| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:SORBS1-FAM135B |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: SORBS1-FAM135B | FusionPDB ID: 85171 | FusionGDB2.0 ID: 85171 | Hgene | Tgene | Gene symbol | SORBS1 | FAM135B | Gene ID | 10580 | 51059 |
| Gene name | sorbin and SH3 domain containing 1 | family with sequence similarity 135 member B | |
| Synonyms | CAP|FLAF2|R85FL|SH3D5|SH3P12|SORB1 | C8ORFK32 | |
| Cytomap | 10q24.1 | 8q24.23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | sorbin and SH3 domain-containing protein 1Fas-ligand associated factor 2SH3 domain protein 5c-Cbl associated proteinponsin | protein FAM135B | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q49AJ0 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000277982, ENST00000306402, ENST00000347291, ENST00000353505, ENST00000354106, ENST00000361941, ENST00000371227, ENST00000371239, ENST00000371241, ENST00000371245, ENST00000371246, ENST00000371247, ENST00000371249, ENST00000393949, ENST00000607232, ENST00000474353, | ENST00000520283, ENST00000395297, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 14 X 12 X 5=840 | 4 X 6 X 4=96 |
| # samples | 16 | 5 | |
| ** MAII score | log2(16/840*10)=-2.39231742277876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/96*10)=-0.941106310946431 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: SORBS1 [Title/Abstract] AND FAM135B [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | SORBS1(97106163)-FAM135B(139255311), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | SORBS1-FAM135B seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. SORBS1-FAM135B seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. SORBS1-FAM135B seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. SORBS1-FAM135B seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. SORBS1-FAM135B seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. SORBS1-FAM135B seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. SORBS1-FAM135B seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across SORBS1 (5'-gene) Fusion gene breakpoints across SORBS1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
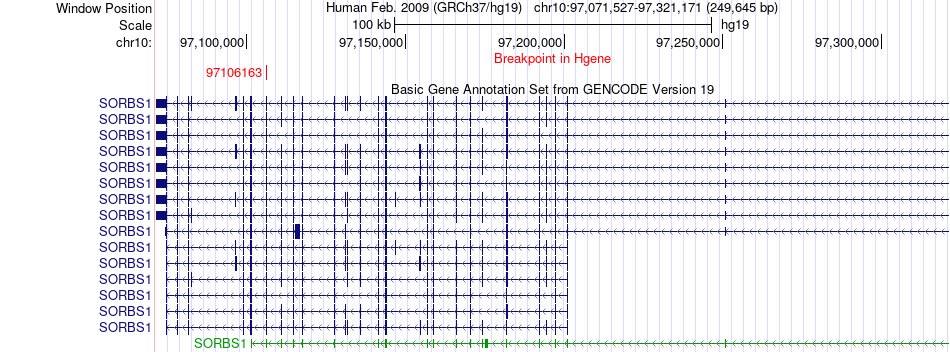 |
 Fusion gene breakpoints across FAM135B (3'-gene) Fusion gene breakpoints across FAM135B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
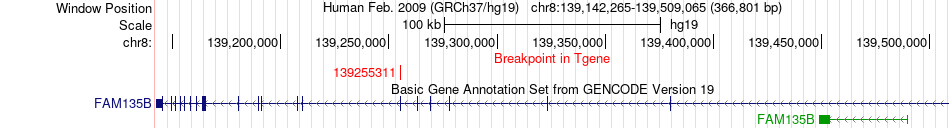 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-3B-A9HY-01A | SORBS1 | chr10 | 97106163 | - | FAM135B | chr8 | 139255311 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000371245 | SORBS1 | chr10 | 97106163 | - | ENST00000395297 | FAM135B | chr8 | 139255311 | - | 8258 | 2009 | 0 | 5687 | 1895 |
| ENST00000361941 | SORBS1 | chr10 | 97106163 | - | ENST00000395297 | FAM135B | chr8 | 139255311 | - | 8705 | 2456 | 0 | 6134 | 2044 |
| ENST00000277982 | SORBS1 | chr10 | 97106163 | - | ENST00000395297 | FAM135B | chr8 | 139255311 | - | 8771 | 2522 | 0 | 6200 | 2066 |
