453346 | + | 3230 | 755 | 529 | 2601 | 690 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000356819 | NRG1 | chr8 | 32453346 | + | 3221 | 755 | 529 | 2592 | 687 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000287845 | NRG1 | chr8 | 32453346 | + | 3119 | 755 | 529 | 2490 | 653 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000341377 | NRG1 | chr8 | 32453346 | + | 3265 | 755 | 1398 | 2636 | 412 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000287842 | NRG1 | chr8 | 32453346 | + | 2678 | 755 | 529 | 2568 | 679 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000521670 | NRG1 | chr8 | 32453346 | + | 2316 | 755 | 529 | 2043 | 504 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000405005 | NRG1 | chr8 | 32453346 | + | 2647 | 755 | 529 | 2577 | 682 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000519301 | NRG1 | chr8 | 32453346 | + | 2472 | 733 | 507 | 2468 | 653 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000520407 | NRG1 | chr8 | 32453346 | + | 1713 | 733 | 507 | 1256 | 249 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000523079 | NRG1 | chr8 | 32453346 | + | 2475 | 733 | 507 | 1895 | 462 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000338921 | NRG1 | chr8 | 32453346 | + | 3208 | 733 | 507 | 2579 | 690 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000356819 | NRG1 | chr8 | 32453346 | + | 3199 | 733 | 507 | 2570 | 687 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000287845 | NRG1 | chr8 | 32453346 | + | 3097 | 733 | 507 | 2468 | 653 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000341377 | NRG1 | chr8 | 32453346 | + | 3243 | 733 | 1376 | 2614 | 412 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000287842 | NRG1 | chr8 | 32453346 | + | 2656 | 733 | 507 | 2546 | 679 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000521670 | NRG1 | chr8 | 32453346 | + | 2294 | 733 | 507 | 2021 | 504 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000405005 | NRG1 | chr8 | 32453346 | + | 2625 | 733 | 507 | 2555 | 682 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000519301 | NRG1 | chr8 | 32453346 | + | 2031 | 292 | 201 | 2027 | 608 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000520407 | NRG1 | chr8 | 32453346 | + | 1272 | 292 | 201 | 815 | 204 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000523079 | NRG1 | chr8 | 32453346 | + | 2034 | 292 | 201 | 1454 | 417 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000338921 | NRG1 | chr8 | 32453346 | + | 2767 | 292 | 201 | 2138 | 645 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000356819 | NRG1 | chr8 | 32453346 | + | 2758 | 292 | 201 | 2129 | 642 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000287845 | NRG1 | chr8 | 32453346 | + | 2656 | 292 | 201 | 2027 | 608 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000341377 | NRG1 | chr8 | 32453346 | + | 2802 | 292 | 935 | 2173 | 412 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000287842 | NRG1 | chr8 | 32453346 | + | 2215 | 292 | 201 | 2105 | 634 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000521670 | NRG1 | chr8 | 32453346 | + | 1853 | 292 | 201 | 1580 | 459 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000405005 | NRG1 | chr8 | 32453346 | + | 2184 | 292 | 201 | 2114 | 637 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000519301 | NRG1 | chr8 | 32453346 | + | 1974 | 235 | 24 | 1970 | 648 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000520407 | NRG1 | chr8 | 32453346 | + | 1215 | 235 | 24 | 758 | 244 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000523079 | NRG1 | chr8 | 32453346 | + | 1977 | 235 | 24 | 1397 | 457 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000338921 | NRG1 | chr8 | 32453346 | + | 2710 | 235 | 24 | 2081 | 685 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000356819 | NRG1 | chr8 | 32453346 | + | 2701 | 235 | 24 | 2072 | 682 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000287845 | NRG1 | chr8 | 32453346 | + | 2599 | 235 | 24 | 1970 | 648 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000341377 | NRG1 | chr8 | 32453346 | + | 2745 | 235 | 878 | 2116 | 412 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000287842 | NRG1 | chr8 | 32453346 | + | 2158 | 235 | 24 | 2048 | 674 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000521670 | NRG1 | chr8 | 32453346 | + | 1796 | 235 | 24 | 1523 | 499 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000405005 | NRG1 | chr8 | 32453346 | + | 2127 | 235 | 24 | 2057 | 677 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000519301 | NRG1 | chr8 | 32453345 | + | 2494 | 755 | 529 | 2490 | 653 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000520407 | NRG1 | chr8 | 32453345 | + | 1735 | 755 | 529 | 1278 | 249 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000523079 | NRG1 | chr8 | 32453345 | + | 2497 | 755 | 529 | 1917 | 462 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000338921 | NRG1 | chr8 | 32453345 | + | 3230 | 755 | 529 | 2601 | 690 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000356819 | NRG1 | chr8 | 32453345 | + | 3221 | 755 | 529 | 2592 | 687 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000287845 | NRG1 | chr8 | 32453345 | + | 3119 | 755 | 529 | 2490 | 653 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000341377 | NRG1 | chr8 | 32453345 | + | 3265 | 755 | 1398 | 2636 | 412 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000287842 | NRG1 | chr8 | 32453345 | + | 2678 | 755 | 529 | 2568 | 679 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000521670 | NRG1 | chr8 | 32453345 | + | 2316 | 755 | 529 | 2043 | 504 |
| ENST00000367816 | ATP1B1 | chr1 | 169080736 | + | ENST00000405005 | NRG1 | chr8 | 32453345 | + | 2647 | 755 | 529 | 2577 | 682 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000519301 | NRG1 | chr8 | 32453345 | + | 2472 | 733 | 507 | 2468 | 653 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000520407 | NRG1 | chr8 | 32453345 | + | 1713 | 733 | 507 | 1256 | 249 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000523079 | NRG1 | chr8 | 32453345 | + | 2475 | 733 | 507 | 1895 | 462 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000338921 | NRG1 | chr8 | 32453345 | + | 3208 | 733 | 507 | 2579 | 690 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000356819 | NRG1 | chr8 | 32453345 | + | 3199 | 733 | 507 | 2570 | 687 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000287845 | NRG1 | chr8 | 32453345 | + | 3097 | 733 | 507 | 2468 | 653 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000341377 | NRG1 | chr8 | 32453345 | + | 3243 | 733 | 1376 | 2614 | 412 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000287842 | NRG1 | chr8 | 32453345 | + | 2656 | 733 | 507 | 2546 | 679 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000521670 | NRG1 | chr8 | 32453345 | + | 2294 | 733 | 507 | 2021 | 504 |
| ENST00000367815 | ATP1B1 | chr1 | 169080736 | + | ENST00000405005 | NRG1 | chr8 | 32453345 | + | 2625 | 733 | 507 | 2555 | 682 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000519301 | NRG1 | chr8 | 32453345 | + | 2031 | 292 | 201 | 2027 | 608 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000520407 | NRG1 | chr8 | 32453345 | + | 1272 | 292 | 201 | 815 | 204 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000523079 | NRG1 | chr8 | 32453345 | + | 2034 | 292 | 201 | 1454 | 417 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000338921 | NRG1 | chr8 | 32453345 | + | 2767 | 292 | 201 | 2138 | 645 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000356819 | NRG1 | chr8 | 32453345 | + | 2758 | 292 | 201 | 2129 | 642 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000287845 | NRG1 | chr8 | 32453345 | + | 2656 | 292 | 201 | 2027 | 608 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000341377 | NRG1 | chr8 | 32453345 | + | 2802 | 292 | 935 | 2173 | 412 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000287842 | NRG1 | chr8 | 32453345 | + | 2215 | 292 | 201 | 2105 | 634 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000521670 | NRG1 | chr8 | 32453345 | + | 1853 | 292 | 201 | 1580 | 459 |
