| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:SUZ12-NF1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: SUZ12-NF1 | FusionPDB ID: 88244 | FusionGDB2.0 ID: 88244 | Hgene | Tgene | Gene symbol | SUZ12 | NF1 | Gene ID | 23512 | 4763 |
| Gene name | SUZ12 polycomb repressive complex 2 subunit | neurofibromin 1 | |
| Synonyms | CHET9|IMMAS|JJAZ1 | NFNS|VRNF|WSS | |
| Cytomap | 17q11.2 | 17q11.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | polycomb protein SUZ12chET 9 proteinchromatin precipitated E2F target 9 proteinjoined to JAZF1 proteinsuppressor of zeste 12 protein homolog | neurofibrominneurofibromatosis 1neurofibromatosis-related protein NF-1truncated neurofibromin 1 | |
| Modification date | 20200313 | 20200322 | |
| UniProtAcc | Q15022 | P21359 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000322652, ENST00000580398, | ENST00000444181, ENST00000356175, ENST00000358273, ENST00000417592, ENST00000431387, ENST00000581113, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 15 X 12 X 10=1800 | 28 X 30 X 15=12600 |
| # samples | 15 | 33 | |
| ** MAII score | log2(15/1800*10)=-3.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(33/12600*10)=-5.25481389902883 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: SUZ12 [Title/Abstract] AND NF1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | SUZ12(30303633)-NF1(29576002), # samples:3 NF1(29533389)-SUZ12(30293166), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | SUZ12-NF1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. SUZ12-NF1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. SUZ12-NF1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. SUZ12-NF1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. NF1-SUZ12 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. NF1-SUZ12 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. NF1-SUZ12 seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. NF1-SUZ12 seems lost the major protein functional domain in Tgene partner, which is a epigenetic factor due to the frame-shifted ORF. NF1-SUZ12 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | NF1 | GO:0043547 | positive regulation of GTPase activity | 2121371 |
 Fusion gene breakpoints across SUZ12 (5'-gene) Fusion gene breakpoints across SUZ12 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
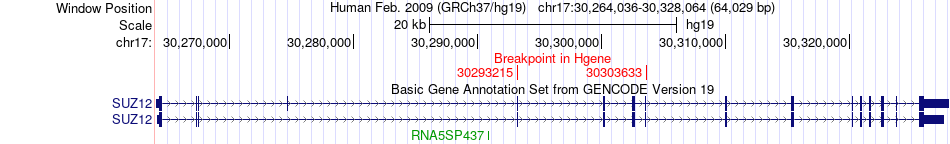 |
 Fusion gene breakpoints across NF1 (3'-gene) Fusion gene breakpoints across NF1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
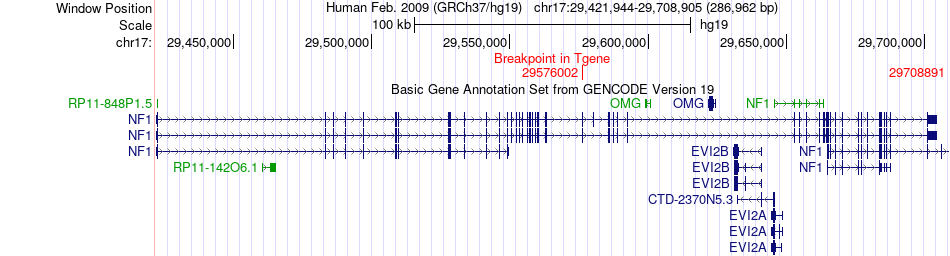 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-EJ-8472-01A | SUZ12 | chr17 | 30293215 | + | NF1 | chr17 | 29708891 | + |
| ChimerDB4 | UCEC | TCGA-BK-A139-01A | SUZ12 | chr17 | 30303633 | - | NF1 | chr17 | 29576002 | + |