| ENST00000354106 | SORBS1 | chr10 | 97106163 | - | ENST00000395297 | FAM135B | chr8 | 139255311 | - | 8588 | 2339 | 0 | 6017 | 2005 |
| ENST00000371239 | SORBS1 | chr10 | 97106163 | - | ENST00000395297 | FAM135B | chr8 | 139255311 | - | 8009 | 1760 | 0 | 5438 | 1812 |
| ENST00000371241 | SORBS1 | chr10 | 97106163 | - | ENST00000395297 | FAM135B | chr8 | 139255311 | - | 7628 | 1379 | 0 | 5057 | 1685 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000371245 | ENST00000395297 | SORBS1 | chr10 | 97106163 | - | FAM135B | chr8 | 139255311 | - | 0.000476899 | 0.99952316 |
| ENST00000361941 | ENST00000395297 | SORBS1 | chr10 | 97106163 | - | FAM135B | chr8 | 139255311 | - | 0.000491572 | 0.9995084 |
| ENST00000277982 | ENST00000395297 | SORBS1 | chr10 | 97106163 | - | FAM135B | chr8 | 139255311 | - | 0.000530543 | 0.99946946 |
| ENST00000354106 | ENST00000395297 | SORBS1 | chr10 | 97106163 | - | FAM135B | chr8 | 139255311 | - | 0.000459149 | 0.9995409 |
| ENST00000371239 | ENST00000395297 | SORBS1 | chr10 | 97106163 | - | FAM135B | chr8 | 139255311 | - | 0.000443262 | 0.9995567 |
| ENST00000371241 | ENST00000395297 | SORBS1 | chr10 | 97106163 | - | FAM135B | chr8 | 139255311 | - | 0.000455635 | 0.9995443 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >85171_85171_1_SORBS1-FAM135B_SORBS1_chr10_97106163_ENST00000277982_FAM135B_chr8_139255311_ENST00000395297_length(amino acids)=2066AA_BP=441 LQTTCPATTMSSECDGGSKAVMNGLAPGSNGQDKATADPLRARSISAVKIIPVKTVKNASGLVLPTDMDLTKICTGKGAVTLRASSSYRE TPSSSPASPQETRQHESKPGLEPEPSSADEWRLSSSADANGNAQPSSLAAKGYRSVHPNLPSDKSQDATSSSAAQPEVIVVPLYLVNTDR GQEGTARPPTPLGPLGCVPTIPATASAASPLTFPTLDDFIPPHLQRWPHHSQPARASGSFAPISQTPPSFSPPPPLVPPAPEDLRRVSEP DLTGAVSSTDSSPLLNEVSSSLIGTDSQAFPSVSKPSSAYPSTTIVNPTIVLLQHNREQQKRLSSLSDPVSERRVGEQDSAPTQEKPTSP GKAIEKRAKDDSRRVVKSTQDLSDVSMDEVGIPLRNTERSKDWYKTMFKQIHKLNRDTPEENPYFPTYKFPELPEIQQTSEETKSCSVMS PRLECSGTVIAHCSLKLLDSSNPPTSASQVAGTADDDSDLYSPRYSFSEDTKSPLSVPRSKSEMSYIDGEKVVKRSATLPLPARSSSLKS SSERNDWEPPDKKVDTRKYRAEPKSIYEYQPGKSSVLTNEKMSRDISPEEIDLKNEPWYKFFSELEFGKPPPKKIWDYTPGDCSILPRED RKTNLDKDLSLCQTELEADLEKMETLNKAPSANVPQSSAISPTPEISSETPGYIYSSNFHAVKRESDGAPGDLTSLENERQIYKSVLEGG DIPLQGLSGLKRPSSSASTKDSESPRHFIPADYLESTEEFIRRRHDDKEKLLADQRRLKREQEEADIAARRHTGVIPTHHQFITNERFGD LLNIDDTAKRKSGSEMRPARAKFDFKAQTLNFTRPGRGSWLGKGGPDTGQEQSIISLENLVFGAGYCKPTSSEGSFYITSENCMQHAHKW HRDLCLLLLHAYRGLRLHFLVIMRDIPELPHTELEALAVEETLSQLCSELQMLNNPEKIAEQISKDLAWLTSHMMTLWTQFLDTVTLHSQ VTTYLTQEHHTLRVRRFSEAFFYMEHQKLAVLTFQENLIQTHSQLSLDIRNSEYLTSMPPLPAECLDIDGDWNTLPVIFEDRYVDCPATG HNLSVYPNFDVPVTSPTIMNLKDKEDNCMVNSNLSFREDLVLSTIKPSQMDSDEEVIRCPEPGENVATQNHMDMCSESQVYISIGEFQNK AGVPEDECWTGQTSDAGTYPVADVDTSRRSPGPEDGQAPVLTYIDVKSSNKNPSRAEPLVAFNAQHESRSSRDKYGLDRTGLSKVVVGGS HQNAISSDKTTLHELSTLGKGIDQEGKMVLLSLKLTPSEPCDPLSSTLREPLDIRSSLKDSHTEEQEELSVLSGVIKRSSSIISDSGIES EPSSVAWSEARSRALELPSDREVLHPFVRRHALHRNSLEGGHTESNTSLPSGIQASLTSISSLPFEEDEREVALTKLTKSVSAPHISSPE EAAEDADTKQQDGGFAEPSDMHSKSQGSPGSCSQLCGDSGTDAGADHPLVEIVLDADNQQGPGYIDIPKGKGKQFDAQGHCLPDGRTENT PGVETKGLNLKIPRVIALENPRTRSLHRALEETPKGMPKDLNVGQQALSNSGISEVEGLSQHQVPELSCTSAADAINRNSTGQQSQSGSP