| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:SQSTM1-ALK |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: SQSTM1-ALK | FusionPDB ID: 86197 | FusionGDB2.0 ID: 86197 | Hgene | Tgene | Gene symbol | SQSTM1 | ALK | Gene ID | 8878 | 238 |
| Gene name | sequestosome 1 | ALK receptor tyrosine kinase | |
| Synonyms | A170|DMRV|FTDALS3|NADGP|OSIL|PDB3|ZIP3|p60|p62|p62B | CD246|NBLST3 | |
| Cytomap | 5q35.3 | 2p23.2-p23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | sequestosome-1EBI3-associated protein of 60 kDaEBI3-associated protein p60EBIAPautophagy receptor p62oxidative stress induced likephosphotyrosine independent ligand for the Lck SH2 domain p62phosphotyrosine-independent ligand for the Lck SH2 domain | ALK tyrosine kinase receptorCD246 antigenanaplastic lymphoma receptor tyrosine kinasemutant anaplastic lymphoma kinase | |
| Modification date | 20200327 | 20200329 | |
| UniProtAcc | Q13501 | Q96BT7 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000360718, ENST00000376929, ENST00000389805, ENST00000402874, ENST00000510187, ENST00000506690, | ENST00000389048, ENST00000431873, ENST00000498037, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 29 X 23 X 12=8004 | 56 X 74 X 20=82880 |
| # samples | 33 | 57 | |
| ** MAII score | log2(33/8004*10)=-4.60018323765993 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(57/82880*10)=-7.18391827352181 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: SQSTM1 [Title/Abstract] AND ALK [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | SQSTM1(179252226)-ALK(29416042), # samples:1 SQSTM1(179252226)-ALK(29446395), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | SQSTM1-ALK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. SQSTM1-ALK seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. SQSTM1-ALK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. SQSTM1-ALK seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | SQSTM1 | GO:0006914 | autophagy | 20452972 |
| Hgene | SQSTM1 | GO:0007032 | endosome organization | 27368102 |
| Hgene | SQSTM1 | GO:0031397 | negative regulation of protein ubiquitination | 20452972 |
| Hgene | SQSTM1 | GO:0061635 | regulation of protein complex stability | 25127057 |
| Hgene | SQSTM1 | GO:1905719 | protein localization to perinuclear region of cytoplasm | 27368102 |
| Tgene | ALK | GO:0016310 | phosphorylation | 9174053 |
| Tgene | ALK | GO:0046777 | protein autophosphorylation | 9174053 |
 Fusion gene breakpoints across SQSTM1 (5'-gene) Fusion gene breakpoints across SQSTM1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
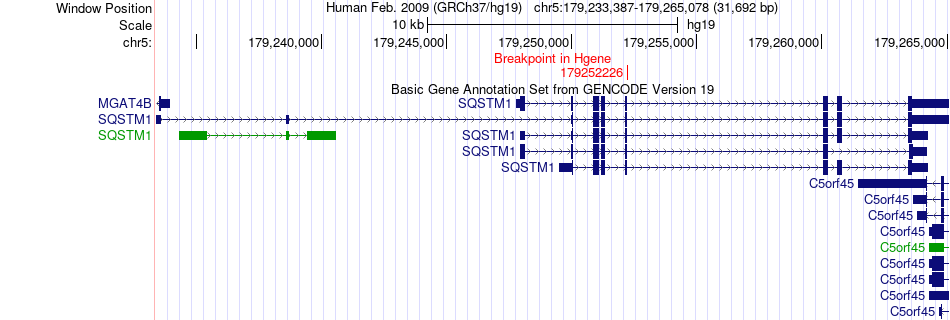 |
 Fusion gene breakpoints across ALK (3'-gene) Fusion gene breakpoints across ALK (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
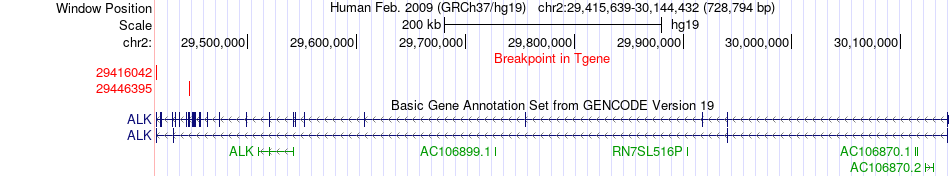 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | diffuse large B cell lymphoma;MALT lymphoma | AB583922 | SQSTM1 | chr5 | 179252226 | ALK | chr2 | 29416042 | ||
| ChimerKB3 | . | . | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29416042 | - |
| ChimerKB3 | . | . | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29446394 | - |
| ChiTaRS5.0 | N/A | AB583922 | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29446395 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000376929 | SQSTM1 | chr5 | 179252226 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3003 | 862 | 303 | 2552 | 749 |
| ENST00000389805 | SQSTM1 | chr5 | 179252226 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3073 | 932 | 178 | 2622 | 814 |
| ENST00000402874 | SQSTM1 | chr5 | 179252226 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2913 | 772 | 18 | 2462 | 814 |
| ENST00000510187 | SQSTM1 | chr5 | 179252226 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 2911 | 770 | 16 | 2460 | 814 |
| ENST00000360718 | SQSTM1 | chr5 | 179252226 | + | ENST00000389048 | ALK | chr2 | 29446394 | - | 3163 | 1022 | 334 | 2712 | 792 |
| ENST00000376929 | SQSTM1 | chr5 | 179252226 | + | ENST00000389048 | ALK | chr2 | 29446395 | - | 3003 | 862 | 303 | 2552 | 749 |
| ENST00000389805 | SQSTM1 | chr5 | 179252226 | + | ENST00000389048 | ALK | chr2 | 29446395 | - | 3073 | 932 | 178 | 2622 | 814 |
| ENST00000402874 | SQSTM1 | chr5 | 179252226 | + | ENST00000389048 | ALK | chr2 | 29446395 | - | 2913 | 772 | 18 | 2462 | 814 |
| ENST00000510187 | SQSTM1 | chr5 | 179252226 | + | ENST00000389048 | ALK | chr2 | 29446395 | - | 2911 | 770 | 16 | 2460 | 814 |
| ENST00000360718 | SQSTM1 | chr5 | 179252226 | + | ENST00000389048 | ALK | chr2 | 29446395 | - | 3163 | 1022 | 334 | 2712 | 792 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000376929 | ENST00000389048 | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29446395 | - | 0.008796504 | 0.99120355 |
| ENST00000389805 | ENST00000389048 | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29446395 | - | 0.002945122 | 0.99705493 |