| ENST00000499679 | ATP1B1 | chr1 | 169080736 | + | ENST00000405005 | NRG1 | chr8 | 32453345 | + | 2184 | 292 | 201 | 2114 | 637 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000519301 | NRG1 | chr8 | 32453345 | + | 1974 | 235 | 24 | 1970 | 648 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000520407 | NRG1 | chr8 | 32453345 | + | 1215 | 235 | 24 | 758 | 244 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000523079 | NRG1 | chr8 | 32453345 | + | 1977 | 235 | 24 | 1397 | 457 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000338921 | NRG1 | chr8 | 32453345 | + | 2710 | 235 | 24 | 2081 | 685 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000356819 | NRG1 | chr8 | 32453345 | + | 2701 | 235 | 24 | 2072 | 682 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000287845 | NRG1 | chr8 | 32453345 | + | 2599 | 235 | 24 | 1970 | 648 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000341377 | NRG1 | chr8 | 32453345 | + | 2745 | 235 | 878 | 2116 | 412 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000287842 | NRG1 | chr8 | 32453345 | + | 2158 | 235 | 24 | 2048 | 674 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000521670 | NRG1 | chr8 | 32453345 | + | 1796 | 235 | 24 | 1523 | 499 |
| ENST00000367813 | ATP1B1 | chr1 | 169080736 | + | ENST00000405005 | NRG1 | chr8 | 32453345 | + | 2127 | 235 | 24 | 2057 | 677 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000367816 | ENST00000519301 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.013678157 | 0.9863218 |
| ENST00000367816 | ENST00000520407 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.028679727 | 0.9713202 |
| ENST00000367816 | ENST00000523079 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.020209659 | 0.97979033 |
| ENST00000367816 | ENST00000338921 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.006510469 | 0.9934895 |
| ENST00000367816 | ENST00000356819 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.004774384 | 0.99522555 |
| ENST00000367816 | ENST00000287845 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.005275538 | 0.99472445 |
| ENST00000367816 | ENST00000341377 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.012058015 | 0.9879419 |
| ENST00000367816 | ENST00000287842 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.008721652 | 0.99127835 |
| ENST00000367816 | ENST00000521670 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.04116651 | 0.9588335 |
| ENST00000367816 | ENST00000405005 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.011588482 | 0.9884115 |
| ENST00000367815 | ENST00000519301 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.011900788 | 0.9880993 |
| ENST00000367815 | ENST00000520407 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.028703861 | 0.97129613 |
| ENST00000367815 | ENST00000523079 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.017748041 | 0.982252 |
| ENST00000367815 | ENST00000338921 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.005681805 | 0.9943182 |
| ENST00000367815 | ENST00000356819 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.004225251 | 0.9957748 |
| ENST00000367815 | ENST00000287845 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.004689646 | 0.99531037 |
| ENST00000367815 | ENST00000341377 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.010759329 | 0.9892407 |
| ENST00000367815 | ENST00000287842 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.007574223 | 0.9924258 |
| ENST00000367815 | ENST00000521670 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.03540205 | 0.964598 |
| ENST00000367815 | ENST00000405005 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.010048028 | 0.989952 |
| ENST00000499679 | ENST00000519301 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.004223153 | 0.9957768 |
| ENST00000499679 | ENST00000520407 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.002537447 | 0.9974625 |
| ENST00000499679 | ENST00000523079 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.004644299 | 0.9953557 |
| ENST00000499679 | ENST00000338921 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.002224845 | 0.99777514 |
| ENST00000499679 | ENST00000356819 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.002537188 | 0.99746275 |
| ENST00000499679 | ENST00000287845 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.00154756 | 0.9984524 |
| ENST00000499679 | ENST00000341377 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.004114232 | 0.9958858 |
| ENST00000499679 | ENST00000287842 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.004960456 | 0.9950395 |
| ENST00000499679 | ENST00000521670 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.014567014 | 0.98543304 |
| ENST00000499679 | ENST00000405005 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.006377821 | 0.99362224 |
| ENST00000367813 | ENST00000519301 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.004125104 | 0.99587494 |
| ENST00000367813 | ENST00000520407 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.003506778 | 0.99649316 |
| ENST00000367813 | ENST00000523079 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.004263957 | 0.995736 |
| ENST00000367813 | ENST00000338921 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.001611841 | 0.9983882 |
| ENST00000367813 | ENST00000356819 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.001608754 | 0.9983912 |
| ENST00000367813 | ENST00000287845 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.001540473 | 0.9984596 |
| ENST00000367813 | ENST00000341377 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.004636632 | 0.99536335 |
| ENST00000367813 | ENST00000287842 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.003373196 | 0.99662685 |
| ENST00000367813 | ENST00000521670 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.013527229 | 0.9864727 |
| ENST00000367813 | ENST00000405005 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453346 | + | 0.003374502 | 0.99662554 |
| ENST00000367816 | ENST00000519301 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.013678157 | 0.9863218 |
| ENST00000367816 | ENST00000520407 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.028679727 | 0.9713202 |
| ENST00000367816 | ENST00000523079 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.020209659 | 0.97979033 |
| ENST00000367816 | ENST00000338921 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.006510469 | 0.9934895 |
| ENST00000367816 | ENST00000356819 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.004774384 | 0.99522555 |
| ENST00000367816 | ENST00000287845 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.005275538 | 0.99472445 |
| ENST00000367816 | ENST00000341377 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.012058015 | 0.9879419 |
| ENST00000367816 | ENST00000287842 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.008721652 | 0.99127835 |
| ENST00000367816 | ENST00000521670 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.04116651 | 0.9588335 |
| ENST00000367816 | ENST00000405005 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.011588482 | 0.9884115 |
| ENST00000367815 | ENST00000519301 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.011900788 | 0.9880993 |
| ENST00000367815 | ENST00000520407 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.028703861 | 0.97129613 |
| ENST00000367815 | ENST00000523079 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.017748041 | 0.982252 |
| ENST00000367815 | ENST00000338921 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.005681805 | 0.9943182 |
| ENST00000367815 | ENST00000356819 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.004225251 | 0.9957748 |
| ENST00000367815 | ENST00000287845 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.004689646 | 0.99531037 |
| ENST00000367815 | ENST00000341377 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.010759329 | 0.9892407 |
| ENST00000367815 | ENST00000287842 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.007574223 | 0.9924258 |
| ENST00000367815 | ENST00000521670 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.03540205 | 0.964598 |
| ENST00000367815 | ENST00000405005 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.010048028 | 0.989952 |
| ENST00000499679 | ENST00000519301 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.004223153 | 0.9957768 |
| ENST00000499679 | ENST00000520407 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.002537447 | 0.9974625 |
| ENST00000499679 | ENST00000523079 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.004644299 | 0.9953557 |
| ENST00000499679 | ENST00000338921 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.002224845 | 0.99777514 |
| ENST00000499679 | ENST00000356819 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.002537188 | 0.99746275 |
| ENST00000499679 | ENST00000287845 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.00154756 | 0.9984524 |
| ENST00000499679 | ENST00000341377 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.004114232 | 0.9958858 |
| ENST00000499679 | ENST00000287842 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.004960456 | 0.9950395 |