CIMDDTAFNRGVNAFPEAKHKAGTVCPTVTHSVHSQVLKNQELKAGTSIMGSHLTSAETFTLDSLKAVEVVNLSVSCTATCLPFSSVPKE TPARAGFSSKQTLFPITHQPLGSFGVVSTHSSTLDEEVSERMFSFYQAKEKFKKELKIEGFLYSDLTVLASDIPYFPPEEEEENLEDGIH LVVCVHGLDGNSADLRLVKTFIELGLPGGKLDFLMSEKNQMDTFADFDTMTDRLLDEIIQHIQLYNLSISRISFIGHSLGNIIIRSVLTR PRFRYYLNKLHTFLSLSGPHLGTLYNNSTLVSTGLWLMQKLKKSGSLLQLTFRDNADLRKCFLYQLSQKTGLQYFKNVVLVASPQDRYVP -------------------------------------------------------------- >85171_85171_2_SORBS1-FAM135B_SORBS1_chr10_97106163_ENST00000354106_FAM135B_chr8_139255311_ENST00000395297_length(amino acids)=2005AA_BP=780 MSSECDGGSKAVMNGLAPGSNGQDKATADPLRARSISAVKIIPVKTVKNASGLVLPTDMDLTKICTGKGAVTLRASSSYRETPSSSPASP QETRQHESKPDEWRLSSSADANGNAQPSSLAAKGYRSVHPNLPSDKSQDATSSSAAQPEVIVVPLYLVNTDRGQEGTARPPTPLGPLGCV PTIPATASAASPLTFPTLDDFIPPHLQRWPHHSQPARASGSFAPISQTPPSFSPPPPLVPPAPEDLRRVSEPDLTGAVSSTDSSPLLNEV SSSLIGTDSQAFPSVSKPSSAYPSTTIVNPTIVLLQHNREQQKRLSSLSDPVSERRVGEQDSAPTQEKPTSPGKAIEKRAKDDSRRVVKS TQDLSDVSMDEVGIPLRNTERSKDWYKTMFKQIHKLNRDTPEENPYFPTYKFPELPEIQQTSEDDDSDLYSPRYSFSEDTKSPLSVPRSK SEMSYIDGEKVVKRSATLPLPARSSSLKSSSERNDWEPPDKKVDTRKYRAEPKSIYEYQPGKSSVLTNEKMSRDISPEEIDLKNEPWYKF FSELEFGKPPPKKIWDYTPGDCSILPREDRKTNLDKDLSLCQTELEADLEKMETLNKAPSANVPQSSAISPTPEISSETPGYIYSSNFHA VKRESDGAPGDLTSLENERQIYKSVLEGGDIPLQGLSGLKRPSSSASTKDSESPRHFIPADYLESTEEFIRRRHDDKEKLLADQRRLKRE QEEADIAARRHTGVIPTHHQFITNERFGDLLNIDDTAKRKSGSEMRPARAKFDFKAQTLNFTRPGRGSWLGKGGPDTGQEQSIISLENLV FGAGYCKPTSSEGSFYITSENCMQHAHKWHRDLCLLLLHAYRGLRLHFLVIMRDIPELPHTELEALAVEETLSQLCSELQMLNNPEKIAE QISKDLAWLTSHMMTLWTQFLDTVTLHSQVTTYLTQEHHTLRVRRFSEAFFYMEHQKLAVLTFQENLIQTHSQLSLDIRNSEYLTSMPPL PAECLDIDGDWNTLPVIFEDRYVDCPATGHNLSVYPNFDVPVTSPTIMNLKDKEDNCMVNSNLSFREDLVLSTIKPSQMDSDEEVIRCPE PGENVATQNHMDMCSESQVYISIGEFQNKAGVPEDECWTGQTSDAGTYPVADVDTSRRSPGPEDGQAPVLTYIDVKSSNKNPSRAEPLVA FNAQHESRSSRDKYGLDRTGLSKVVVGGSHQNAISSDKTTLHELSTLGKGIDQEGKMVLLSLKLTPSEPCDPLSSTLREPLDIRSSLKDS HTEEQEELSVLSGVIKRSSSIISDSGIESEPSSVAWSEARSRALELPSDREVLHPFVRRHALHRNSLEGGHTESNTSLPSGIQASLTSIS SLPFEEDEREVALTKLTKSVSAPHISSPEEAAEDADTKQQDGGFAEPSDMHSKSQGSPGSCSQLCGDSGTDAGADHPLVEIVLDADNQQG PGYIDIPKGKGKQFDAQGHCLPDGRTENTPGVETKGLNLKIPRVIALENPRTRSLHRALEETPKGMPKDLNVGQQALSNSGISEVEGLSQ HQVPELSCTSAADAINRNSTGQQSQSGSPCIMDDTAFNRGVNAFPEAKHKAGTVCPTVTHSVHSQVLKNQELKAGTSIMGSHLTSAETFT LDSLKAVEVVNLSVSCTATCLPFSSVPKETPARAGFSSKQTLFPITHQPLGSFGVVSTHSSTLDEEVSERMFSFYQAKEKFKKELKIEGF LYSDLTVLASDIPYFPPEEEEENLEDGIHLVVCVHGLDGNSADLRLVKTFIELGLPGGKLDFLMSEKNQMDTFADFDTMTDRLLDEIIQH IQLYNLSISRISFIGHSLGNIIIRSVLTRPRFRYYLNKLHTFLSLSGPHLGTLYNNSTLVSTGLWLMQKLKKSGSLLQLTFRDNADLRKC FLYQLSQKTGLQYFKNVVLVASPQDRYVPFHSARIEMCKTALKDRHTGPVYAEMINNLLGPLVEAKDCTLIRHNVFHALPNTANTLIGRA -------------------------------------------------------------- >85171_85171_3_SORBS1-FAM135B_SORBS1_chr10_97106163_ENST00000361941_FAM135B_chr8_139255311_ENST00000395297_length(amino acids)=2044AA_BP=819 LQTTCPATTMSSECDGGSKAVMNGLAPGSNGQDKATADPLRARSISAVKIIPVKTVKNASGLVLPTDMDLTKICTGKGAVTLRASSSYRE TPSSSPASPQETRQHESKPGLEPEPSSADEWRLSSSADANGNAQPSSLAAKGYRSVHPNLPSDKSQDATSSSAAQPEVIVVPLYLVNTDR GQEGTARPPTPLGPLGCVPTIPATASAASPLTFPTLDDFIPPHLQRWPHHSQPARASGSFAPISQTPPSFSPPPPLVPPAPEDLRRVSEP