| ENST00000402874 | ENST00000389048 | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29446395 | - | 0.002964661 | 0.9970354 |
| ENST00000510187 | ENST00000389048 | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29446395 | - | 0.002938603 | 0.99706143 |
| ENST00000360718 | ENST00000389048 | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29446395 | - | 0.007438368 | 0.99256164 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >86197_86197_1_SQSTM1-ALK_SQSTM1_chr5_179252226_ENST00000360718_ALK_chr2_29446394_ENST00000389048_length(amino acids)=792AA_BP=221 MPSVAQVGVFVYSPVSVPFILASAHSSSLCPAENPWVLTCCLLNNLDEDGDLVAFSSDEELTMAMSYVKDDIFRIYIKEKKECRRDHRPP CAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCEGKGLHRGHTKLAFPSPFGHLSEGFSHSRWLRKVKHGHFGWPGWEMGP PGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVAAALSPLVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCF AGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQ SLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIY RASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQH QPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTA AEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATG -------------------------------------------------------------- >86197_86197_2_SQSTM1-ALK_SQSTM1_chr5_179252226_ENST00000360718_ALK_chr2_29446395_ENST00000389048_length(amino acids)=792AA_BP=221 MPSVAQVGVFVYSPVSVPFILASAHSSSLCPAENPWVLTCCLLNNLDEDGDLVAFSSDEELTMAMSYVKDDIFRIYIKEKKECRRDHRPP CAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCEGKGLHRGHTKLAFPSPFGHLSEGFSHSRWLRKVKHGHFGWPGWEMGP PGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVAAALSPLVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCF AGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQ SLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIY RASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQH QPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTA AEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATG -------------------------------------------------------------- >86197_86197_3_SQSTM1-ALK_SQSTM1_chr5_179252226_ENST00000376929_ALK_chr2_29446394_ENST00000389048_length(amino acids)=749AA_BP=178 MRKDEDGDLVAFSSDEELTMAMSYVKDDIFRIYIKEKKECRRDHRPPCAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCE GKGLHRGHTKLAFPSPFGHLSEGFSHSRWLRKVKHGHFGWPGWEMGPPGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVA AALSPLVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPND PSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVAR DIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWE IFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKV PVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTS LWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGL -------------------------------------------------------------- >86197_86197_4_SQSTM1-ALK_SQSTM1_chr5_179252226_ENST00000376929_ALK_chr2_29446395_ENST00000389048_length(amino acids)=749AA_BP=178 MRKDEDGDLVAFSSDEELTMAMSYVKDDIFRIYIKEKKECRRDHRPPCAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCE GKGLHRGHTKLAFPSPFGHLSEGFSHSRWLRKVKHGHFGWPGWEMGPPGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVA AALSPLVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPND PSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVAR DIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWE IFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKV PVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTS LWNPTYGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGL -------------------------------------------------------------- >86197_86197_5_SQSTM1-ALK_SQSTM1_chr5_179252226_ENST00000389805_ALK_chr2_29446394_ENST00000389048_length(amino acids)=814AA_BP=243 MASLTVKAYLLGKEDAAREIRRFSFCCSPEPEAEAEAAAGPGPCERLLSRVAALFPALRPGGFQAHYRDEDGDLVAFSSDEELTMAMSYV KDDIFRIYIKEKKECRRDHRPPCAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCEGKGLHRGHTKLAFPSPFGHLSEGFS HSRWLRKVKHGHFGWPGWEMGPPGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVAAALSPLVYRRKHQELQAMQMELQSP EYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLM EALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCL LTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGG RMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEER SPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKE PHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMN -------------------------------------------------------------- >86197_86197_6_SQSTM1-ALK_SQSTM1_chr5_179252226_ENST00000389805_ALK_chr2_29446395_ENST00000389048_length(amino acids)=814AA_BP=243 MASLTVKAYLLGKEDAAREIRRFSFCCSPEPEAEAEAAAGPGPCERLLSRVAALFPALRPGGFQAHYRDEDGDLVAFSSDEELTMAMSYV KDDIFRIYIKEKKECRRDHRPPCAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCEGKGLHRGHTKLAFPSPFGHLSEGFS HSRWLRKVKHGHFGWPGWEMGPPGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVAAALSPLVYRRKHQELQAMQMELQSP EYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLM EALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCL LTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGG RMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEER SPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKE PHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMN -------------------------------------------------------------- >86197_86197_7_SQSTM1-ALK_SQSTM1_chr5_179252226_ENST00000402874_ALK_chr2_29446394_ENST00000389048_length(amino acids)=814AA_BP=243 MASLTVKAYLLGKEDAAREIRRFSFCCSPEPEAEAEAAAGPGPCERLLSRVAALFPALRPGGFQAHYRDEDGDLVAFSSDEELTMAMSYV KDDIFRIYIKEKKECRRDHRPPCAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCEGKGLHRGHTKLAFPSPFGHLSEGFS HSRWLRKVKHGHFGWPGWEMGPPGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVAAALSPLVYRRKHQELQAMQMELQSP EYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLM EALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCL LTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGG RMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEER SPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKE PHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMN -------------------------------------------------------------- >86197_86197_8_SQSTM1-ALK_SQSTM1_chr5_179252226_ENST00000402874_ALK_chr2_29446395_ENST00000389048_length(amino acids)=814AA_BP=243 MASLTVKAYLLGKEDAAREIRRFSFCCSPEPEAEAEAAAGPGPCERLLSRVAALFPALRPGGFQAHYRDEDGDLVAFSSDEELTMAMSYV KDDIFRIYIKEKKECRRDHRPPCAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCEGKGLHRGHTKLAFPSPFGHLSEGFS HSRWLRKVKHGHFGWPGWEMGPPGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVAAALSPLVYRRKHQELQAMQMELQSP EYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLM EALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCL LTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGG RMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEER SPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKE PHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMN -------------------------------------------------------------- >86197_86197_9_SQSTM1-ALK_SQSTM1_chr5_179252226_ENST00000510187_ALK_chr2_29446394_ENST00000389048_length(amino acids)=814AA_BP=243 MASLTVKAYLLGKEDAAREIRRFSFCCSPEPEAEAEAAAGPGPCERLLSRVAALFPALRPGGFQAHYRDEDGDLVAFSSDEELTMAMSYV KDDIFRIYIKEKKECRRDHRPPCAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCEGKGLHRGHTKLAFPSPFGHLSEGFS HSRWLRKVKHGHFGWPGWEMGPPGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVAAALSPLVYRRKHQELQAMQMELQSP EYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLM EALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCL LTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGG RMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEER SPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKE PHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMN -------------------------------------------------------------- >86197_86197_10_SQSTM1-ALK_SQSTM1_chr5_179252226_ENST00000510187_ALK_chr2_29446395_ENST00000389048_length(amino acids)=814AA_BP=243 MASLTVKAYLLGKEDAAREIRRFSFCCSPEPEAEAEAAAGPGPCERLLSRVAALFPALRPGGFQAHYRDEDGDLVAFSSDEELTMAMSYV KDDIFRIYIKEKKECRRDHRPPCAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCEGKGLHRGHTKLAFPSPFGHLSEGFS HSRWLRKVKHGHFGWPGWEMGPPGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVAAALSPLVYRRKHQELQAMQMELQSP EYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLM EALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCL LTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGG RMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQQAKREEER SPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKE PHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYEDTILKSKNSMN -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:179252226/chr2:29416042) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| SQSTM1 | ALK |
| FUNCTION: Autophagy receptor required for selective macroautophagy (aggrephagy). Functions as a bridge between polyubiquitinated cargo and autophagosomes. Interacts directly with both the cargo to become degraded and an autophagy modifier of the MAP1 LC3 family (PubMed:16286508, PubMed:20168092, PubMed:24128730, PubMed:28404643, PubMed:22622177). Along with WDFY3, involved in the formation and autophagic degradation of cytoplasmic ubiquitin-containing inclusions (p62 bodies, ALIS/aggresome-like induced structures). Along with WDFY3, required to recruit ubiquitinated proteins to PML bodies in the nucleus (PubMed:24128730, PubMed:20168092). May regulate the activation of NFKB1 by TNF-alpha, nerve growth factor (NGF) and interleukin-1. May play a role in titin/TTN downstream signaling in muscle cells. May regulate signaling cascades through ubiquitination. Adapter that mediates the interaction between TRAF6 