| ChimerDB4 | UCEC | TCGA-BK-A139-01A | SUZ12 | chr17 | 30303633 | + | NF1 | chr17 | 29576002 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000322652 | SUZ12 | chr17 | 30303633 | + | ENST00000356175 | NF1 | chr17 | 29576002 | + | 9151 | 1146 | 37 | 5628 | 1863 |
| ENST00000322652 | SUZ12 | chr17 | 30303633 | + | ENST00000358273 | NF1 | chr17 | 29576002 | + | 9214 | 1146 | 37 | 5691 | 1884 |
| ENST00000580398 | SUZ12 | chr17 | 30303633 | + | ENST00000356175 | NF1 | chr17 | 29576002 | + | 9033 | 1028 | 108 | 5510 | 1800 |
| ENST00000580398 | SUZ12 | chr17 | 30303633 | + | ENST00000358273 | NF1 | chr17 | 29576002 | + | 9096 | 1028 | 108 | 5573 | 1821 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000322652 | ENST00000356175 | SUZ12 | chr17 | 30303633 | + | NF1 | chr17 | 29576002 | + | 0.000176275 | 0.99982375 |
| ENST00000322652 | ENST00000358273 | SUZ12 | chr17 | 30303633 | + | NF1 | chr17 | 29576002 | + | 0.00017642 | 0.99982363 |
| ENST00000580398 | ENST00000356175 | SUZ12 | chr17 | 30303633 | + | NF1 | chr17 | 29576002 | + | 0.000167305 | 0.99983263 |
| ENST00000580398 | ENST00000358273 | SUZ12 | chr17 | 30303633 | + | NF1 | chr17 | 29576002 | + | 0.00016609 | 0.99983394 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >88244_88244_1_SUZ12-NF1_SUZ12_chr17_30303633_ENST00000322652_NF1_chr17_29576002_ENST00000356175_length(amino acids)=1863AA_BP=5 MRRHFFFPPSFPPLLLPSLPLSSPLSSFPPRSAGACWGERLVLQALALRGRPAGSWRGEEAGTAMAPQKHGGGGGGGSGPSAGSGGGGFG GSAAVAAATASGGKSGGGSCGGGGSYSASSSSSAAAAAGAAVLPVKKPKMEHVQADHELFLQAFEKPTQIYRFLRTRNLIAPIFLHRTLT YMSHRNSRTNIKRKTFKVDDMLSKVEKMKGEQESHSLSAHLQLTFTGFFHKNDKPSPNSENEQNSVTLEVLLVKVCHKKRKDVSCPIRQV PTGKKQVPLNPDLNQTKPGNFPSLAVSSNEFEPSNSHMVKSYSLLFRVTRPGRREFNGMINGETNENIDVNEELPARRKRNREDGEKTFV AQMTVFDKNRLEPSESLEENQRNLLQMTEKFFHAIISSSSEFPPQLRSVCHCLYQVVSQRFPQNSIGAVGSAMFLRFINPAIVSPYEAGI LDKKPPPRIERGLKLMSKILQSIANHVLFTKEEHMRPFNDFVKSNFDAARRFFLDIASDCPTSDAVNHSLSFISDGNVLALHRLLWNNQE KIGQYLSSNRDHKAVGRRPFDKMATLLAYLGPPEHKPVADTHWSSLNLTSSKFEEFMTRHQVHEKEEFKALKTLSIFYQAGTSKAGNPIF YYVARRFKTGQINGDLLIYHVLLTLKPYYAKPYEIVVDLTHTGPSNRFKTDFLSKWFVVFPGFAYDNVSAVYIYNCNSWVREYTKYHERL LTGLKGSKRLVFIDCPGKLAEHIEHEQQKLPAATLALEEDLKVFHNALKLAHKDTKVSIKVGSTAVQVTSAERTKVLGQSVFLNDIYYAS EIEEICLVDENQFTLTIANQGTPLTFMHQECEAIVQSIIHIRTRWELSQPDSIPQHTKIRPKDVPGTLLNIALLNLGSSDPSLRSAAYNL LCALTCTFNLKIEGQLLETSGLCIPANNTLFIVSISKTLAANEPHLTLEFLEECISGFSKSSIELKHLCLEYMTPWLSNLVRFCKHNDDA KRQRVTAILDKLITMTINEKQMYPSIQAKIWGSLGQITDLLDVVLDSFIKTSATGGLGSIKAEVMADTAVALASGNVKLVSSKVIGRMCK IIDKTCLSPTPTLEQHLMWDDIAILARYMLMLSFNNSLDVAAHLPYLFHVVTFLVATGPLSLRASTHGLVINIIHSLCTCSQLHFSEETK QVLRLSLTEFSLPKFYLLFGISKVKSAAVIAFRSSYRDRSFSPGSYERETFALTSLETVTEALLEIMEACMRDIPTCKWLDQWTELAQRF AFQYNPSLQPRALVVFGCISKRVSHGQIKQIIRILSKALESCLKGPDTYNSQVLIEATVIALTKLQPLLNKDSPLHKALFWVAVAVLQLD EVNLYSAGTALLEQNLHTLDSLRIFNDKSPEEVFMAIRNPLEWHCKQMDHFVGLNFNSNFNFALVGHLLKGYRHPSPAIVARTVRILHTL LTLVNKHRNCDKFEVNTQSVAYLAALLTVSEEVRSRCSLKHRKSLLLTDISMENVPMDTYPIHHGDPSYRTLKETQPWSSPKGSEGYLAA