DLTGAVSSTDSSPLLNEVSSSLIGTDSQAFPSVSKPSSAYPSTTIVNPTIVLLQHNREQQKRLSSLSDPVSERRVGEQDSAPTQEKPTSP GKAIEKRAKDDSRRVVKSTQDLSDVSMDEVGIPLRNTERSKDWYKTMFKQIHKLNRDTPEENPYFPTYKFPELPEIQQTSEEDNPYTPTY QFPASTPSPKSEDDDSDLYSPRYSFSEDTKSPLSVPRSKSEMSYIDGEKVVKRSATLPLPARSSSLKSSSERNDWEPPDKKVDTRKYRAE PKSIYEYQPGKSSVLTNEKMSRDISPEEIDLKNEPWYKFFSELEFGKPPPKKIWDYTPGDCSILPREDRKTNLDKDLSLCQTELEADLEK METLNKAPSANVPQSSAISPTPEISSETPGYIYSSNFHAVKRESDGAPGDLTSLENERQIYKSVLEGGDIPLQGLSGLKRPSSSASTKDS ESPRHFIPADYLESTEEFIRRRHDDKEKLLADQRRLKREQEEADIAARRHTGVIPTHHQFITNERFGDLLNIDDTAKRKSGSEMRPARAK FDFKAQTLNFTRPGRGSWLGKGGPDTGQEQSIISLENLVFGAGYCKPTSSEGSFYITSENCMQHAHKWHRDLCLLLLHAYRGLRLHFLVI MRDIPELPHTELEALAVEETLSQLCSELQMLNNPEKIAEQISKDLAWLTSHMMTLWTQFLDTVTLHSQVTTYLTQEHHTLRVRRFSEAFF YMEHQKLAVLTFQENLIQTHSQLSLDIRNSEYLTSMPPLPAECLDIDGDWNTLPVIFEDRYVDCPATGHNLSVYPNFDVPVTSPTIMNLK DKEDNCMVNSNLSFREDLVLSTIKPSQMDSDEEVIRCPEPGENVATQNHMDMCSESQVYISIGEFQNKAGVPEDECWTGQTSDAGTYPVA DVDTSRRSPGPEDGQAPVLTYIDVKSSNKNPSRAEPLVAFNAQHESRSSRDKYGLDRTGLSKVVVGGSHQNAISSDKTTLHELSTLGKGI DQEGKMVLLSLKLTPSEPCDPLSSTLREPLDIRSSLKDSHTEEQEELSVLSGVIKRSSSIISDSGIESEPSSVAWSEARSRALELPSDRE VLHPFVRRHALHRNSLEGGHTESNTSLPSGIQASLTSISSLPFEEDEREVALTKLTKSVSAPHISSPEEAAEDADTKQQDGGFAEPSDMH SKSQGSPGSCSQLCGDSGTDAGADHPLVEIVLDADNQQGPGYIDIPKGKGKQFDAQGHCLPDGRTENTPGVETKGLNLKIPRVIALENPR TRSLHRALEETPKGMPKDLNVGQQALSNSGISEVEGLSQHQVPELSCTSAADAINRNSTGQQSQSGSPCIMDDTAFNRGVNAFPEAKHKA GTVCPTVTHSVHSQVLKNQELKAGTSIMGSHLTSAETFTLDSLKAVEVVNLSVSCTATCLPFSSVPKETPARAGFSSKQTLFPITHQPLG SFGVVSTHSSTLDEEVSERMFSFYQAKEKFKKELKIEGFLYSDLTVLASDIPYFPPEEEEENLEDGIHLVVCVHGLDGNSADLRLVKTFI ELGLPGGKLDFLMSEKNQMDTFADFDTMTDRLLDEIIQHIQLYNLSISRISFIGHSLGNIIIRSVLTRPRFRYYLNKLHTFLSLSGPHLG TLYNNSTLVSTGLWLMQKLKKSGSLLQLTFRDNADLRKCFLYQLSQKTGLQYFKNVVLVASPQDRYVPFHSARIEMCKTALKDRHTGPVY -------------------------------------------------------------- >85171_85171_4_SORBS1-FAM135B_SORBS1_chr10_97106163_ENST00000371239_FAM135B_chr8_139255311_ENST00000395297_length(amino acids)=1812AA_BP=587 MSSECDGGSKAVMNGLAPGSNGQDKDMDLTKICTGKGAVTLRASSSYRETPSSSPASPQETRQHESKPGLEPEPSSADEWRLSSSADANG NAQPSSLAAKGYRSVHPNLPSDKSQDSSPLLNEVSSSLIGTDSQAFPSVSKPSSAYPSTTIVNPTIVLLQHNREQQKRLSSLSDPVSERR VGEQDSAPTQEKPTSPGKAIEKRAKDDSRRVVKSTQDLSDVSMDEVGIPLRNTERSKDWYKTMFKQIHKLNRDDDSDLYSPRYSFSEDTK SPLSVPRSKSEMSYIDGEKVVKRSATLPLPARSSSLKSSSERNDWEPPDKKVDTRKYRAEPKSIYEYQPGKSSVLTNEKMSRDISPEEID LKNEPWYKFFSELEFGKPTNLDKDLSLCQTELEADLEKMETLNKAPSANVPQSSAISPTPEISSETPGYIYSSNFHAVKRESDGAPGDLT SLENERQIYKSVLEGGDIPLQGLSGLKRPSSSASTKDSESPRHFIPADYLESTEEFIRRRHDDKEKLLADQRRLKREQEEADIAARRHTG VIPTHHQFITNERFGDLLNIDDTAKRKSGSEMRPARAKFDFKAQTLNFTRPGRGSWLGKGGPDTGQEQSIISLENLVFGAGYCKPTSSEG SFYITSENCMQHAHKWHRDLCLLLLHAYRGLRLHFLVIMRDIPELPHTELEALAVEETLSQLCSELQMLNNPEKIAEQISKDLAWLTSHM MTLWTQFLDTVTLHSQVTTYLTQEHHTLRVRRFSEAFFYMEHQKLAVLTFQENLIQTHSQLSLDIRNSEYLTSMPPLPAECLDIDGDWNT LPVIFEDRYVDCPATGHNLSVYPNFDVPVTSPTIMNLKDKEDNCMVNSNLSFREDLVLSTIKPSQMDSDEEVIRCPEPGENVATQNHMDM CSESQVYISIGEFQNKAGVPEDECWTGQTSDAGTYPVADVDTSRRSPGPEDGQAPVLTYIDVKSSNKNPSRAEPLVAFNAQHESRSSRDK