and CYLD (By similarity). May be involved in cell differentiation, apoptosis, immune response and regulation of K(+) channels. Involved in endosome organization by retaining vesicles in the perinuclear cloud: following ubiquitination by RNF26, attracts specific vesicle-associated adapters, forming a molecular bridge that restrains cognate vesicles in the perinuclear region and organizes the endosomal pathway for efficient cargo transport (PubMed:27368102). Promotes relocalization of 'Lys-63'-linked ubiquitinated STING1 to autophagosomes (PubMed:29496741). Acts as an activator of the NFE2L2/NRF2 pathway via interaction with KEAP1: interaction inactivates the BCR(KEAP1) complex, promoting nuclear accumulation of NFE2L2/NRF2 and subsequent expression of cytoprotective genes (PubMed:20452972, PubMed:28380357, PubMed:33393215). {ECO:0000250|UniProtKB:O08623, ECO:0000250|UniProtKB:Q64337, ECO:0000269|PubMed:10356400, ECO:0000269|PubMed:10747026, ECO:0000269|PubMed:11244088, ECO:0000269|PubMed:12471037, ECO:0000269|PubMed:15340068, ECO:0000269|PubMed:15802564, ECO:0000269|PubMed:15911346, ECO:0000269|PubMed:15953362, ECO:0000269|PubMed:16079148, ECO:0000269|PubMed:16286508, ECO:0000269|PubMed:19931284, ECO:0000269|PubMed:20168092, ECO:0000269|PubMed:20452972, ECO:0000269|PubMed:22622177, ECO:0000269|PubMed:24128730, ECO:0000269|PubMed:27368102, ECO:0000269|PubMed:28380357, ECO:0000269|PubMed:28404643, ECO:0000269|PubMed:29496741, ECO:0000269|PubMed:33393215}. | FUNCTION: Catalyzes the methylation of 5-carboxymethyl uridine to 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its methyltransferase domain (PubMed:20123966, PubMed:20308323, PubMed:31079898). Catalyzes the last step in the formation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in target tRNA (PubMed:20123966, PubMed:20308323). Has a preference for tRNA(Arg) and tRNA(Glu), and does not bind tRNA(Lys)(PubMed:20308323). Binds tRNA and catalyzes the iron and alpha-ketoglutarate dependent hydroxylation of 5-methylcarboxymethyl uridine at the wobble position of the anticodon loop in tRNA via its dioxygenase domain, giving rise to 5-(S)-methoxycarbonylhydroxymethyluridine; has a preference for tRNA(Gly) (PubMed:21285950). Required for normal survival after DNA damage (PubMed:20308323). May inhibit apoptosis and promote cell survival and angiogenesis (PubMed:19293182). {ECO:0000269|PubMed:19293182, ECO:0000269|PubMed:20123966, ECO:0000269|PubMed:20308323, ECO:0000269|PubMed:21285950, ECO:0000269|PubMed:31079898}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 3_102 | 167.33333333333334 | 357.0 | Domain | PB1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 3_102 | 167.33333333333334 | 357.0 | Domain | PB1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000389805 | + | 5 | 8 | 3_102 | 251.33333333333334 | 441.0 | Domain | PB1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 3_102 | 167.33333333333334 | 357.0 | Domain | PB1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000389805 | + | 5 | 8 | 228_233 | 251.33333333333334 | 441.0 | Motif | Note=TRAF6-binding |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000389805 | + | 5 | 8 | 170_220 | 251.33333333333334 | 441.0 | Region | Note=LIM protein-binding (LB) |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 122_167 | 167.33333333333334 | 357.0 | Zinc finger | ZZ-type |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 122_167 | 167.33333333333334 | 357.0 | Zinc finger | ZZ-type |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000389805 | + | 5 | 8 | 122_167 | 251.33333333333334 | 441.0 | Zinc finger | ZZ-type |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 122_167 | 167.33333333333334 | 357.0 | Zinc finger | ZZ-type |
| Tgene | ALK | chr5:179252226 | chr2:29446395 | ENST00000389048 | 18 | 29 | 1116_1392 | 1057.3333333333333 | 1621.0 | Domain | Protein kinase | |
| Tgene | ALK | chr5:179252226 | chr2:29446395 | ENST00000389048 | 18 | 29 | 1197_1199 | 1057.3333333333333 | 1621.0 | Region | Note=Inhibitor binding | |
| Tgene | ALK | chr5:179252226 | chr2:29446395 | ENST00000389048 | 18 | 29 | 1060_1620 | 1057.3333333333333 | 1621.0 | Topological domain | Cytoplasmic |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 272_294 | 167.33333333333334 | 357.0 | Compositional bias | Note=Ser-rich |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 272_294 | 167.33333333333334 | 357.0 | Compositional bias | Note=Ser-rich |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000389805 | + | 5 | 8 | 272_294 | 251.33333333333334 | 441.0 | Compositional bias | Note=Ser-rich |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 272_294 | 167.33333333333334 | 357.0 | Compositional bias | Note=Ser-rich |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 389_434 | 167.33333333333334 | 357.0 | Domain | UBA |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 389_434 | 167.33333333333334 | 357.0 | Domain | UBA |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000389805 | + | 5 | 8 | 389_434 | 251.33333333333334 | 441.0 | Domain | UBA |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 389_434 | 167.33333333333334 | 357.0 | Domain | UBA |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 228_233 | 167.33333333333334 | 357.0 | Motif | Note=TRAF6-binding |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 336_341 | 167.33333333333334 | 357.0 | Motif | Note=LIR |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 228_233 | 167.33333333333334 | 357.0 | Motif | Note=TRAF6-binding |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 336_341 | 167.33333333333334 | 357.0 | Motif | Note=LIR |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000389805 | + | 5 | 8 | 336_341 | 251.33333333333334 | 441.0 | Motif | Note=LIR |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 228_233 | 167.33333333333334 | 357.0 | Motif | Note=TRAF6-binding |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 336_341 | 167.33333333333334 | 357.0 | Motif | Note=LIR |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 170_220 | 167.33333333333334 | 357.0 | Region | Note=LIM protein-binding (LB) |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 321_342 | 167.33333333333334 | 357.0 | Region | MAP1LC3B-binding |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 170_220 | 167.33333333333334 | 357.0 | Region | Note=LIM protein-binding (LB) |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 321_342 | 167.33333333333334 | 357.0 | Region | MAP1LC3B-binding |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000389805 | + | 5 | 8 | 321_342 | 251.33333333333334 | 441.0 | Region | MAP1LC3B-binding |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 170_220 | 167.33333333333334 | 357.0 | Region | Note=LIM protein-binding (LB) |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 321_342 | 167.33333333333334 | 357.0 | Region | MAP1LC3B-binding |
| Tgene | ALK | chr5:179252226 | chr2:29446395 | ENST00000389048 | 18 | 29 | 816_940 | 1057.3333333333333 | 1621.0 | Compositional bias | Note=Gly-rich | |
| Tgene | ALK | chr5:179252226 | chr2:29446395 | ENST00000389048 | 18 | 29 | 264_427 | 1057.3333333333333 | 1621.0 | Domain | MAM 1 | |
| Tgene | ALK | chr5:179252226 | chr2:29446395 | ENST00000389048 | 18 | 29 | 437_473 | 1057.3333333333333 | 1621.0 | Domain | Note=LDL-receptor class A | |
| Tgene | ALK | chr5:179252226 | chr2:29446395 | ENST00000389048 | 18 | 29 | 478_636 | 1057.3333333333333 | 1621.0 | Domain | MAM 2 | |
| Tgene | ALK | chr5:179252226 | chr2:29446395 | ENST00000389048 | 18 | 29 | 19_1038 | 1057.3333333333333 | 1621.0 | Topological domain | Extracellular | |
| Tgene | ALK | chr5:179252226 | chr2:29446395 | ENST00000389048 | 18 | 29 | 1039_1059 | 1057.3333333333333 | 1621.0 | Transmembrane | Helical |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (612) >>>612.pdbFusion protein BP residue: 178 CIF file (612) >>>612.cif | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29446394 | - | MRKDEDGDLVAFSSDEELTMAMSYVKDDIFRIYIKEKKECRRDHRPPCAQ EAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCEGKGLHRGHTK LAFPSPFGHLSEGFSHSRWLRKVKHGHFGWPGWEMGPPGNWSPRPPRAGE ARPGPTAESASGPSEDPSVNFLKNVGESVAAALSPLVYRRKHQELQAMQM ELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRG LGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALII SKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSL AMLDLLHVARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDF GMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWE IFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPE DRPNFAIILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGV PPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAV EGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNP IAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPL | 749 |
| 3D view using mol* of 612 (AA BP:178) | ||||||||||
 | ||||||||||
| PDB file (647) >>>647.pdbFusion protein BP residue: 221 CIF file (647) >>>647.cif | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29446394 | - | MPSVAQVGVFVYSPVSVPFILASAHSSSLCPAENPWVLTCCLLNNLDEDG DLVAFSSDEELTMAMSYVKDDIFRIYIKEKKECRRDHRPPCAQEAPRNMV HPNVICDGCNGPVVGTRYKCSVCPDYDLCSVCEGKGLHRGHTKLAFPSPF GHLSEGFSHSRWLRKVKHGHFGWPGWEMGPPGNWSPRPPRAGEARPGPTA ESASGPSEDPSVNFLKNVGESVAAALSPLVYRRKHQELQAMQMELQSPEY KLSKLRTSTIMTDYNPNYCFAGKTSSISDLKEVPRKNITLIRGLGHGAFG EVYEGQVSGMPNDPSPLQVAVKTLPEVCSEQDELDFLMEALIISKFNHQN IVRCIGVSLQSLPRFILLELMAGGDLKSFLRETRPRPSQPSSLAMLDLLH VARDIACGCQYLEENHFIHRDIAARNCLLTCPGPGRVAKIGDFGMARDIY RASYYRKGGCAMLPVKWMPPEAFMEGIFTSKTDTWSFGVLLWEIFSLGYM PYPSKSNQEVLEFVTSGGRMDPPKNCPGPVYRIMTQCWQHQPEDRPNFAI ILERIEYCTQDPDVINTALPIEYGPLVEEEEKVPVRPKDPEGVPPLLVSQ QAKREEERSPAAPPPLPTTSSGKAAKKPTAAEISVRVPRGPAVEGGHVNM AFSQSNPPSELHKVHGSRNKPTSLWNPTYGSWFTEKPTKKNNPIAKKEPH DRGNLGLEGSCTVPPNVATGRLPGASLLLEPSSLTANMKEVPLFRLRHFP | 792 |