| ENST00000499679 | ENST00000521670 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.014567014 | 0.98543304 |
| ENST00000499679 | ENST00000405005 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.006377821 | 0.99362224 |
| ENST00000367813 | ENST00000519301 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.004125104 | 0.99587494 |
| ENST00000367813 | ENST00000520407 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.003506778 | 0.99649316 |
| ENST00000367813 | ENST00000523079 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.004263957 | 0.995736 |
| ENST00000367813 | ENST00000338921 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.001611841 | 0.9983882 |
| ENST00000367813 | ENST00000356819 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.001608754 | 0.9983912 |
| ENST00000367813 | ENST00000287845 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.001540473 | 0.9984596 |
| ENST00000367813 | ENST00000341377 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.004636632 | 0.99536335 |
| ENST00000367813 | ENST00000287842 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.003373196 | 0.99662685 |
| ENST00000367813 | ENST00000521670 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.013527229 | 0.9864727 |
| ENST00000367813 | ENST00000405005 | ATP1B1 | chr1 | 169080736 | + | NRG1 | chr8 | 32453345 | + | 0.003374502 | 0.99662554 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >7804_7804_1_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453345_ENST00000287842_length(amino acids)=674AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAE ELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVER EAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYR DSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPA HDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLA -------------------------------------------------------------- >7804_7804_2_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453345_ENST00000287845_length(amino acids)=648AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTS HLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVV AYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPS HSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTP SSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYE PAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQE -------------------------------------------------------------- >7804_7804_3_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453345_ENST00000338921_length(amino acids)=685AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQE KHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSK NVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFL RHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHP QQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGED -------------------------------------------------------------- >7804_7804_4_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453345_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_5_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453345_ENST00000356819_length(amino acids)=682AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHL GIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_6_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453345_ENST00000405005_length(amino acids)=677AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQE KAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHI VEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPD SYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHH NPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQN -------------------------------------------------------------- >7804_7804_7_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453345_ENST00000519301_length(amino acids)=648AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTS HLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVV AYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPS HSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTP SSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYE PAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQE -------------------------------------------------------------- >7804_7804_8_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453345_ENST00000520407_length(amino acids)=244AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTS -------------------------------------------------------------- >7804_7804_9_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453345_ENST00000521670_length(amino acids)=499AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQE KAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHI VEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPD -------------------------------------------------------------- >7804_7804_10_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453345_ENST00000523079_length(amino acids)=457AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAE ELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVER EAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYR -------------------------------------------------------------- >7804_7804_11_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453346_ENST00000287842_length(amino acids)=674AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAE ELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVER EAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYR DSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPA HDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLA -------------------------------------------------------------- >7804_7804_12_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453346_ENST00000287845_length(amino acids)=648AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTS HLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVV AYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPS HSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTP SSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYE PAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQE -------------------------------------------------------------- >7804_7804_13_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453346_ENST00000338921_length(amino acids)=685AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQE KHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSK NVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFL RHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHP QQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGED -------------------------------------------------------------- >7804_7804_14_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453346_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_15_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453346_ENST00000356819_length(amino acids)=682AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHL GIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_16_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453346_ENST00000405005_length(amino acids)=677AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQE KAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHI VEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPD SYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHH NPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQN -------------------------------------------------------------- >7804_7804_17_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453346_ENST00000519301_length(amino acids)=648AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTS HLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVV AYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPS HSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTP SSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYE PAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQE -------------------------------------------------------------- >7804_7804_18_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453346_ENST00000520407_length(amino acids)=244AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTS -------------------------------------------------------------- >7804_7804_19_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453346_ENST00000521670_length(amino acids)=499AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQE KAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHI VEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPD -------------------------------------------------------------- >7804_7804_20_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367813_NRG1_chr8_32453346_ENST00000523079_length(amino acids)=457AA_BP=70 MLTMGKRKCSPASQSLFLFQFQKMELKFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKL VLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPAST EGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAE ELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVER EAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYR -------------------------------------------------------------- >7804_7804_21_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453345_ENST00000287842_length(amino acids)=679AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSE HIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARET PDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSF HHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGI -------------------------------------------------------------- >7804_7804_22_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453345_ENST00000287845_length(amino acids)=653AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS TTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVG IMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTV TQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPV DFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYET TQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGR -------------------------------------------------------------- >7804_7804_23_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453345_ENST00000338921_length(amino acids)=690AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVN QYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRE CNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKK FDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDE -------------------------------------------------------------- >7804_7804_24_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453345_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_25_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453345_ENST00000356819_length(amino acids)=687AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYV SKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNS FLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDH HPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVG -------------------------------------------------------------- >7804_7804_26_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453345_ENST00000405005_length(amino acids)=682AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_27_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453345_ENST00000519301_length(amino acids)=653AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS TTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVG IMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTV TQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPV DFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYET TQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGR -------------------------------------------------------------- >7804_7804_28_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453345_ENST00000520407_length(amino acids)=249AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS -------------------------------------------------------------- >7804_7804_29_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453345_ENST00000521670_length(amino acids)=504AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA -------------------------------------------------------------- >7804_7804_30_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453345_ENST00000523079_length(amino acids)=462AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSE HIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARET -------------------------------------------------------------- >7804_7804_31_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453346_ENST00000287842_length(amino acids)=679AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSE HIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARET PDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSF HHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGI -------------------------------------------------------------- >7804_7804_32_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453346_ENST00000287845_length(amino acids)=653AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS TTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVG IMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTV TQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPV DFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYET TQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGR -------------------------------------------------------------- >7804_7804_33_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453346_ENST00000338921_length(amino acids)=690AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVN QYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRE CNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKK FDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDE -------------------------------------------------------------- >7804_7804_34_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453346_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_35_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453346_ENST00000356819_length(amino acids)=687AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYV SKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNS FLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDH HPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVG -------------------------------------------------------------- >7804_7804_36_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453346_ENST00000405005_length(amino acids)=682AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_37_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453346_ENST00000519301_length(amino acids)=653AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS TTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVG IMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTV TQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPV DFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYET TQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGR -------------------------------------------------------------- >7804_7804_38_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453346_ENST00000520407_length(amino acids)=249AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS -------------------------------------------------------------- >7804_7804_39_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453346_ENST00000521670_length(amino acids)=504AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA -------------------------------------------------------------- >7804_7804_40_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367815_NRG1_chr8_32453346_ENST00000523079_length(amino acids)=462AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSE HIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARET -------------------------------------------------------------- >7804_7804_41_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453345_ENST00000287842_length(amino acids)=679AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSE HIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARET PDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSF HHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGI -------------------------------------------------------------- >7804_7804_42_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453345_ENST00000287845_length(amino acids)=653AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS TTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVG IMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTV TQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPV DFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYET TQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGR -------------------------------------------------------------- >7804_7804_43_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453345_ENST00000338921_length(amino acids)=690AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVN QYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRE CNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKK FDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDE -------------------------------------------------------------- >7804_7804_44_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453345_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_45_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453345_ENST00000356819_length(amino acids)=687AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYV SKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNS FLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDH HPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVG -------------------------------------------------------------- >7804_7804_46_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453345_ENST00000405005_length(amino acids)=682AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_47_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453345_ENST00000519301_length(amino acids)=653AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS TTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVG IMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTV TQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPV DFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYET TQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGR -------------------------------------------------------------- >7804_7804_48_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453345_ENST00000520407_length(amino acids)=249AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS -------------------------------------------------------------- >7804_7804_49_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453345_ENST00000521670_length(amino acids)=504AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA -------------------------------------------------------------- >7804_7804_50_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453345_ENST00000523079_length(amino acids)=462AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSE HIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARET -------------------------------------------------------------- >7804_7804_51_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453346_ENST00000287842_length(amino acids)=679AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSE HIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARET PDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSF HHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGI -------------------------------------------------------------- >7804_7804_52_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453346_ENST00000287845_length(amino acids)=653AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS TTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVG IMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTV TQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPV DFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYET TQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGR -------------------------------------------------------------- >7804_7804_53_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453346_ENST00000338921_length(amino acids)=690AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVN QYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRE CNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKK FDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDE -------------------------------------------------------------- >7804_7804_54_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453346_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_55_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453346_ENST00000356819_length(amino acids)=687AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYV SKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNS FLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDH HPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVG -------------------------------------------------------------- >7804_7804_56_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453346_ENST00000405005_length(amino acids)=682AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_57_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453346_ENST00000519301_length(amino acids)=653AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS TTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVG IMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTV TQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPV DFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYET TQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGR -------------------------------------------------------------- >7804_7804_58_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453346_ENST00000520407_length(amino acids)=249AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTS -------------------------------------------------------------- >7804_7804_59_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453346_ENST00000521670_length(amino acids)=504AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA -------------------------------------------------------------- >7804_7804_60_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000367816_NRG1_chr8_32453346_ENST00000523079_length(amino acids)=462AA_BP=75 MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMAS FYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSE HIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARET -------------------------------------------------------------- >7804_7804_61_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453345_ENST00000287842_length(amino acids)=634AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHD RLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSE SHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPV SSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRR AKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARLSSVIANQD -------------------------------------------------------------- >7804_7804_62_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453345_ENST00000287845_length(amino acids)=608AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFT GDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPP PENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRL NGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVT PPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSN -------------------------------------------------------------- >7804_7804_63_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453345_ENST00000338921_length(amino acids)=645AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYC KTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSW SNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSP KSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQ EPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQ -------------------------------------------------------------- >7804_7804_64_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453345_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_65_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453345_ENST00000356819_length(amino acids)=642AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTK KQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNG HTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSP PSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPV KKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARL -------------------------------------------------------------- >7804_7804_66_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453345_ENST00000405005_length(amino acids)=637AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKK LHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESI LSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMS PPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLAN SRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARLSSVIA -------------------------------------------------------------- >7804_7804_67_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453345_ENST00000519301_length(amino acids)=608AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFT GDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPP PENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRL NGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVT PPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSN -------------------------------------------------------------- >7804_7804_68_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453345_ENST00000520407_length(amino acids)=204AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFT -------------------------------------------------------------- >7804_7804_69_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453345_ENST00000521670_length(amino acids)=459AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKK LHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESI LSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERHNLIAELRRNKAHRSKCMQIQLSATHLRSS -------------------------------------------------------------- >7804_7804_70_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453345_ENST00000523079_length(amino acids)=417AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHD RLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSE -------------------------------------------------------------- >7804_7804_71_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453346_ENST00000287842_length(amino acids)=634AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHD RLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSE SHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPV SSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRR AKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARLSSVIANQD -------------------------------------------------------------- >7804_7804_72_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453346_ENST00000287845_length(amino acids)=608AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFT GDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPP PENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRL NGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVT PPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSN -------------------------------------------------------------- >7804_7804_73_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453346_ENST00000338921_length(amino acids)=645AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYC KTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSW SNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSP KSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQ EPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQ -------------------------------------------------------------- >7804_7804_74_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453346_ENST00000341377_length(amino acids)=412AA_BP= MASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVI SSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHA RETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQF SSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPF -------------------------------------------------------------- >7804_7804_75_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453346_ENST00000356819_length(amino acids)=642AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTK KQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNG HTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSP PSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPV KKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARL -------------------------------------------------------------- >7804_7804_76_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453346_ENST00000405005_length(amino acids)=637AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKK LHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESI LSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMS PPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLAN SRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDERVGEDTPFLGIQNPLAASLEATPAFRLADSRTNPAGRFSTQEEIQARLSSVIA -------------------------------------------------------------- >7804_7804_77_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453346_ENST00000519301_length(amino acids)=608AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFT GDRCQNYVMASFYKHLGIEFMEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPP PENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRL NGTGGPRECNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVT PPRLREKKFDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSN -------------------------------------------------------------- >7804_7804_78_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453346_ENST00000520407_length(amino acids)=204AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNATSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFT -------------------------------------------------------------- >7804_7804_79_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453346_ENST00000521670_length(amino acids)=459AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMKVQNQEKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKK LHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESI LSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRECNSFLRHARETPDSYRDSPHSERHNLIAELRRNKAHRSKCMQIQLSATHLRSS -------------------------------------------------------------- >7804_7804_80_ATP1B1-NRG1_ATP1B1_chr1_169080736_ENST00000499679_NRG1_chr8_32453346_ENST00000523079_length(amino acids)=417AA_BP=30 MAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESAAGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPG KSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITGMPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCA EKEKTFCVNGGECFMVKDLSNPSRYLCKCPNEFTGDRCQNYVMASFYKAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHD RLRQSLRSERNNMMNIANGPHHPNPPPENVQLVNQYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSE -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:169080736/chr8:32453346) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ATP1B1 | NRG1 |
| FUNCTION: This is the non-catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of Na(+) and K(+) ions across the plasma membrane. The beta subunit regulates, through assembly of alpha/beta heterodimers, the number of sodium pumps transported to the plasma membrane. {ECO:0000269|PubMed:19694409}.; FUNCTION: Involved in cell adhesion and establishing epithelial cell polarity. {ECO:0000269|PubMed:19694409}. | FUNCTION: Direct ligand for ERBB3 and ERBB4 tyrosine kinase receptors. Concomitantly recruits ERBB1 and ERBB2 coreceptors, resulting in ligand-stimulated tyrosine phosphorylation and activation of the ERBB receptors. The multiple isoforms perform diverse functions such as inducing growth and differentiation of epithelial, glial, neuronal, and skeletal muscle cells; inducing expression of acetylcholine receptor in synaptic vesicles during the formation of the neuromuscular junction; stimulating lobuloalveolar budding and milk production in the mammary gland and inducing differentiation of mammary tumor cells; stimulating Schwann cell proliferation; implication in the development of the myocardium such as trabeculation of the developing heart. Isoform 10 may play a role in motor and sensory neuron development. Binds to ERBB4 (PubMed:10867024, PubMed:7902537). Binds to ERBB3 (PubMed:20682778). Acts as a ligand for integrins and binds (via EGF domain) to integrins ITGAV:ITGB3 or ITGA6:ITGB4. Its binding to integrins and subsequent ternary complex formation with integrins and ERRB3 are essential for NRG1-ERBB signaling. Induces the phosphorylation and activation of MAPK3/ERK1, MAPK1/ERK2 and AKT1 (PubMed:20682778). Ligand-dependent ERBB4 endocytosis is essential for the NRG1-mediated activation of these kinases in neurons (By similarity). {ECO:0000250|UniProtKB:P43322, ECO:0000269|PubMed:10867024, ECO:0000269|PubMed:1348215, ECO:0000269|PubMed:20682778, ECO:0000269|PubMed:7902537}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453345 | ENST00000367815 | + | 2 | 6 | 1_34 | 75.33333333333333 | 304.0 | Topological domain | Cytoplasmic |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453345 | ENST00000367816 | + | 3 | 7 | 1_34 | 75.33333333333333 | 304.0 | Topological domain | Cytoplasmic |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453346 | ENST00000367815 | + | 2 | 6 | 1_34 | 75.33333333333333 | 304.0 | Topological domain | Cytoplasmic |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453346 | ENST00000367816 | + | 3 | 7 | 1_34 | 75.33333333333333 | 304.0 | Topological domain | Cytoplasmic |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453345 | ENST00000367815 | + | 2 | 6 | 35_62 | 75.33333333333333 | 304.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453345 | ENST00000367816 | + | 3 | 7 | 35_62 | 75.33333333333333 | 304.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453346 | ENST00000367815 | + | 2 | 6 | 35_62 | 75.33333333333333 | 304.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453346 | ENST00000367816 | + | 3 | 7 | 35_62 | 75.33333333333333 | 304.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000287842 | 0 | 12 | 165_177 | 33.333333333333336 | 638.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000341377 | 0 | 13 | 165_177 | 33.333333333333336 | 525.3333333333334 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000356819 | 0 | 13 | 165_177 | 33.333333333333336 | 646.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000405005 | 0 | 12 | 165_177 | 33.333333333333336 | 641.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000519301 | 0 | 11 | 165_177 | 12.333333333333334 | 591.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520502 | 0 | 3 | 165_177 | 0.0 | 297.