YGLDRTGLSKVVVGGSHQNAISSDKTTLHELSTLGKGIDQEGKMVLLSLKLTPSEPCDPLSSTLREPLDIRSSLKDSHTEEQEELSVLSG VIKRSSSIISDSGIESEPSSVAWSEARSRALELPSDREVLHPFVRRHALHRNSLEGGHTESNTSLPSGIQASLTSISSLPFEEDEREVAL TKLTKSVSAPHISSPEEAAEDADTKQQDGGFAEPSDMHSKSQGSPGSCSQLCGDSGTDAGADHPLVEIVLDADNQQGPGYIDIPKGKGKQ FDAQGHCLPDGRTENTPGVETKGLNLKIPRVIALENPRTRSLHRALEETPKGMPKDLNVGQQALSNSGISEVEGLSQHQVPELSCTSAAD AINRNSTGQQSQSGSPCIMDDTAFNRGVNAFPEAKHKAGTVCPTVTHSVHSQVLKNQELKAGTSIMGSHLTSAETFTLDSLKAVEVVNLS VSCTATCLPFSSVPKETPARAGFSSKQTLFPITHQPLGSFGVVSTHSSTLDEEVSERMFSFYQAKEKFKKELKIEGFLYSDLTVLASDIP YFPPEEEEENLEDGIHLVVCVHGLDGNSADLRLVKTFIELGLPGGKLDFLMSEKNQMDTFADFDTMTDRLLDEIIQHIQLYNLSISRISF IGHSLGNIIIRSVLTRPRFRYYLNKLHTFLSLSGPHLGTLYNNSTLVSTGLWLMQKLKKSGSLLQLTFRDNADLRKCFLYQLSQKTGLQY FKNVVLVASPQDRYVPFHSARIEMCKTALKDRHTGPVYAEMINNLLGPLVEAKDCTLIRHNVFHALPNTANTLIGRAAHIAVLDSELFLE -------------------------------------------------------------- >85171_85171_5_SORBS1-FAM135B_SORBS1_chr10_97106163_ENST00000371241_FAM135B_chr8_139255311_ENST00000395297_length(amino acids)=1685AA_BP=460 MSSECDGGSKAVMNGLAPGSNGQDKDMDLTKICTGKGAVTLRASSSYRETPSSSPASPQETRQHESKPDEWRLSSSADANGNAQPSSLAA KGYRSVHPNLPSDKSQDSSPLLNEVSSSLIGTDSQAFPSVSKPSSAYPSTTIVNPTIVLLQHNREQQKRLSSLSDPVSERRVGEQDSAPT QEKPTSPGKAIEKRAKDDSRRVVKSTQDLSDVSMDEVGIPLRNTERSKDWYKTMFKQIHKLNRDDDSDLYSPRYSFSEDTKSPLSVPRSK SEMSYIDGEKVVKRSATLPLPARSSSLKSSSERNDWEPPDKKVDTRKYRAEPKSIYEYQPGKSSVLTNEKMSSAISPTPEISSETPGYIY SSNFHAVKRESDGAPGDLTSLENERQIYKSVLEGGDIPLQGLSGLKRPSSSASTKDSESPRHFIPADYLESTEEFIRRRHDDKEMRPARA KFDFKAQTLNFTRPGRGSWLGKGGPDTGQEQSIISLENLVFGAGYCKPTSSEGSFYITSENCMQHAHKWHRDLCLLLLHAYRGLRLHFLV IMRDIPELPHTELEALAVEETLSQLCSELQMLNNPEKIAEQISKDLAWLTSHMMTLWTQFLDTVTLHSQVTTYLTQEHHTLRVRRFSEAF FYMEHQKLAVLTFQENLIQTHSQLSLDIRNSEYLTSMPPLPAECLDIDGDWNTLPVIFEDRYVDCPATGHNLSVYPNFDVPVTSPTIMNL KDKEDNCMVNSNLSFREDLVLSTIKPSQMDSDEEVIRCPEPGENVATQNHMDMCSESQVYISIGEFQNKAGVPEDECWTGQTSDAGTYPV ADVDTSRRSPGPEDGQAPVLTYIDVKSSNKNPSRAEPLVAFNAQHESRSSRDKYGLDRTGLSKVVVGGSHQNAISSDKTTLHELSTLGKG IDQEGKMVLLSLKLTPSEPCDPLSSTLREPLDIRSSLKDSHTEEQEELSVLSGVIKRSSSIISDSGIESEPSSVAWSEARSRALELPSDR EVLHPFVRRHALHRNSLEGGHTESNTSLPSGIQASLTSISSLPFEEDEREVALTKLTKSVSAPHISSPEEAAEDADTKQQDGGFAEPSDM HSKSQGSPGSCSQLCGDSGTDAGADHPLVEIVLDADNQQGPGYIDIPKGKGKQFDAQGHCLPDGRTENTPGVETKGLNLKIPRVIALENP RTRSLHRALEETPKGMPKDLNVGQQALSNSGISEVEGLSQHQVPELSCTSAADAINRNSTGQQSQSGSPCIMDDTAFNRGVNAFPEAKHK AGTVCPTVTHSVHSQVLKNQELKAGTSIMGSHLTSAETFTLDSLKAVEVVNLSVSCTATCLPFSSVPKETPARAGFSSKQTLFPITHQPL GSFGVVSTHSSTLDEEVSERMFSFYQAKEKFKKELKIEGFLYSDLTVLASDIPYFPPEEEEENLEDGIHLVVCVHGLDGNSADLRLVKTF IELGLPGGKLDFLMSEKNQMDTFADFDTMTDRLLDEIIQHIQLYNLSISRISFIGHSLGNIIIRSVLTRPRFRYYLNKLHTFLSLSGPHL GTLYNNSTLVSTGLWLMQKLKKSGSLLQLTFRDNADLRKCFLYQLSQKTGLQYFKNVVLVASPQDRYVPFHSARIEMCKTALKDRHTGPV -------------------------------------------------------------- >85171_85171_6_SORBS1-FAM135B_SORBS1_chr10_97106163_ENST00000371245_FAM135B_chr8_139255311_ENST00000395297_length(amino acids)=1895AA_BP=670 LQTTCPATTMSSECDGGSKAVMNGLAPGSNGQDKATADPLRARSISAVKIIPVKTVKNASGLVLPTDMDLTKICTGKGAVTLRASSSYRE TPSSSPASPQETRQHESKPGLEPEPSSADEWRLSSSADANGNAQPSSLAAKGYRSVHPNLPSDKSQRWPHHSQPARASGSFAPISQTPPS