| 3D view using mol* of 647 (AA BP:221) | ||||||||||
 | ||||||||||
| PDB file (659) | SQSTM1 | chr5 | 179252226 | + | ALK | chr2 | 29446394 | - | MASLTVKAYLLGKEDAAREIRRFSFCCSPEPEAEAEAAAGPGPCERLLSR VAALFPALRPGGFQAHYRDEDGDLVAFSSDEELTMAMSYVKDDIFRIYIK EKKECRRDHRPPCAQEAPRNMVHPNVICDGCNGPVVGTRYKCSVCPDYDL CSVCEGKGLHRGHTKLAFPSPFGHLSEGFSHSRWLRKVKHGHFGWPGWEM GPPGNWSPRPPRAGEARPGPTAESASGPSEDPSVNFLKNVGESVAAALSP LVYRRKHQELQAMQMELQSPEYKLSKLRTSTIMTDYNPNYCFAGKTSSIS DLKEVPRKNITLIRGLGHGAFGEVYEGQVSGMPNDPSPLQVAVKTLPEVC SEQDELDFLMEALIISKFNHQNIVRCIGVSLQSLPRFILLELMAGGDLKS FLRETRPRPSQPSSLAMLDLLHVARDIACGCQYLEENHFIHRDIAARNCL LTCPGPGRVAKIGDFGMARDIYRASYYRKGGCAMLPVKWMPPEAFMEGIF TSKTDTWSFGVLLWEIFSLGYMPYPSKSNQEVLEFVTSGGRMDPPKNCPG PVYRIMTQCWQHQPEDRPNFAIILERIEYCTQDPDVINTALPIEYGPLVE EEEKVPVRPKDPEGVPPLLVSQQAKREEERSPAAPPPLPTTSSGKAAKKP TAAEISVRVPRGPAVEGGHVNMAFSQSNPPSELHKVHGSRNKPTSLWNPT YGSWFTEKPTKKNNPIAKKEPHDRGNLGLEGSCTVPPNVATGRLPGASLL LEPSSLTANMKEVPLFRLRHFPCGNVNYGYQQQGLPLEAATAPGAGHYED | 814 |
Top |
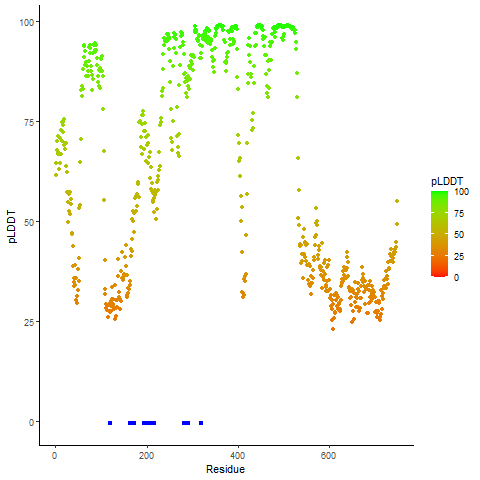
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
SQSTM1_pLDDT.png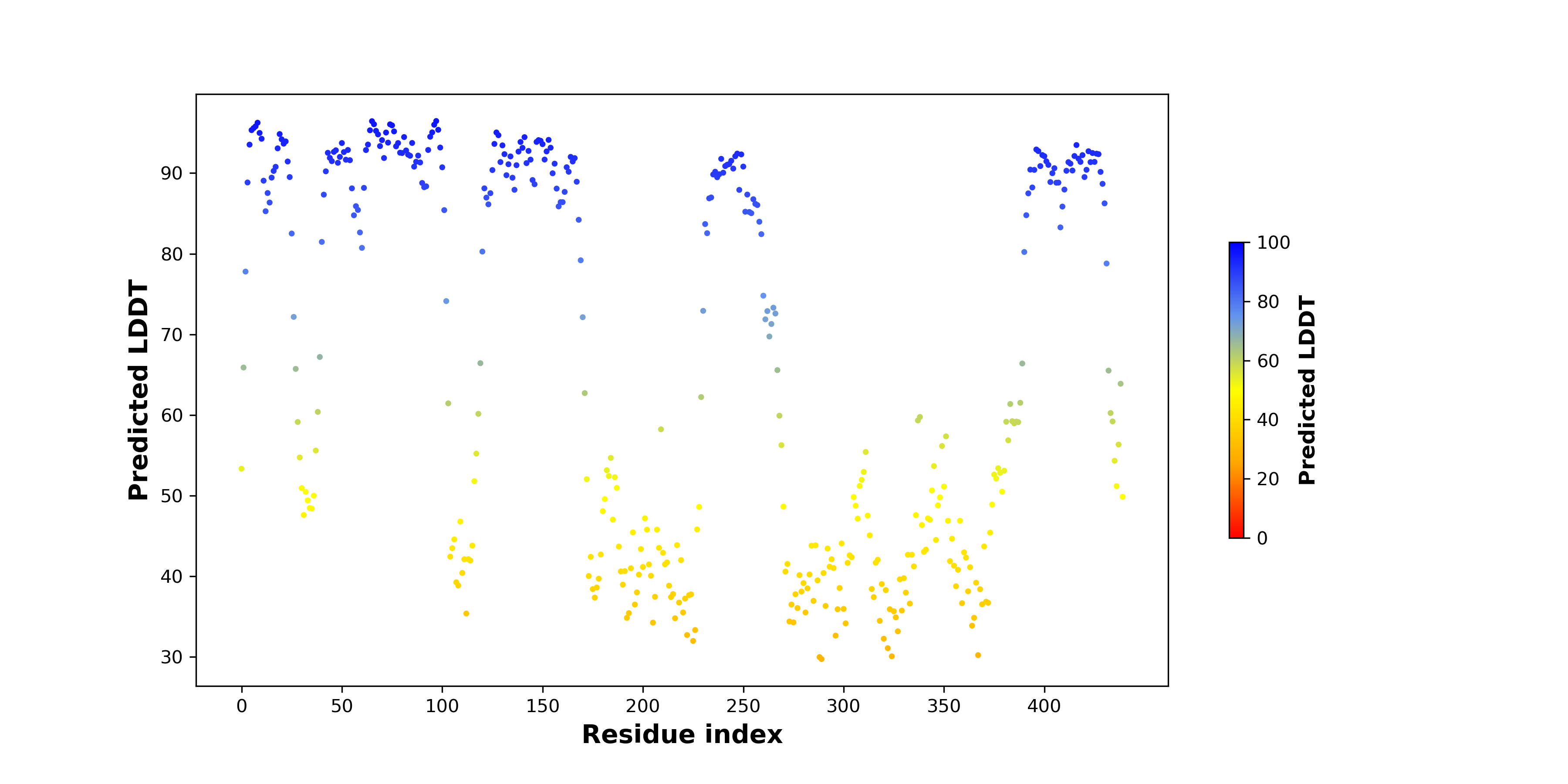 |
ALK_pLDDT.png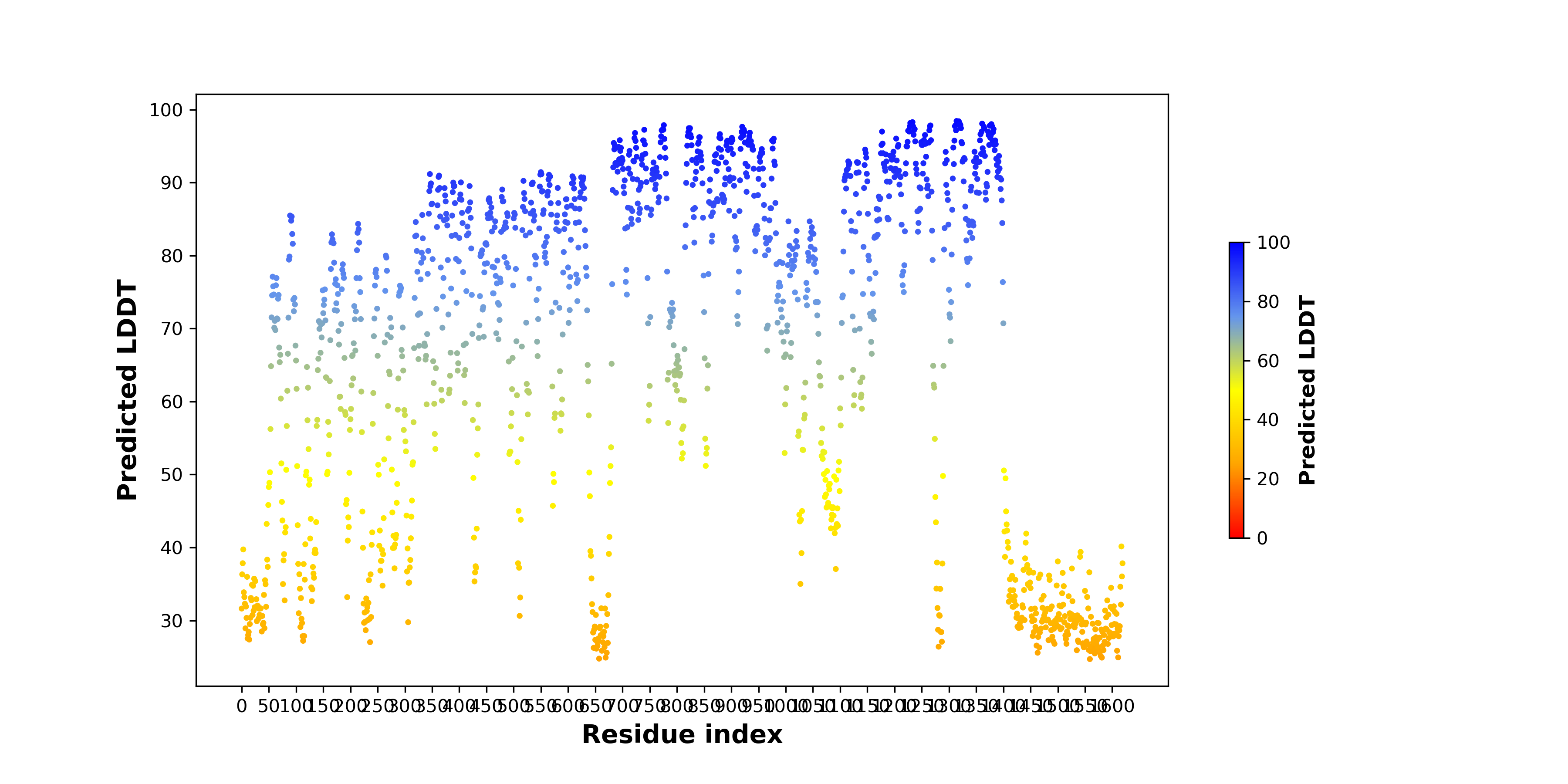 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
| SQSTM1_ALK_612.png |
 |
| SQSTM1_ALK_647.png |
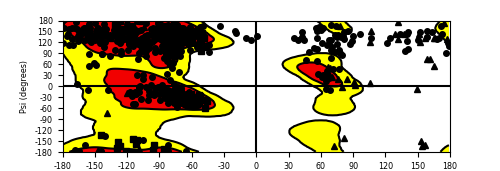 |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 612 | 1.051 | 111 | 1.11 | 359.807 | 0.578 | 0.69 | 0.913 | 0.995 | 0.749 | 1.329 | 1.419 | Chain A: 119,164,165,166,168,169,172,195,198,199,2 01,202,203,204,206,207,209,210,211,213,214,217,281 ,283,284,285,286,287,290,319 |
| 647 | 1.053 | 191 | 1.085 | 560.805 | 0.524 | 0.745 | 0.965 | 0.888 | 0.925 | 0.959 | 0.91 | Chain A: 292,294,295,296,297,298,299,302,304,320,3 22,335,339,352,368,369,370,371,372,373,374,375,378 ,379,382,419,421,425,426,428,441,442,443,444,445,4 46,447,448,451,462,463,464,467,675,677,678,679 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| SQSTM1 | GABARAPL2, RAD23A, GABARAPL1, SQSTM1, LINC00341, MAP1LC3B, TRAF6, PRKCZ, IRAK1, PRKCI, NTRK1, RIPK1, NTRK2, NTRK3, LCK, MAP1LC3A, Shank1, Dlg4, Grin2a, Gria1, Chrna7, KEAP1, GABARAP, C10orf2, ABHD10, CEP78, DIP2B, BLOC1S5, EMILIN3, NSUN4, OSBPL8, INA, NIPSNAP1, GBAS, LLGL1, HADHA, GPC4, HADHB, CTNND1, ASPH, ENPP1, TRMT61B, GLG1, MRPL38, NEFM, GTF3C3, SRRM2, MYD88, MYC, MAP3K3, MAP2K5, IRF8, TRIM21, CYLD, Cep78, Poc1b, CALM1, BNIP1, NFE2L2, CUL3, PIK3CA, PPHLN1, BPTF, UBC, DAZAP2, CDC37, TRIM13, CASP8, PCK1, STAT5A, PDE4A, MAPT, ATG4B, CHMP2B, TARDBP, CUL1, CUL2, HDAC6, ATG7, FUS, CSNK2A1, ZFAND5, TUBA1A, WDFY3, ATG8, BAG3, NBR1, SNCA, TRIM63, TRIM55, TTN, RARA, PSMC2, PSMD4, MAP1LC3C, TRIM50, ARHGEF28, TGM2, PAWR, RPTOR, MTOR, AKT1S1, MLST8, RPS6KB1, RRAGC, RRAGB, CDK9, tat, SPRED2, TRIM5, AJUBA, LIMD1, MAPK14, PARP10, vif, nef, TNFRSF10A, HTT, HNRNPA2B1, SDHA, CAV1, BID, FAS, SESN2, SESN1, MALT1, IKBKG, FHOD3, NOD2, SYNPO2, PYCARD, MLH1, PARK2, BCL2, PAN2, NPM1, MBP, YWHAZ, STXBP1, FKBP4, MEIS2, TKT, HSPA4, RELN, CAMK2A, ULK2, NCOR1, NGFR, env, HIV2gp7, CALCOCO2, Prkci, CDK1, CCNB1, PRKCD, MAPK1, SMAD3, EEF1D, RPL37, GEMIN4, DVL2, CHAF1A, TP53, CASP9, HIF1A, PSMD12, PSMD3, FLNB, PSMD10, TXNL1, TNK2, LGALS3, EDEM1, CFTR, CSNK2A2, DCP2, EGLN3, EPAS1, ULK1, TOLLIP, OPTN, ATXN3, STUB1, ISG15, MOV10, NXF1, CUL7, PIK3R1, PIK3R2, AGAP1, INSR, Ubc, TBK1, CRHBP, SCCPDH, DNAI1, DNAI2, TP53INP1, LLGL2, HSPB1, Ripk3, Nr2f2, IFI16, KIAA0753, MED4, CEP135, CNTRL, SASS6, TMEM17, XPO1, CD44, KIF5B, PML, Mgp, Poc1a, Fgfr1op, ENC1, SKI, TRIB3, RBM45, MAPK13, CHDH, EPDR1, GAS6, NDUFS2, NDUFS3, GFM2, NDUFA5, CD48, LDHA, RCN2, TRAF1, GBP2, KLHL3, EPM2A, RAD54L2, HSPA5, GRIA1, UBXN1, PEX5, BCL10, ATG5, CSNK1A1, VANGL2, SERPINA1, RNF168, LRRK2, VHL, TCEB2, TCEB1, RNF166, NOTCH1, RETN, KERA, INSL5, TMX1, VWCE, CD96, HTR3A, RNF26, INO80B, DLST, TRIM25, PLIN1, TES, WDR81, PSMA6, TRIM23, PGI1, UBE2D2, UBE2D3, ID1, ID2, TAX1BP1, Map1lc3b, UBD, RMND5A, RIPK4, MAPK6, TMPO, AIM2, TRIM11, TNFAIP3, VDR, NEDD4, CNOT2, WDR77, KRAS, RXRA, TGFB1, TRIP4, C19orf80, CCNF, VIM, FLNA, RAD51, NR4A1, RAD18, REL, RELA, RNF4, RNF31, PARK7, MYO6, XIAP, DDX5, CCND1, BPLF1, ESR2, BBC3, CDC6, BRCA1, CRYAB, DNAJC10, PADI1, CALCR, USP14, DDX58, Gabarapl1, MB21D1, SMN1, CRBN, FBL, ELAVL1, KIAA1429, AMBRA1, VCP, LAMP2, ATG16L1, LRRK1, PHB, BECN1, SOX2, PARP1, USP10, MTDH, FUNDC1, GBF1, MECOM, DCAF15, asp, SHMT2, CCT7, SLIRP, SND1, FADD, BIRC2, DYRK1A, PINK1, RB1CC1, IFITM3, P2RY6, GCH1, NUB1, PPP1CA, Casp8, FBXW7, BIRC3, LMBR1L, IGF2BP1, SERPINE2, MYO5A, FN1, CTHRC1, CD59, ITIH5, TNC, SNTB1, ROBO1, TUBB2A, GDF15, CNP, RAI14, ACSL3, CEP170, TAOK2, CNST, SSR3, SLC25A11, SPATA20, FARP1, UTRN, DHX33, CHCHD3, HS2ST1, PTRH2, SOD3, BCKDK, CCDC77, IQGAP1, FKBP11, CEP350, SCO1, SNTB2, MTX2, MYO1B, PWP1, SRPR, ATAD3A, ACTR1A, SLC25A1, MOGS, GOLGA2, SMG8, DYNC1LI2, UQCRH, SRPK2, CAPZA2, POLRMT, SMARCD1, GNB1, HSD17B12, DYNC1LI1, AIFM1, DARS2, GNAI2, TUBA1C, TUBA1B, MRPS28, CEP131, WDR82, PAFAH1B1, TUBB, MRPS34, ATP5A1, NOL9, ZC3H4, ATP5B, MRPS26, MRPS18B, ERAL1, PNKP, MIB1, SRP14, SLC25A3, MRPS22, CEP250, CCDC61, CKAP4, RPS16, BICD1, RPS15, ATP5H, AP2B1, NFIX, CDK5RAP2, RRBP1, SLC25A4, RPS5, TPX2, AP2A2, IK, PPP2R1A, GOLGB1, MPRIP, AHNAK, PCNT, RPS24, CETN2, HNRNPUL1, PRRC2A, DAP3, PIBF1, TJP1, RFC4, NOL8, CEP290, MAGI1, ZNF384, IQSEC1, TAOK1, PGAM5, SMG1, GAPVD1, MRPS9, ATAD3B, CSTF3, FRYL, EPRS, ARHGEF18, SNAP23, PRPF38A, RBM12B, SLC25A10, UTP23, FGFR1OP, SMU1, RAPGEF2, OFD1, PARD3, S100A9, EIF4B, EIF5B, IGF2BP3, ATXN1, HCVgp1, IKBKB, SKP2, PHB2, PLN, MAVS, APAF1, ECSIT, G3BP1, G3BP2, BNIP3, BNIP3L, CALR, DAXX, LINC01554, HMOX1, KAT5, PDPK1, PXN, RAF1, SH3BGRL, ORF7a, E, nsp6, ORF6, ORF8, ESR1, TRIM32, RQCD1, CIT, ANLN, AURKB, CHMP4B, ECT2, KIF14, KIF20A, KIF23, PRC1, PDCD4, BRD1, FIS1, MARCH5, MFN2, OCIAD1, ABL1, Rnf183, AGR2, HULC, NLRP3, NUPR1, USP8, GSK3B, UCHL5, FAM20C, GJA1, CCDC88C, PRDM9, PPIA, USP20, USP19, OTUD7B, JOSD1, IRF3, STK17A, OGT, BAG5, PSMB2, PSMD14, TRIM37, AR, MCL1, AMOT, B3GAT1, GJD3, GOLGA1, IMPDH2, KRT8, PXMP2, AMER1, ARHGAP32, BCR, C1orf198, CC2D1A, CCDC85C, CDC27, CENPB, CEP85, CNOT1, CNOT10, CSTF2T, DNAJA2, EDRF1, FAM193A, FARSA, FLJ44635, GGA3, HGS, HIST1H2BD, HIST1H3F, HSPA1B, IARS2, IFT172, IFT74, IQGAP2, JADE3, KIF7, KPNA3, LZTS2, MAGED1, NAE1, NAV1, NUDT19, OCRL, PAPD7, PDLIM7, PEBP1, PIK3C2B, PJA2, PPP1R13B, PPP2R3B, PPP2R5D, RC3H1, RIPK2, SCYL2, SEC16A, SIRT2, SMG7, SNRPE, SYNJ1, TNIP1, TNRC6A, TNRC6B, TP53BP2, TRIM26, TRIP6, TTK, UBB, VWA8, ZCCHC14, ZCCHC2, ZFP36L2, ABLIM1, KIAA1244, ASNS, CEP152, DIAPH3, DNAAF2, DRG1, DVL3, EXOC4, KIAA1671, LPP, NELFCD, NRBF2, PAK4, PDLIM3, PDLIM5, RPAP2, SDCCAG3, SIPA1L3, SLAIN2, SYNJ2, TAB2, TANC1, TTC28, TTF2, UCHL1, WRAP73, YEATS2, SYNE3, VASP, JUN, UBQLN2, CCDC50, IFIH1, VDAC1, MOAP1, CHRNB4, ZCCHC10, PRR27, IL7R, IL17F, BAGE2, HFE, CST8, POGLUT1, EDDM3B, LY6G5C, ST14, TNFSF18, GREM2, C1QTNF1, CST11, HLA-DPB1, FIBIN, ADAMTS12, GLIPR1L2, FAM46C, BTF3, ZRANB1, UXT, WBP4, IFITM1, KDR, NEDD8, KDM1A, AARS, ACLY, ACTB, ARF4, ATG9A, ATP2A2, BZW1, CACYBP, CAD, CALCOCO1, CCAR2, CCPG1, CCT2, CCT3, CCT4, CCT5, CCT6A, CEP112, CKB, CLTC, CTPS1, DDX3X, DNM1L, DYNC1H1, EEF2, EIF2S3, EIF3A, EIF4A1, ERLIN1, FASN, FASTKD2, FTL, GART, GPC1, HEXB, HIST1H2BN, HMGB1, HSP90AA1, HSP90AB1, HSPA8, HSPH1, IPO5, IPO7, KLHL8, KPNA2, KPNA4, KPNB1, LPL, LRRC59, MAT2A, MFF, MTHFD1, MTMR4, NCOA4, NPR3, NUP155, PABPC1, PAICS, PAIP1, PDCD6, PFKM, PFKP, PGD, PHGDH, PRKACA, PRKAR1A, PSMC1, PSMC3, PSMC4, PSMC5, PSMC6, PSMD2, RAB18, RAB1A, RAB21, RAB2A, RAB5C, RAB7A, RAB8A, RHEB, RPS17, RRM1, RTN4, S100A6, SAR1B, SDC2, SDC4, SDCBP, SEPT7, SMC4, SPG20, ST13P5, STIP1, SYNGR2, TAGLN2, TBC1D15, TCP1, TFRC, TM9SF2, TMEM59, TNPO1, TPP1, TRAF2, TRAF3, TRIP13, TUBB4B, VAPA, VAPB, YBX1, YWHAB, YWHAE, TRIO, HECTD1, WNK1, SPATA5L1, CTNNB1, LUZP1, MTCL1, STAM, TMEM160, POLR2B, TNKS1BP1, PPP6R3, KIF11, RTN3, ADAR, RACGAP1, BUB3, CORO1B, PABPC4, MAP7, DCXR, TSR1, DNMT1, ZC3HAV1, RPS27A, RAB6A, ADRM1, DHX9, UBA1, GSTP1, ACTG1, CISD2, DNAJA4, SETX, KIAA0101, TYSND1, STARD7, RNF114, ALKBH5, LGALS1, ARPC1A, TPM2, MRPS11, NT5C2, ACOT1, MRPS21, ARHGEF1, UBAP2L, SLC38A2, RPP38, MAP7D3, BAG1, MMS19, EARS2, SPATS2L, LGALS3BP, OXSR1, ARHGEF40, IGF2R, TUBG1, NUP188, TNPO3, PTRF, TDRD3, MORF4L2, NIF3L1, NIPBL, FTH1, CHEK2, FBXO3, RPL37A, METAP2, MRPS23, RPS19, SART1, APRT, WBSCR22, HMGB2, RPL12, PSIP1, RPS7, STRBP, ZC3H15, MAGT1, DHX37, RFC1, SRP72, PRPF31, SEMA3C, HNRNPA0, SCAMP3, SRPK1, UPF3B, ZFR, SYNCRIP, ILF2, SLK, AP3D1, ARPC4-TTLL3, RPS26, SSBP1, HNRNPAB, TMEM192, SCAP, CNR2, KLF15, SAMM50, IMMT, MTX1, MAP1LC3B2, VPS29, TOMM40, PSMB4, TRIM44, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| SQSTM1 |  |
| ALK |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 122_224 | 167.33333333333334 | 357.0 | GABRR3 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 122_224 | 167.33333333333334 | 357.0 | GABRR3 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 122_224 | 167.33333333333334 | 357.0 | GABRR3 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 347_352 | 167.33333333333334 | 357.0 | KEAP1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 347_352 | 167.33333333333334 | 357.0 | KEAP1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000389805 | + | 5 | 8 | 347_352 | 251.33333333333334 | 441.0 | KEAP1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 347_352 | 167.33333333333334 | 357.0 | KEAP1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000360718 | + | 4 | 7 | 269_440 | 167.33333333333334 | 357.0 | NTRK1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000376929 | + | 6 | 9 | 269_440 | 167.33333333333334 | 357.0 | NTRK1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000389805 | + | 5 | 8 | 269_440 | 251.33333333333334 | 441.0 | NTRK1 |