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000521670 | 0 | 13 | 165_177 | 33.333333333333336 | 477.3333333333333 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000523079 | 0 | 11 | 165_177 | 33.333333333333336 | 421.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000287842 | 0 | 12 | 165_177 | 33.333333333333336 | 638.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000341377 | 0 | 13 | 165_177 | 33.333333333333336 | 525.3333333333334 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000356819 | 0 | 13 | 165_177 | 33.333333333333336 | 646.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000405005 | 0 | 12 | 165_177 | 33.333333333333336 | 641.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000519301 | 0 | 11 | 165_177 | 12.333333333333334 | 591.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520502 | 0 | 3 | 165_177 | 0.0 | 297.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000521670 | 0 | 13 | 165_177 | 33.333333333333336 | 477.3333333333333 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000523079 | 0 | 11 | 165_177 | 33.333333333333336 | 421.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000287842 | 0 | 12 | 178_222 | 33.333333333333336 | 638.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000287842 | 0 | 12 | 37_128 | 33.333333333333336 | 638.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000341377 | 0 | 13 | 178_222 | 33.333333333333336 | 525.3333333333334 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000341377 | 0 | 13 | 37_128 | 33.333333333333336 | 525.3333333333334 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000356819 | 0 | 13 | 178_222 | 33.333333333333336 | 646.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000356819 | 0 | 13 | 37_128 | 33.333333333333336 | 646.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000405005 | 0 | 12 | 178_222 | 33.333333333333336 | 641.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000405005 | 0 | 12 | 37_128 | 33.333333333333336 | 641.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000519301 | 0 | 11 | 178_222 | 12.333333333333334 | 591.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000519301 | 0 | 11 | 37_128 | 12.333333333333334 | 591.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520502 | 0 | 3 | 178_222 | 0.0 | 297.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520502 | 0 | 3 | 37_128 | 0.0 | 297.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000521670 | 0 | 13 | 178_222 | 33.333333333333336 | 477.3333333333333 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000521670 | 0 | 13 | 37_128 | 33.333333333333336 | 477.3333333333333 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000523079 | 0 | 11 | 178_222 | 33.333333333333336 | 421.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000523079 | 0 | 11 | 37_128 | 33.333333333333336 | 421.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000287842 | 0 | 12 | 178_222 | 33.333333333333336 | 638.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000287842 | 0 | 12 | 37_128 | 33.333333333333336 | 638.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000341377 | 0 | 13 | 178_222 | 33.333333333333336 | 525.3333333333334 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000341377 | 0 | 13 | 37_128 | 33.333333333333336 | 525.3333333333334 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000356819 | 0 | 13 | 178_222 | 33.333333333333336 | 646.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000356819 | 0 | 13 | 37_128 | 33.333333333333336 | 646.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000405005 | 0 | 12 | 178_222 | 33.333333333333336 | 641.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000405005 | 0 | 12 | 37_128 | 33.333333333333336 | 641.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000519301 | 0 | 11 | 178_222 | 12.333333333333334 | 591.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000519301 | 0 | 11 | 37_128 | 12.333333333333334 | 591.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520502 | 0 | 3 | 178_222 | 0.0 | 297.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520502 | 0 | 3 | 37_128 | 0.0 | 297.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000521670 | 0 | 13 | 178_222 | 33.333333333333336 | 477.3333333333333 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000521670 | 0 | 13 | 37_128 | 33.333333333333336 | 477.3333333333333 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000523079 | 0 | 11 | 178_222 | 33.333333333333336 | 421.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000523079 | 0 | 11 | 37_128 | 33.333333333333336 | 421.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000287842 | 0 | 12 | 266_640 | 33.333333333333336 | 638.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000341377 | 0 | 13 | 266_640 | 33.333333333333336 | 525.3333333333334 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000356819 | 0 | 13 | 266_640 | 33.333333333333336 | 646.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000405005 | 0 | 12 | 266_640 | 33.333333333333336 | 641.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000519301 | 0 | 11 | 20_242 | 12.333333333333334 | 591.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000519301 | 0 | 11 | 266_640 | 12.333333333333334 | 591.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520407 | 0 | 5 | 266_640 | 248.33333333333334 | 423.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520502 | 0 | 3 | 20_242 | 0.0 | 297.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520502 | 0 | 3 | 266_640 | 0.0 | 297.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000521670 | 0 | 13 | 266_640 | 33.333333333333336 | 477.3333333333333 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000523079 | 0 | 11 | 266_640 | 33.333333333333336 | 421.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000287842 | 0 | 12 | 266_640 | 33.333333333333336 | 638.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000341377 | 0 | 13 | 266_640 | 33.333333333333336 | 525.3333333333334 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000356819 | 0 | 13 | 266_640 | 33.333333333333336 | 646.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000405005 | 0 | 12 | 266_640 | 33.333333333333336 | 641.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000519301 | 0 | 11 | 20_242 | 12.333333333333334 | 591.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000519301 | 0 | 11 | 266_640 | 12.333333333333334 | 591.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520407 | 0 | 5 | 266_640 | 248.33333333333334 | 423.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520502 | 0 | 3 | 20_242 | 0.0 | 297.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520502 | 0 | 3 | 266_640 | 0.0 | 297.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000521670 | 0 | 13 | 266_640 | 33.333333333333336 | 477.3333333333333 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000523079 | 0 | 11 | 266_640 | 33.333333333333336 | 421.0 | Topological domain | Cytoplasmic | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000287842 | 0 | 12 | 243_265 | 33.333333333333336 | 638.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000341377 | 0 | 13 | 243_265 | 33.333333333333336 | 525.3333333333334 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000356819 | 0 | 13 | 243_265 | 33.333333333333336 | 646.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000405005 | 0 | 12 | 243_265 | 33.333333333333336 | 641.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000519301 | 0 | 11 | 243_265 | 12.333333333333334 | 591.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520502 | 0 | 3 | 243_265 | 0.0 | 297.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000521670 | 0 | 13 | 243_265 | 33.333333333333336 | 477.3333333333333 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000523079 | 0 | 11 | 243_265 | 33.333333333333336 | 421.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000287842 | 0 | 12 | 243_265 | 33.333333333333336 | 638.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000341377 | 0 | 13 | 243_265 | 33.333333333333336 | 525.3333333333334 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000356819 | 0 | 13 | 243_265 | 33.333333333333336 | 646.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000405005 | 0 | 12 | 243_265 | 33.333333333333336 | 641.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000519301 | 0 | 11 | 243_265 | 12.333333333333334 | 591.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520502 | 0 | 3 | 243_265 | 0.0 | 297.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000521670 | 0 | 13 | 243_265 | 33.333333333333336 | 477.3333333333333 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000523079 | 0 | 11 | 243_265 | 33.333333333333336 | 421.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453345 | ENST00000367815 | + | 2 | 6 | 191_303 | 75.33333333333333 | 304.0 | Region | Note=immunoglobulin-like |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453345 | ENST00000367816 | + | 3 | 7 | 191_303 | 75.33333333333333 | 304.0 | Region | Note=immunoglobulin-like |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453346 | ENST00000367815 | + | 2 | 6 | 191_303 | 75.33333333333333 | 304.0 | Region | Note=immunoglobulin-like |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453346 | ENST00000367816 | + | 3 | 7 | 191_303 | 75.33333333333333 | 304.0 | Region | Note=immunoglobulin-like |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453345 | ENST00000367815 | + | 2 | 6 | 63_303 | 75.33333333333333 | 304.0 | Topological domain | Extracellular |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453345 | ENST00000367816 | + | 3 | 7 | 63_303 | 75.33333333333333 | 304.0 | Topological domain | Extracellular |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453346 | ENST00000367815 | + | 2 | 6 | 63_303 | 75.33333333333333 | 304.0 | Topological domain | Extracellular |