FSPPPPLVPPAPEDLRRVSEPDLTGAVSSTDSSPLLNEVSSSLIGTDSQAFPSVSKPSSAYPSTTIVNPTIVLLQHNREQQKRLSSLSDP VSERRVGEQDSAPTQEKPTSPGKAIEKRAKDDSRRVVKSTQDLSDVSMDEVGIPLRNTERSKDWYKTMFKQIHKLNRDDDSDLYSPRYSF SEDTKSPLSVPRSKSEMSYIDGEKVVKRSATLPLPARSSSLKSSSERNDWEPPDKKVDTRKYRAEPKSIYEYQPGKSSVLTNEKMSRDIS PEEIDLKNEPWYKFFSELEFGKPPPKKIWDYTPGDCSILPREDRKSSAISPTPEISSETPGYIYSSNFHAVKRESDGAPGDLTSLENERQ IYKSVLEGGDIPLQGLSGLKRPSSSASTKDSESPRHFIPADYLESTEEFIRRRHDDKEKLLADQRRLKREQEEADIAARRHTGVIPTHHQ FITNERFGDLLNIDDTAKRKSGSEMRPARAKFDFKAQTLNFTRPGRGSWLGKGGPDTGQEQSIISLENLVFGAGYCKPTSSEGSFYITSE NCMQHAHKWHRDLCLLLLHAYRGLRLHFLVIMRDIPELPHTELEALAVEETLSQLCSELQMLNNPEKIAEQISKDLAWLTSHMMTLWTQF LDTVTLHSQVTTYLTQEHHTLRVRRFSEAFFYMEHQKLAVLTFQENLIQTHSQLSLDIRNSEYLTSMPPLPAECLDIDGDWNTLPVIFED RYVDCPATGHNLSVYPNFDVPVTSPTIMNLKDKEDNCMVNSNLSFREDLVLSTIKPSQMDSDEEVIRCPEPGENVATQNHMDMCSESQVY ISIGEFQNKAGVPEDECWTGQTSDAGTYPVADVDTSRRSPGPEDGQAPVLTYIDVKSSNKNPSRAEPLVAFNAQHESRSSRDKYGLDRTG LSKVVVGGSHQNAISSDKTTLHELSTLGKGIDQEGKMVLLSLKLTPSEPCDPLSSTLREPLDIRSSLKDSHTEEQEELSVLSGVIKRSSS IISDSGIESEPSSVAWSEARSRALELPSDREVLHPFVRRHALHRNSLEGGHTESNTSLPSGIQASLTSISSLPFEEDEREVALTKLTKSV SAPHISSPEEAAEDADTKQQDGGFAEPSDMHSKSQGSPGSCSQLCGDSGTDAGADHPLVEIVLDADNQQGPGYIDIPKGKGKQFDAQGHC LPDGRTENTPGVETKGLNLKIPRVIALENPRTRSLHRALEETPKGMPKDLNVGQQALSNSGISEVEGLSQHQVPELSCTSAADAINRNST GQQSQSGSPCIMDDTAFNRGVNAFPEAKHKAGTVCPTVTHSVHSQVLKNQELKAGTSIMGSHLTSAETFTLDSLKAVEVVNLSVSCTATC LPFSSVPKETPARAGFSSKQTLFPITHQPLGSFGVVSTHSSTLDEEVSERMFSFYQAKEKFKKELKIEGFLYSDLTVLASDIPYFPPEEE EENLEDGIHLVVCVHGLDGNSADLRLVKTFIELGLPGGKLDFLMSEKNQMDTFADFDTMTDRLLDEIIQHIQLYNLSISRISFIGHSLGN IIIRSVLTRPRFRYYLNKLHTFLSLSGPHLGTLYNNSTLVSTGLWLMQKLKKSGSLLQLTFRDNADLRKCFLYQLSQKTGLQYFKNVVLV ASPQDRYVPFHSARIEMCKTALKDRHTGPVYAEMINNLLGPLVEAKDCTLIRHNVFHALPNTANTLIGRAAHIAVLDSELFLEKFFLVAG -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr10:97106163/chr8:139255311) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | FAM135B |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000277982 | - | 24 | 31 | 366_469 | 831.6666666666666 | 1152.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000306402 | - | 20 | 26 | 366_469 | 556.6666666666666 | 782.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000347291 | - | 22 | 28 | 366_469 | 621.6666666666666 | 847.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000353505 | - | 23 | 30 | 366_469 | 660.6666666666666 | 906.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000354106 | - | 22 | 28 | 366_469 | 779.6666666666666 | 1005.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000361941 | - | 24 | 31 | 366_469 | 809.6666666666666 | 1293.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371227 | - | 24 | 32 | 366_469 | 763.6666666666666 | 1267.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371239 | - | 19 | 25 | 366_469 | 586.6666666666666 | 812.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371245 | - | 21 | 28 | 366_469 | 660.6666666666666 | 906.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371246 | - | 26 | 33 | 366_469 | 831.6666666666666 | 1152.