| Hgene | SQSTM1 | chr5:179252226 | chr2:29446395 | ENST00000402874 | + | 5 | 8 | 269_440 | 167.33333333333334 | 357.0 | NTRK1 |
Top |
Related Drugs to SQSTM1-ALK |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
| SQSTM1 | ALK | Alk Inhibitor Alectinib | PubMed | 30790150 |
Top |
Related Diseases to SQSTM1-ALK |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| SQSTM1 | ALK | Inflammatory Myofibroblastic Tumor (Imt) | PubMed | 30790150 |
| SQSTM1 | ALK | Lung Adenocarcinoma | MyCancerGenome | |
| SQSTM1 | ALK | Colon Adenocarcinoma | MyCancerGenome | |
| SQSTM1 | ALK | Acute Myeloid Leukemia | MyCancerGenome | |
| SQSTM1 | ALK | Cancer Of Unknown Primary | MyCancerGenome | |
| SQSTM1 | ALK | Diffuse Large B-Cell Lymphoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | SQSTM1 | C4085252 | PAGET DISEASE OF BONE 3 | 9 | GENOMICS_ENGLAND;UNIPROT |
| Hgene | SQSTM1 | C0002736 | Amyotrophic Lateral Sclerosis | 5 | CTD_human;ORPHANET |
| Hgene | SQSTM1 | C4225326 | FRONTOTEMPORAL DEMENTIA AND/OR AMYOTROPHIC LATERAL SCLEROSIS 3 | 4 | CTD_human;UNIPROT |
| Hgene | SQSTM1 | C0029463 | Osteosarcoma | 2 | GENOMICS_ENGLAND |
| Hgene | SQSTM1 | C0221054 | Welander Distal Myopathy | 1 | ORPHANET |
| Hgene | SQSTM1 | C0242383 | Age related macular degeneration | 1 | CTD_human |
| Hgene | SQSTM1 | C0393554 | Amyotrophic Lateral Sclerosis With Dementia | 1 | CTD_human |
| Hgene | SQSTM1 | C0543859 | Amyotrophic Lateral Sclerosis, Guam Form | 1 | CTD_human |
| Hgene | SQSTM1 | C1853926 | NONAKA MYOPATHY | 1 | CTD_human;GENOMICS_ENGLAND |
| Hgene | SQSTM1 | C2931290 | Welander distal myopathy, Swedish type | 1 | ORPHANET |
| Hgene | SQSTM1 | C3888102 | Frontotemporal Dementia With Motor Neuron Disease | 1 | ORPHANET |
| Hgene | SQSTM1 | C4011788 | Behavioral variant of frontotemporal dementia | 1 | ORPHANET |
| Tgene | ALK | C0007131 | Non-Small Cell Lung Carcinoma | 28 | CGI;CTD_human |
| Tgene | ALK | C0027819 | Neuroblastoma | 13 | CGI;CTD_human;ORPHANET |
| Tgene | ALK | C0152013 | Adenocarcinoma of lung (disorder) | 8 | CGI;CTD_human |
| Tgene | ALK | C2751681 | NEUROBLASTOMA, SUSCEPTIBILITY TO, 3 | 8 | CLINGEN;UNIPROT |
| Tgene | ALK | C0206180 | Ki-1+ Anaplastic Large Cell Lymphoma | 6 | CGI;CTD_human |
| Tgene | ALK | C0334121 | Inflammatory Myofibroblastic Tumor | 4 | CGI;CTD_human;ORPHANET |
| Tgene | ALK | C0018199 | Granuloma, Plasma Cell | 3 | CTD_human |
| Tgene | ALK | C0007621 | Neoplastic Cell Transformation | 2 | CTD_human |
| Tgene | ALK | C0027627 | Neoplasm Metastasis | 2 | CTD_human |
| Tgene | ALK | C0238463 | Papillary thyroid carcinoma | 2 | ORPHANET |
| Tgene | ALK | C0001973 | Alcoholic Intoxication, Chronic | 1 | PSYGENET |
| Tgene | ALK | C0006118 | Brain Neoplasms | 1 | CGI;CTD_human |
| Tgene | ALK | C0006142 | Malignant neoplasm of breast | 1 | CTD_human |
| Tgene | ALK | C0007134 | Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0011570 | Mental Depression | 1 | PSYGENET |
| Tgene | ALK | C0011581 | Depressive disorder | 1 | PSYGENET |
| Tgene | ALK | C0027643 | Neoplasm Recurrence, Local | 1 | CTD_human |
| Tgene | ALK | C0036341 | Schizophrenia | 1 | PSYGENET |
| Tgene | ALK | C0079744 | Diffuse Large B-Cell Lymphoma | 1 | CTD_human |
| Tgene | ALK | C0085269 | Plasma Cell Granuloma, Pulmonary | 1 | CTD_human |
| Tgene | ALK | C0153633 | Malignant neoplasm of brain | 1 | CGI;CTD_human |
| Tgene | ALK | C0278601 | Inflammatory Breast Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0279702 | Conventional (Clear Cell) Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0496899 | Benign neoplasm of brain, unspecified | 1 | CTD_human |
| Tgene | ALK | C0678222 | Breast Carcinoma | 1 | CTD_human |
| Tgene | ALK | C0750974 | Brain Tumor, Primary | 1 | CTD_human |
| Tgene | ALK | C0750977 | Recurrent Brain Neoplasm | 1 | CTD_human |
| Tgene | ALK | C0750979 | Primary malignant neoplasm of brain | 1 | CTD_human |
| Tgene | ALK | C1257931 | Mammary Neoplasms, Human | 1 | CTD_human |
| Tgene | ALK | C1266042 | Chromophobe Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1266043 | Sarcomatoid Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1266044 | Collecting Duct Carcinoma of the Kidney | 1 | CTD_human |
| Tgene | ALK | C1306837 | Papillary Renal Cell Carcinoma | 1 | CTD_human |
| Tgene | ALK | C1332079 | Anaplastic Large Cell Lymphoma, ALK-Positive | 1 | ORPHANET |
| Tgene | ALK | C1458155 | Mammary Neoplasms | 1 | CTD_human |
| Tgene | ALK | C1527390 | Neoplasms, Intracranial | 1 | CTD_human |
| Tgene | ALK | C2931189 | Neural crest tumor | 1 | ORPHANET |
| Tgene | ALK | C3899155 | hereditary neuroblastoma | 1 | GENOMICS_ENGLAND |
| Tgene | ALK | C4704874 | Mammary Carcinoma, Human | 1 | CTD_human |