| Hgene | ATP1B1 | chr1:169080736 | chr8:32453346 | ENST00000367816 | + | 3 | 7 | 63_303 | 75.33333333333333 | 304.0 | Topological domain | Extracellular |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520407 | 0 | 5 | 165_177 | 248.33333333333334 | 423.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520407 | 0 | 5 | 165_177 | 248.33333333333334 | 423.0 | Compositional bias | Note=Ser/Thr-rich | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520407 | 0 | 5 | 178_222 | 248.33333333333334 | 423.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520407 | 0 | 5 | 37_128 | 248.33333333333334 | 423.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520407 | 0 | 5 | 178_222 | 248.33333333333334 | 423.0 | Domain | EGF-like | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520407 | 0 | 5 | 37_128 | 248.33333333333334 | 423.0 | Domain | Note=Ig-like C2-type | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000287842 | 0 | 12 | 20_242 | 33.333333333333336 | 638.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000341377 | 0 | 13 | 20_242 | 33.333333333333336 | 525.3333333333334 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000356819 | 0 | 13 | 20_242 | 33.333333333333336 | 646.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000405005 | 0 | 12 | 20_242 | 33.333333333333336 | 641.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520407 | 0 | 5 | 20_242 | 248.33333333333334 | 423.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000521670 | 0 | 13 | 20_242 | 33.333333333333336 | 477.3333333333333 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000523079 | 0 | 11 | 20_242 | 33.333333333333336 | 421.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000287842 | 0 | 12 | 20_242 | 33.333333333333336 | 638.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000341377 | 0 | 13 | 20_242 | 33.333333333333336 | 525.3333333333334 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000356819 | 0 | 13 | 20_242 | 33.333333333333336 | 646.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000405005 | 0 | 12 | 20_242 | 33.333333333333336 | 641.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520407 | 0 | 5 | 20_242 | 248.33333333333334 | 423.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000521670 | 0 | 13 | 20_242 | 33.333333333333336 | 477.3333333333333 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000523079 | 0 | 11 | 20_242 | 33.333333333333336 | 421.0 | Topological domain | Extracellular | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453345 | ENST00000520407 | 0 | 5 | 243_265 | 248.33333333333334 | 423.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence | |
| Tgene | NRG1 | chr1:169080736 | chr8:32453346 | ENST00000520407 | 0 | 5 | 243_265 | 248.33333333333334 | 423.0 | Transmembrane | Helical%3B Note%3DInternal signal sequence |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file >>>1413_ATP1B1_169080736_NRG1_32453346_ranked_0.pdb | ATP1B1 | 169080736 | 169080736 | ENST00000405005 | NRG1 | chr8 | 32453346 | + | MARGKAKEEGSWKKFIWNSEKKEFLGRTGGSWFKILLFYVIFYGCLAGIFIGTIQVMLLTISEFKPTYQDRVAPPALPPRLKEMKSQESA AGSKLVLRCETSSEYSSLRFKWFKNGNELNRKNKPQNIKIQKKPGKSELRINKASLADSGEYMCKVISKLGNDSASANITIVESNEIITG MPASTEGAYVSSESPIRISVSTEGANTSSSTSTSTTGTSHLVKCAEKEKTFCVNGGECFMVKDLSNPSRYLCKCQPGFTGARCTENVPMK VQNQEKHSGEPFPEAEELYQKRVLTITGICIALLVVGIMCVVAYCKTKKQRKKLHDRLRQSLRSERNNMMNIANGPHHPNPPPENVQLVN QYVSKNVISSEHIVEREAETSFSTSHYTSTAHHSTTVTQTPSHSWSNGHTESILSESHSVIVMSSVENSRHSSPTGGPRGRLNGTGGPRE CNSFLRHARETPDSYRDSPHSERYVSAMTTPARMSPVDFHTPSSPKSPPSEMSPPVSSMTVSMPSMAVSPFMEEERPLLLVTPPRLREKK FDHHPQQFSSFHHNPAHDSNSLPASPLRIVEDEEYETTQEYEPAQEPVKKLANSRRAKRTKPNGHIANRLEVDSNTSSQSSNSESETEDE | 690 |
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
ATP1B1_pLDDT.png |
NRG1_pLDDT.png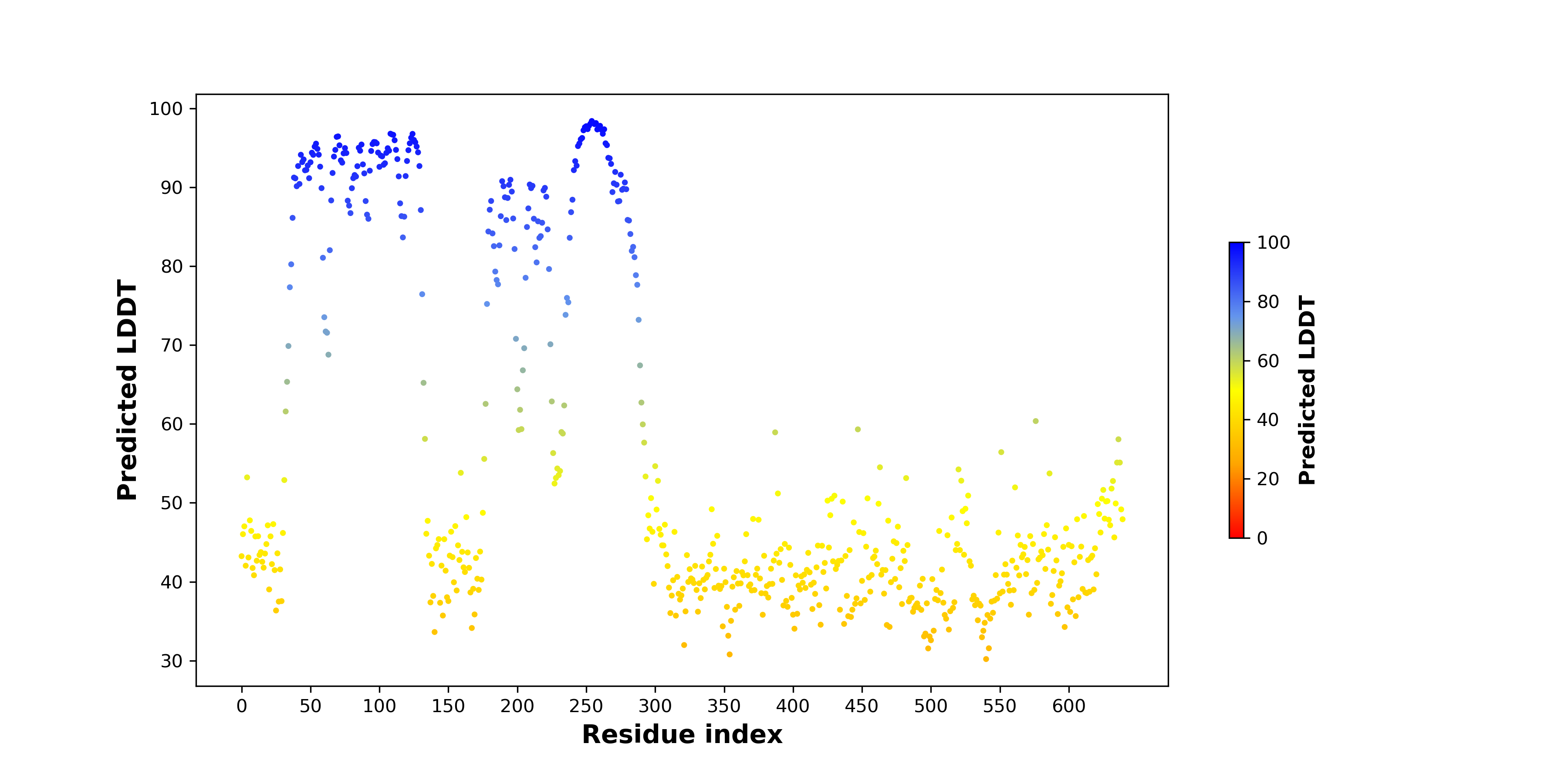 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
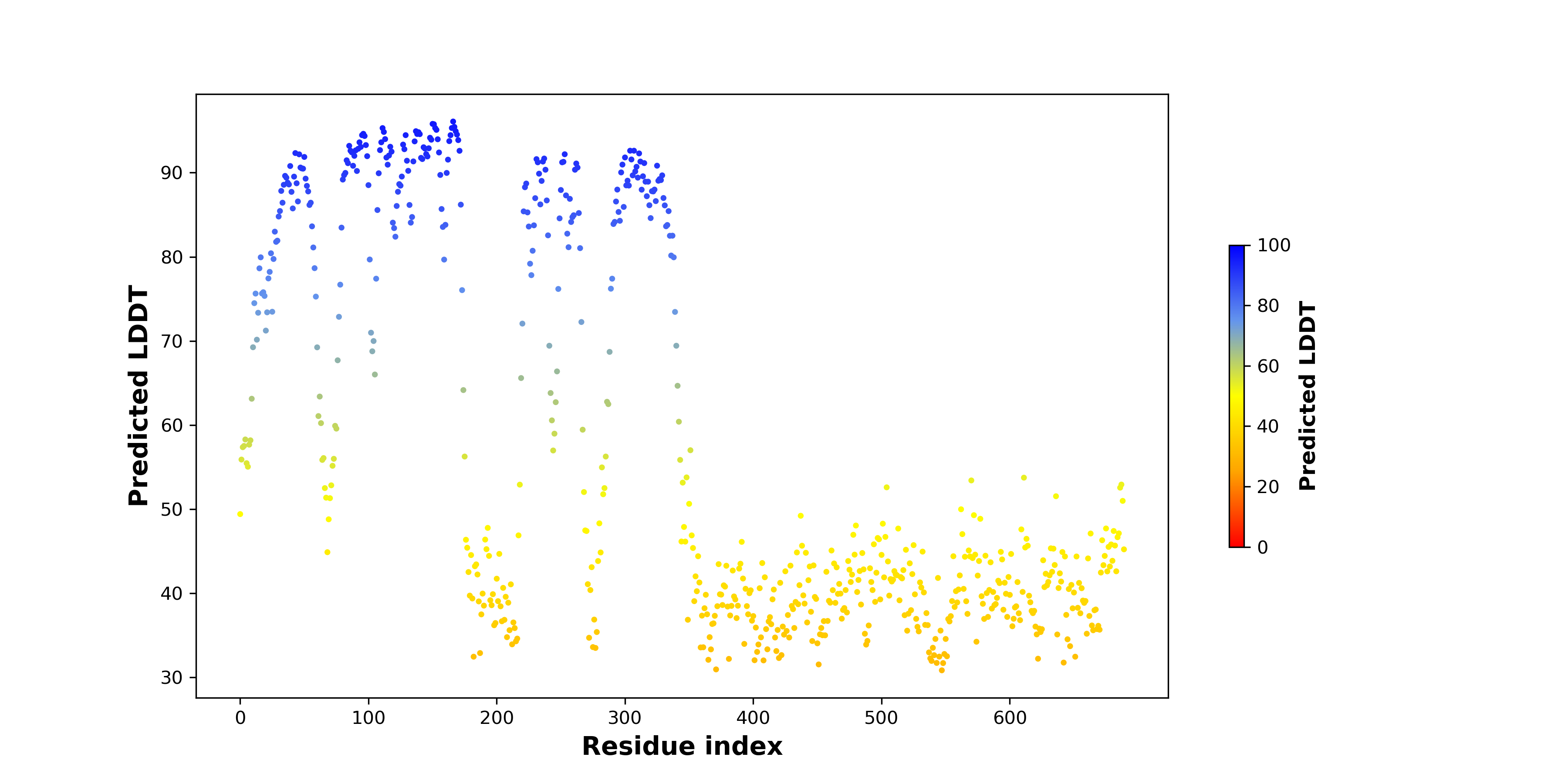 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| ATP1B1 | |
| NRG1 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to ATP1B1-NRG1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to ATP1B1-NRG1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Tgene | NRG1 | C0036341 | Schizophrenia | 7 | CTD_human |
| Tgene | NRG1 | C0005586 | Bipolar Disorder | 5 | PSYGENET |
| Tgene | NRG1 | C0024809 | Marijuana Abuse | 3 | PSYGENET |
| Tgene | NRG1 | C0011570 | Mental Depression | 2 | PSYGENET |
| Tgene | NRG1 | C0011581 | Depressive disorder | 2 | PSYGENET |
| Tgene | NRG1 | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | NRG1 | C0006870 | Cannabis Dependence | 1 | PSYGENET |
| Tgene | NRG1 | C0007621 | Neoplastic Cell Transformation | 1 | CTD_human |
| Tgene | NRG1 | C0011616 | Contact Dermatitis | 1 | CTD_human |
| Tgene | NRG1 | C0018801 | Heart failure | 1 | CTD_human |
| Tgene | NRG1 | C0018802 | Congestive heart failure | 1 | CTD_human |
| Tgene | NRG1 | C0019569 | Hirschsprung Disease | 1 | CTD_human |
| Tgene | NRG1 | C0023212 | Left-Sided Heart Failure | 1 | CTD_human |
| Tgene | NRG1 | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Tgene | NRG1 | C0024121 | Lung Neoplasms | 1 | CTD_human |
| Tgene | NRG1 | C0026650 | Movement Disorders | 1 | CTD_human |
| Tgene | NRG1 | C0027626 | Neoplasm Invasiveness | 1 | CTD_human |
| Tgene | NRG1 | C0030193 | Pain | 1 | CTD_human |
| Tgene | NRG1 | C0032460 | Polycystic Ovary Syndrome | 1 | CTD_human |
| Tgene | NRG1 | C0038358 | Gastric ulcer | 1 | CTD_human |
| Tgene | NRG1 | C0085758 | Aganglionosis, Colonic | 1 | CTD_human |
| Tgene | NRG1 | C0087031 | Juvenile-Onset Still Disease | 1 | CTD_human |
| Tgene | NRG1 | C0162351 | Contact hypersensitivity | 1 | CTD_human |
| Tgene | NRG1 | C0234230 | Pain, Burning | 1 | CTD_human |
| Tgene | NRG1 | C0234238 | Ache | 1 | CTD_human |
| Tgene | NRG1 | C0234254 | Radiating pain | 1 | CTD_human |
| Tgene | NRG1 | C0235527 | Heart Failure, Right-Sided | 1 | CTD_human |
| Tgene | NRG1 | C0236733 | Amphetamine-Related Disorders | 1 | CTD_human |
| Tgene | NRG1 | C0236804 | Amphetamine Addiction | 1 | CTD_human |
| Tgene | NRG1 | C0236807 | Amphetamine Abuse | 1 | CTD_human |
| Tgene | NRG1 | C0242379 | Malignant neoplasm of lung | 1 | CTD_human |
| Tgene | NRG1 | C0266487 | Etat Marbre | 1 | CTD_human |
| Tgene | NRG1 | C0458257 | Pain, Splitting | 1 | CTD_human |
| Tgene | NRG1 | C0458259 | Pain, Crushing | 1 | CTD_human |
| Tgene | NRG1 | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | NRG1 | C0751407 | Pain, Migratory | 1 | CTD_human |
| Tgene | NRG1 | C0751408 | Suffering, Physical | 1 | CTD_human |
| Tgene | NRG1 | C0876994 | Cardiotoxicity | 1 | CTD_human |
| Tgene | NRG1 | C1136382 | Sclerocystic Ovaries | 1 | CTD_human |
| Tgene | NRG1 | C1257840 | Aganglionosis, Rectosigmoid Colon | 1 | CTD_human |
| Tgene | NRG1 | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | NRG1 | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | NRG1 | C1959583 | Myocardial Failure | 1 | CTD_human |
| Tgene | NRG1 | C1961112 | Heart Decompensation | 1 | CTD_human |
| Tgene | NRG1 | C3495559 | Juvenile arthritis | 1 | CTD_human |
| Tgene | NRG1 | C3661523 | Congenital Intestinal Aganglionosis | 1 | CTD_human |
| Tgene | NRG1 | C3714758 | Juvenile psoriatic arthritis | 1 | CTD_human |
| Tgene | NRG1 | C4552091 | Polyarthritis, Juvenile, Rheumatoid Factor Negative | 1 | CTD_human |
| Tgene | NRG1 | C4704862 | Polyarthritis, Juvenile, Rheumatoid Factor Positive | 1 | CTD_human |
| Tgene | NRG1 | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |