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371247 | - | 26 | 33 | 366_469 | 809.6666666666666 | 1293.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371249 | - | 19 | 25 | 366_469 | 591.6666666666666 | 817.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000393949 | - | 24 | 30 | 366_469 | 779.6666666666666 | 1005.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000607232 | - | 21 | 27 | 366_469 | 1069.6666666666667 | 1295.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000607232 | - | 21 | 27 | 793_852 | 1069.6666666666667 | 1295.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000607232 | - | 21 | 27 | 867_928 | 1069.6666666666667 | 1295.0 | Domain | SH3 2 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000277982 | - | 24 | 31 | 1231_1292 | 831.6666666666666 | 1152.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000277982 | - | 24 | 31 | 793_852 | 831.6666666666666 | 1152.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000277982 | - | 24 | 31 | 867_928 | 831.6666666666666 | 1152.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000306402 | - | 20 | 26 | 1231_1292 | 556.6666666666666 | 782.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000306402 | - | 20 | 26 | 793_852 | 556.6666666666666 | 782.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000306402 | - | 20 | 26 | 867_928 | 556.6666666666666 | 782.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000347291 | - | 22 | 28 | 1231_1292 | 621.6666666666666 | 847.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000347291 | - | 22 | 28 | 793_852 | 621.6666666666666 | 847.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000347291 | - | 22 | 28 | 867_928 | 621.6666666666666 | 847.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000353505 | - | 23 | 30 | 1231_1292 | 660.6666666666666 | 906.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000353505 | - | 23 | 30 | 793_852 | 660.6666666666666 | 906.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000353505 | - | 23 | 30 | 867_928 | 660.6666666666666 | 906.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000354106 | - | 22 | 28 | 1231_1292 | 779.6666666666666 | 1005.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000354106 | - | 22 | 28 | 793_852 | 779.6666666666666 | 1005.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000354106 | - | 22 | 28 | 867_928 | 779.6666666666666 | 1005.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000361941 | - | 24 | 31 | 1231_1292 | 809.6666666666666 | 1293.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000361941 | - | 24 | 31 | 793_852 | 809.6666666666666 | 1293.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000361941 | - | 24 | 31 | 867_928 | 809.6666666666666 | 1293.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371227 | - | 24 | 32 | 1231_1292 | 763.6666666666666 | 1267.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371227 | - | 24 | 32 | 793_852 | 763.6666666666666 | 1267.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371227 | - | 24 | 32 | 867_928 | 763.6666666666666 | 1267.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371239 | - | 19 | 25 | 1231_1292 | 586.6666666666666 | 812.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371239 | - | 19 | 25 | 793_852 | 586.6666666666666 | 812.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371239 | - | 19 | 25 | 867_928 | 586.6666666666666 | 812.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371241 | - | 15 | 21 | 1231_1292 | 459.6666666666667 | 685.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371241 | - | 15 | 21 | 366_469 | 459.6666666666667 | 685.0 | Domain | SoHo |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371241 | - | 15 | 21 | 793_852 | 459.6666666666667 | 685.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371241 | - | 15 | 21 | 867_928 | 459.6666666666667 | 685.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371245 | - | 21 | 28 | 1231_1292 | 660.6666666666666 | 906.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371245 | - | 21 | 28 | 793_852 | 660.6666666666666 | 906.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371245 | - | 21 | 28 | 867_928 | 660.6666666666666 | 906.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371246 | - | 26 | 33 | 1231_1292 | 831.6666666666666 | 1152.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371246 | - | 26 | 33 | 793_852 | 831.6666666666666 | 1152.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371246 | - | 26 | 33 | 867_928 | 831.6666666666666 | 1152.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371247 | - | 26 | 33 | 1231_1292 | 809.6666666666666 | 1293.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371247 | - | 26 | 33 | 793_852 | 809.6666666666666 | 1293.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371247 | - | 26 | 33 | 867_928 | 809.6666666666666 | 1293.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371249 | - | 19 | 25 | 1231_1292 | 591.6666666666666 | 817.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371249 | - | 19 | 25 | 793_852 | 591.6666666666666 | 817.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000371249 | - | 19 | 25 | 867_928 | 591.6666666666666 | 817.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000393949 | - | 24 | 30 | 1231_1292 | 779.6666666666666 | 1005.0 | Domain | SH3 3 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000393949 | - | 24 | 30 | 793_852 | 779.6666666666666 | 1005.0 | Domain | SH3 1 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000393949 | - | 24 | 30 | 867_928 | 779.6666666666666 | 1005.0 | Domain | SH3 2 |
| Hgene | SORBS1 | chr10:97106163 | chr8:139255311 | ENST00000607232 | - | 21 | 27 | 1231_1292 | 1069.6666666666667 | 1295.0 | Domain | SH3 3 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| SORBS1 | |
| FAM135B |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to SORBS1-FAM135B |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to SORBS1-FAM135B |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