TYPTVGQTSPRARKSMSLDMGQPSQANTKKLLGTRKSFDHLISDTKAPKRQEMESGITTPPKMRRVAETDYEMETQRISSSQQHPHLRKV SVSESNVLLDEEVLTDPKIQALLLTVLATLVKYTTDEFDQRILYEYLAEASVVFPKVFPVVHNLLDSKINTLLSLCQDPNLLNPIHGIVQ SVVYHEESPPQYQTSYLQSFGFNGLWRFAGPFSKQTQIPDYAELIVKFLDALIDTYLPGIDEETSEESLLTPTSPYPPALQSQLSITANL -------------------------------------------------------------- >88244_88244_2_SUZ12-NF1_SUZ12_chr17_30303633_ENST00000322652_NF1_chr17_29576002_ENST00000358273_length(amino acids)=1884AA_BP=5 MRRHFFFPPSFPPLLLPSLPLSSPLSSFPPRSAGACWGERLVLQALALRGRPAGSWRGEEAGTAMAPQKHGGGGGGGSGPSAGSGGGGFG GSAAVAAATASGGKSGGGSCGGGGSYSASSSSSAAAAAGAAVLPVKKPKMEHVQADHELFLQAFEKPTQIYRFLRTRNLIAPIFLHRTLT YMSHRNSRTNIKRKTFKVDDMLSKVEKMKGEQESHSLSAHLQLTFTGFFHKNDKPSPNSENEQNSVTLEVLLVKVCHKKRKDVSCPIRQV PTGKKQVPLNPDLNQTKPGNFPSLAVSSNEFEPSNSHMVKSYSLLFRVTRPGRREFNGMINGETNENIDVNEELPARRKRNREDGEKTFV AQMTVFDKNRLEPSESLEENQRNLLQMTEKFFHAIISSSSEFPPQLRSVCHCLYQATCHSLLNKATVKEKKENKKSVVSQRFPQNSIGAV GSAMFLRFINPAIVSPYEAGILDKKPPPRIERGLKLMSKILQSIANHVLFTKEEHMRPFNDFVKSNFDAARRFFLDIASDCPTSDAVNHS LSFISDGNVLALHRLLWNNQEKIGQYLSSNRDHKAVGRRPFDKMATLLAYLGPPEHKPVADTHWSSLNLTSSKFEEFMTRHQVHEKEEFK ALKTLSIFYQAGTSKAGNPIFYYVARRFKTGQINGDLLIYHVLLTLKPYYAKPYEIVVDLTHTGPSNRFKTDFLSKWFVVFPGFAYDNVS AVYIYNCNSWVREYTKYHERLLTGLKGSKRLVFIDCPGKLAEHIEHEQQKLPAATLALEEDLKVFHNALKLAHKDTKVSIKVGSTAVQVT SAERTKVLGQSVFLNDIYYASEIEEICLVDENQFTLTIANQGTPLTFMHQECEAIVQSIIHIRTRWELSQPDSIPQHTKIRPKDVPGTLL NIALLNLGSSDPSLRSAAYNLLCALTCTFNLKIEGQLLETSGLCIPANNTLFIVSISKTLAANEPHLTLEFLEECISGFSKSSIELKHLC LEYMTPWLSNLVRFCKHNDDAKRQRVTAILDKLITMTINEKQMYPSIQAKIWGSLGQITDLLDVVLDSFIKTSATGGLGSIKAEVMADTA VALASGNVKLVSSKVIGRMCKIIDKTCLSPTPTLEQHLMWDDIAILARYMLMLSFNNSLDVAAHLPYLFHVVTFLVATGPLSLRASTHGL VINIIHSLCTCSQLHFSEETKQVLRLSLTEFSLPKFYLLFGISKVKSAAVIAFRSSYRDRSFSPGSYERETFALTSLETVTEALLEIMEA CMRDIPTCKWLDQWTELAQRFAFQYNPSLQPRALVVFGCISKRVSHGQIKQIIRILSKALESCLKGPDTYNSQVLIEATVIALTKLQPLL NKDSPLHKALFWVAVAVLQLDEVNLYSAGTALLEQNLHTLDSLRIFNDKSPEEVFMAIRNPLEWHCKQMDHFVGLNFNSNFNFALVGHLL KGYRHPSPAIVARTVRILHTLLTLVNKHRNCDKFEVNTQSVAYLAALLTVSEEVRSRCSLKHRKSLLLTDISMENVPMDTYPIHHGDPSY RTLKETQPWSSPKGSEGYLAATYPTVGQTSPRARKSMSLDMGQPSQANTKKLLGTRKSFDHLISDTKAPKRQEMESGITTPPKMRRVAET DYEMETQRISSSQQHPHLRKVSVSESNVLLDEEVLTDPKIQALLLTVLATLVKYTTDEFDQRILYEYLAEASVVFPKVFPVVHNLLDSKI NTLLSLCQDPNLLNPIHGIVQSVVYHEESPPQYQTSYLQSFGFNGLWRFAGPFSKQTQIPDYAELIVKFLDALIDTYLPGIDEETSEESL -------------------------------------------------------------- >88244_88244_3_SUZ12-NF1_SUZ12_chr17_30303633_ENST00000580398_NF1_chr17_29576002_ENST00000356175_length(amino acids)=1800AA_BP=1 MVLQALALRGRPAGSWRGEEAGTAMAPQKHGGGGGGGSGPSAGSGGGGFGGSAAVAAATASGGKSGGGSCGGGGSYSASSSSSAAAAAGA AVLPVKKPKMEHVQADHELFLQAFEKPTQIYRFLRTRNLIAPIFLHRTLTYMSHRNSRTNIKSLSAHLQLTFTGFFHKNDKPSPNSENEQ NSVTLEVLLVKVCHKKRKDVSCPIRQVPTGKKQVPLNPDLNQTKPGNFPSLAVSSNEFEPSNSHMVKSYSLLFRVTRPGRREFNGMINGE TNENIDVNEELPARRKRNREDGEKTFVAQMTVFDKNRLEPSESLEENQRNLLQMTEKFFHAIISSSSEFPPQLRSVCHCLYQVVSQRFPQ NSIGAVGSAMFLRFINPAIVSPYEAGILDKKPPPRIERGLKLMSKILQSIANHVLFTKEEHMRPFNDFVKSNFDAARRFFLDIASDCPTS DAVNHSLSFISDGNVLALHRLLWNNQEKIGQYLSSNRDHKAVGRRPFDKMATLLAYLGPPEHKPVADTHWSSLNLTSSKFEEFMTRHQVH EKEEFKALKTLSIFYQAGTSKAGNPIFYYVARRFKTGQINGDLLIYHVLLTLKPYYAKPYEIVVDLTHTGPSNRFKTDFLSKWFVVFPGF AYDNVSAVYIYNCNSWVREYTKYHERLLTGLKGSKRLVFIDCPGKLAEHIEHEQQKLPAATLALEEDLKVFHNALKLAHKDTKVSIKVGS TAVQVTSAERTKVLGQSVFLNDIYYASEIEEICLVDENQFTLTIANQGTPLTFMHQECEAIVQSIIHIRTRWELSQPDSIPQHTKIRPKD VPGTLLNIALLNLGSSDPSLRSAAYNLLCALTCTFNLKIEGQLLETSGLCIPANNTLFIVSISKTLAANEPHLTLEFLEECISGFSKSSI ELKHLCLEYMTPWLSNLVRFCKHNDDAKRQRVTAILDKLITMTINEKQMYPSIQAKIWGSLGQITDLLDVVLDSFIKTSATGGLGSIKAE VMADTAVALASGNVKLVSSKVIGRMCKIIDKTCLSPTPTLEQHLMWDDIAILARYMLMLSFNNSLDVAAHLPYLFHVVTFLVATGPLSLR ASTHGLVINIIHSLCTCSQLHFSEETKQVLRLSLTEFSLPKFYLLFGISKVKSAAVIAFRSSYRDRSFSPGSYERETFALTSLETVTEAL LEIMEACMRDIPTCKWLDQWTELAQRFAFQYNPSLQPRALVVFGCISKRVSHGQIKQIIRILSKALESCLKGPDTYNSQVLIEATVIALT KLQPLLNKDSPLHKALFWVAVAVLQLDEVNLYSAGTALLEQNLHTLDSLRIFNDKSPEEVFMAIRNPLEWHCKQMDHFVGLNFNSNFNFA LVGHLLKGYRHPSPAIVARTVRILHTLLTLVNKHRNCDKFEVNTQSVAYLAALLTVSEEVRSRCSLKHRKSLLLTDISMENVPMDTYPIH HGDPSYRTLKETQPWSSPKGSEGYLAATYPTVGQTSPRARKSMSLDMGQPSQANTKKLLGTRKSFDHLISDTKAPKRQEMESGITTPPKM RRVAETDYEMETQRISSSQQHPHLRKVSVSESNVLLDEEVLTDPKIQALLLTVLATLVKYTTDEFDQRILYEYLAEASVVFPKVFPVVHN LLDSKINTLLSLCQDPNLLNPIHGIVQSVVYHEESPPQYQTSYLQSFGFNGLWRFAGPFSKQTQIPDYAELIVKFLDALIDTYLPGIDEE TSEESLLTPTSPYPPALQSQLSITANLNLSNSMTSLATSQHSPGIDKENVELSPTTGHCNSGRTRHGSASQVQKQRSAGSFKRNSIKKIV -------------------------------------------------------------- >88244_88244_4_SUZ12-NF1_SUZ12_chr17_30303633_ENST00000580398_NF1_chr17_29576002_ENST00000358273_length(amino acids)=1821AA_BP=1 MVLQALALRGRPAGSWRGEEAGTAMAPQKHGGGGGGGSGPSAGSGGGGFGGSAAVAAATASGGKSGGGSCGGGGSYSASSSSSAAAAAGA AVLPVKKPKMEHVQADHELFLQAFEKPTQIYRFLRTRNLIAPIFLHRTLTYMSHRNSRTNIKSLSAHLQLTFTGFFHKNDKPSPNSENEQ NSVTLEVLLVKVCHKKRKDVSCPIRQVPTGKKQVPLNPDLNQTKPGNFPSLAVSSNEFEPSNSHMVKSYSLLFRVTRPGRREFNGMINGE TNENIDVNEELPARRKRNREDGEKTFVAQMTVFDKNRLEPSESLEENQRNLLQMTEKFFHAIISSSSEFPPQLRSVCHCLYQATCHSLLN KATVKEKKENKKSVVSQRFPQNSIGAVGSAMFLRFINPAIVSPYEAGILDKKPPPRIERGLKLMSKILQSIANHVLFTKEEHMRPFNDFV KSNFDAARRFFLDIASDCPTSDAVNHSLSFISDGNVLALHRLLWNNQEKIGQYLSSNRDHKAVGRRPFDKMATLLAYLGPPEHKPVADTH WSSLNLTSSKFEEFMTRHQVHEKEEFKALKTLSIFYQAGTSKAGNPIFYYVARRFKTGQINGDLLIYHVLLTLKPYYAKPYEIVVDLTHT GPSNRFKTDFLSKWFVVFPGFAYDNVSAVYIYNCNSWVREYTKYHERLLTGLKGSKRLVFIDCPGKLAEHIEHEQQKLPAATLALEEDLK VFHNALKLAHKDTKVSIKVGSTAVQVTSAERTKVLGQSVFLNDIYYASEIEEICLVDENQFTLTIANQGTPLTFMHQECEAIVQSIIHIR TRWELSQPDSIPQHTKIRPKDVPGTLLNIALLNLGSSDPSLRSAAYNLLCALTCTFNLKIEGQLLETSGLCIPANNTLFIVSISKTLAAN EPHLTLEFLEECISGFSKSSIELKHLCLEYMTPWLSNLVRFCKHNDDAKRQRVTAILDKLITMTINEKQMYPSIQAKIWGSLGQITDLLD VVLDSFIKTSATGGLGSIKAEVMADTAVALASGNVKLVSSKVIGRMCKIIDKTCLSPTPTLEQHLMWDDIAILARYMLMLSFNNSLDVAA HLPYLFHVVTFLVATGPLSLRASTHGLVINIIHSLCTCSQLHFSEETKQVLRLSLTEFSLPKFYLLFGISKVKSAAVIAFRSSYRDRSFS PGSYERETFALTSLETVTEALLEIMEACMRDIPTCKWLDQWTELAQRFAFQYNPSLQPRALVVFGCISKRVSHGQIKQIIRILSKALESC LKGPDTYNSQVLIEATVIALTKLQPLLNKDSPLHKALFWVAVAVLQLDEVNLYSAGTALLEQNLHTLDSLRIFNDKSPEEVFMAIRNPLE WHCKQMDHFVGLNFNSNFNFALVGHLLKGYRHPSPAIVARTVRILHTLLTLVNKHRNCDKFEVNTQSVAYLAALLTVSEEVRSRCSLKHR KSLLLTDISMENVPMDTYPIHHGDPSYRTLKETQPWSSPKGSEGYLAATYPTVGQTSPRARKSMSLDMGQPSQANTKKLLGTRKSFDHLI SDTKAPKRQEMESGITTPPKMRRVAETDYEMETQRISSSQQHPHLRKVSVSESNVLLDEEVLTDPKIQALLLTVLATLVKYTTDEFDQRI LYEYLAEASVVFPKVFPVVHNLLDSKINTLLSLCQDPNLLNPIHGIVQSVVYHEESPPQYQTSYLQSFGFNGLWRFAGPFSKQTQIPDYA ELIVKFLDALIDTYLPGIDEETSEESLLTPTSPYPPALQSQLSITANLNLSNSMTSLATSQHSPGIDKENVELSPTTGHCNSGRTRHGSA -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:30303633/chr17:29576002) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| SUZ12 | NF1 |
| FUNCTION: Polycomb group (PcG) protein. Component of the PRC2 complex, which methylates 'Lys-9' (H3K9me) and 'Lys-27' (H3K27me) of histone H3, leading to transcriptional repression of the affected target gene (PubMed:15225548, PubMed:15231737, PubMed:15385962, PubMed:16618801, PubMed:17344414, PubMed:18285464, PubMed:28229514, PubMed:29499137, PubMed:31959557). The PRC2 complex may also serve as a recruiting platform for DNA methyltransferases, thereby linking two epigenetic repression systems (PubMed:12435631, PubMed:12351676, PubMed:15385962, PubMed:15099518, PubMed:15225548, PubMed:15684044, PubMed:16431907, PubMed:18086877, PubMed:18285464). Genes repressed by the PRC2 complex include HOXC8, HOXA9, MYT1 and CDKN2A (PubMed:15231737, PubMed:16618801, PubMed:17200670, PubMed:31959557). {ECO:0000269|PubMed:12351676, ECO:0000269|PubMed:12435631, ECO:0000269|PubMed:15099518, ECO:0000269|PubMed:15225548, ECO:0000269|PubMed:15231737, ECO:0000269|PubMed:15385962, ECO:0000269|PubMed:15684044, ECO:0000269|PubMed:16431907, ECO:0000269|PubMed:16618801, ECO:0000269|PubMed:17200670, ECO:0000269|PubMed:17344414, ECO:0000269|PubMed:18086877, ECO:0000269|PubMed:18285464, ECO:0000269|PubMed:28229514, ECO:0000269|PubMed:29499137, ECO:0000269|PubMed:31959557}. | FUNCTION: Stimulates the GTPase activity of Ras. NF1 shows greater affinity for Ras GAP, but lower specific activity. May be a regulator of Ras activity. {ECO:0000269|PubMed:2121371, ECO:0000269|PubMed:8417346}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SUZ12 | chr17:30303633 | chr17:29576002 | ENST00000322652 | + | 8 | 16 | 51_59 | 305.6666666666667 | 740.0 | Compositional bias | Note=Poly-Ser |
| Hgene | SUZ12 | chr17:30303633 | chr17:29576002 | ENST00000322652 | + | 8 | 16 | 60_67 | 305.6666666666667 | 740.0 | Compositional bias | Note=Poly-Ala |
| Hgene | SUZ12 | chr17:30303633 | chr17:29576002 | ENST00000322652 | + | 8 | 16 | 7_50 | 305.6666666666667 | 740.0 | Compositional bias | Note=Gly-rich |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000356175 | 28 | 57 | 1352_1355 | 1324.6666666666667 | 2819.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000358273 | 28 | 58 | 1352_1355 | 1324.6666666666667 | 2840.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000431387 | 0 | 15 | 1352_1355 | 0 | 594.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000356175 | 28 | 57 | 1580_1738 | 1324.6666666666667 | 2819.0 | Domain | CRAL-TRIO | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000358273 | 28 | 58 | 1580_1738 | 1324.6666666666667 | 2840.0 | Domain | CRAL-TRIO | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000431387 | 0 | 15 | 1235_1451 | 0 | 594.0 | Domain | Ras-GAP | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000431387 | 0 | 15 | 1580_1738 | 0 | 594.0 | Domain | CRAL-TRIO | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000356175 | 28 | 57 | 2555_2571 | 1324.6666666666667 | 2819.0 | Motif | Note=Bipartite nuclear localization signal | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000358273 | 28 | 58 | 2555_2571 | 1324.6666666666667 | 2840.0 | Motif | Note=Bipartite nuclear localization signal | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000431387 | 0 | 15 | 2555_2571 | 0 | 594.0 | Motif | Note=Bipartite nuclear localization signal | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000356175 | 28 | 57 | 1580_1837 | 1324.6666666666667 | 2819.0 | Region | Note=Lipid binding | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000358273 | 28 | 58 | 1580_1837 | 1324.6666666666667 | 2840.0 | Region | Note=Lipid binding | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000431387 | 0 | 15 | 1580_1837 | 0 | 594.0 | Region | Note=Lipid binding |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SUZ12 | chr17:30303633 | chr17:29576002 | ENST00000322652 | + | 8 | 16 | 563_639 | 305.6666666666667 | 740.0 | Region | Note=VEFS-box |
| Hgene | SUZ12 | chr17:30303633 | chr17:29576002 | ENST00000322652 | + | 8 | 16 | 448_471 | 305.6666666666667 | 740.0 | Zinc finger | Note=C2H2-type |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000356175 | 28 | 57 | 1235_1451 | 1324.6666666666667 | 2819.0 | Domain | Ras-GAP | |
| Tgene | NF1 | chr17:30303633 | chr17:29576002 | ENST00000358273 | 28 | 58 | 1235_1451 | 1324.6666666666667 | 2840.0 | Domain | Ras-GAP |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| NF1 | HRAS, SMARCA4, POLR2A, TIRAP, SIRT7, VCP, NSFL1C, CAV1, YWHAB, FAF2, NXF1, CCDC8, HLA-DPA1, EFNB2, CD274, TNFSF13B, CA14, SLAMF1, VSIG4, TGOLN2, HTR6, NOSIP, KRAS, CLK1, CDC5L, APP, YWHAH, PML, TRAF6, PTEN, SDC2, EGFR, CD79B, P4HA3, SCN3B, VSIG1, SIGLECL1, EPHA1, FAM174A, KCTD3, KIF13B, ZBTB21, KSR1, CGN, GIGYF1, LRFN1, RTKN, MAST3, DENND1A, SH3PXD2A, SRGAP2, DENND4C, PPM1H, EIF4E2, SIPA1L1, LIMA1, RALGPS2, MAGI1, CBY1, TESK2, CDC25C, DCLK1, GIGYF2, HDAC4, LPIN3, ZNF638, CAMSAP2, NADK, MAPKAP1, SRSF12, RASAL2, SYDE1, KIAA1804, TIAM1, AGAP1, CDC25B, CDK16, SH3RF3, PHLDB2, PLEKHA7, KIF1C, OSBPL6, FAM110B, USP21, NAV1, TANC2, PTPN13, FAM53C, ANKRD34A, STARD13, PTPN14, GAB2, DENND4A, INPP5E, TOP1, TOP2A, TOP3A, NRAS, ZUFSP, ESR2, LZTR1, SPRED1, MYC, LIMK2, KIAA1429, CTDP1, DYRK1A, F12, AKT1, EPHA2, MAP2K3, RAF1, SOX4, BIRC3, ORF7a, LCK, NOLC1, N, nsp7, KIF14, PRC1, MKI67, FASN, NR3C1, DDRGK1, FLOT1, LYN, PFN1, RHOB, ATG7, EFS, C19orf38, CDHR2, ITFG3, CD80, SEMA4C, DGCR2, EFNA4, PTGES2, LRRC25, EVI2A, EFNB1, TPST2, C3orf18, GYPA, HCST, TACSTD2, OPALIN, IL2RA, CD226, SIRT6, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| SUZ12 | 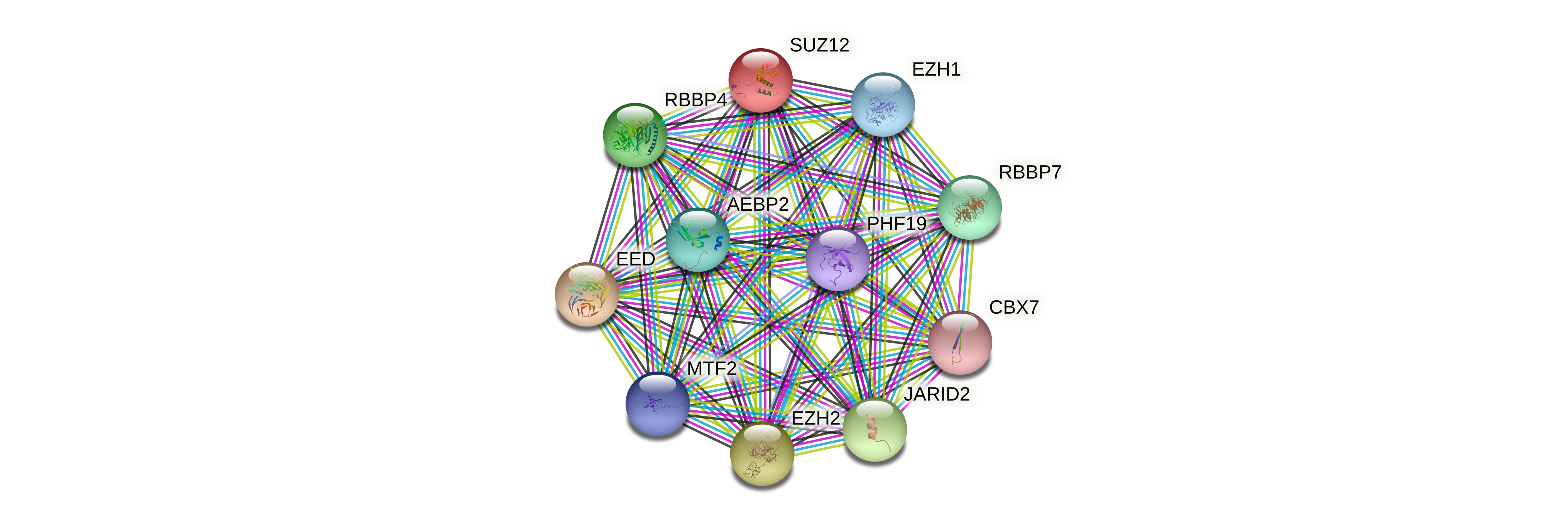 |
| NF1 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | SUZ12 | chr17:30303633 | chr17:29576002 | ENST00000322652 | + | 8 | 16 | 146_363 | 305.6666666666667 | 740.0 | AEBP2 and PHF19 |
Top |
Related Drugs to SUZ12-NF1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to SUZ12-NF1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | SUZ12 | C0206727 | Nerve Sheath Tumors | 2 | CTD_human |
| Hgene | SUZ12 | C0751689 | Peripheral Nerve Sheath Neoplasm | 2 | CTD_human |
| Hgene | SUZ12 | C0751691 | Perineurioma | 2 | CTD_human |
| Hgene | SUZ12 | C0001430 | Adenoma | 1 | CTD_human |
| Hgene | SUZ12 | C0014170 | Endometrial Neoplasms | 1 | CTD_human |
| Hgene | SUZ12 | C0017636 | Glioblastoma | 1 | CTD_human |
| Hgene | SUZ12 | C0017638 | Glioma | 1 | CTD_human |
| Hgene | SUZ12 | C0025202 | melanoma | 1 | CTD_human |
| Hgene | SUZ12 | C0027809 | Neurilemmoma | 1 | CTD_human |
| Hgene | SUZ12 | C0027830 | neurofibroma | 1 | CTD_human |
| Hgene | SUZ12 | C0027962 | Melanocytic nevus | 1 | CTD_human |
| Hgene | SUZ12 | C0042063 | Urogenital Abnormalities | 1 | GENOMICS_ENGLAND |
| Hgene | SUZ12 | C0205646 | Adenoma, Basal Cell | 1 | CTD_human |
| Hgene | SUZ12 | C0205647 | Follicular adenoma | 1 | CTD_human |
| Hgene | SUZ12 | C0205648 | Adenoma, Microcystic | 1 | CTD_human |
| Hgene | SUZ12 | C0205649 | Adenoma, Monomorphic | 1 | CTD_human |
| Hgene | SUZ12 | C0205650 | Papillary adenoma | 1 | CTD_human |
| Hgene | SUZ12 | C0205651 | Adenoma, Trabecular | 1 | CTD_human |
| Hgene | SUZ12 | C0206630 | Endometrial Stromal Sarcoma | 1 | ORPHANET |
| Hgene | SUZ12 | C0259783 | mixed gliomas | 1 | CTD_human |
| Hgene | SUZ12 | C0265210 | Weaver syndrome | 1 | ORPHANET |
| Hgene | SUZ12 | C0334588 | Giant Cell Glioblastoma | 1 | CTD_human |
| Hgene | SUZ12 | C0476089 | Endometrial Carcinoma | 1 | CTD_human |
| Hgene | SUZ12 | C0545053 | Advanced bone age | 1 | GENOMICS_ENGLAND |
| Hgene | SUZ12 | C0555198 | Malignant Glioma | 1 | CTD_human |
| Hgene | SUZ12 | C0557874 | Global developmental delay | 1 | GENOMICS_ENGLAND |
| Hgene | SUZ12 | C0751374 | Schwannomatosis, Plexiform | 1 | CTD_human |
| Hgene | SUZ12 | C1621958 | Glioblastoma Multiforme | 1 | CTD_human |
| Hgene | SUZ12 | C1842581 | Abnormal corpus callosum morphology | 1 | GENOMICS_ENGLAND |
| Hgene | SUZ12 | C1849265 | Overgrowth | 1 | GENOMICS_ENGLAND |
| Hgene | SUZ12 | C3714756 | Intellectual Disability | 1 | GENOMICS_ENGLAND |
| Hgene | SUZ12 | C4018871 | Abnormality of the respiratory system | 1 | GENOMICS_ENGLAND |
| Hgene | SUZ12 | C4021664 | Abnormality of the abdominal wall | 1 | GENOMICS_ENGLAND |
| Hgene | SUZ12 | C4021790 | Abnormality of the skeletal system | 1 | GENOMICS_ENGLAND |
| Tgene | NF1 | C0027831 | Neurofibromatosis 1 | 44 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | NF1 | C1708353 | Hereditary Paraganglioma-Pheochromocytoma Syndrome | 10 | CLINGEN |
| Tgene | NF1 | C0349639 | Juvenile Myelomonocytic Leukemia | 7 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | NF1 | C2931482 | Neurofibromatosis-Noonan syndrome | 6 | CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Tgene | NF1 | C0553586 | Cafe-au-lait macules with pulmonary stenosis | 5 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | NF1 | C0162678 | Neurofibromatoses | 3 | CGI;CTD_human;GENOMICS_ENGLAND |
| Tgene | NF1 | C0004114 | Astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0023467 | Leukemia, Myelocytic, Acute | 2 | CTD_human |
| Tgene | NF1 | C0025202 | melanoma | 2 | CGI;CTD_human |
| Tgene | NF1 | C0026998 | Acute Myeloid Leukemia, M1 | 2 | CTD_human |
| Tgene | NF1 | C0205768 | Subependymal Giant Cell Astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0206727 | Nerve Sheath Tumors | 2 | CTD_human |
| Tgene | NF1 | C0280783 | Juvenile Pilocytic Astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0280785 | Diffuse Astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0334579 | Anaplastic astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0334580 | Protoplasmic astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0334581 | Gemistocytic astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0334582 | Fibrillary Astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0334583 | Pilocytic Astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0338070 | Childhood Cerebral Astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0547065 | Mixed oligoastrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0750935 | Cerebral Astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0750936 | Intracranial Astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C0751689 | Peripheral Nerve Sheath Neoplasm | 2 | CTD_human |
| Tgene | NF1 | C0751691 | Perineurioma | 2 | CTD_human |
| Tgene | NF1 | C1704230 | Grade I Astrocytoma | 2 | CTD_human |
| Tgene | NF1 | C1834235 | NEUROFIBROMATOSIS, FAMILIAL SPINAL | 2 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | NF1 | C1879321 | Acute Myeloid Leukemia (AML-M2) | 2 | CTD_human |
| Tgene | NF1 | C0001430 | Adenoma | 1 | CTD_human |
| Tgene | NF1 | C0004352 | Autistic Disorder | 1 | CTD_human |
| Tgene | NF1 | C0016057 | Fibrosarcoma | 1 | CTD_human |
| Tgene | NF1 | C0017636 | Glioblastoma | 1 | CTD_human |
| Tgene | NF1 | C0017638 | Glioma | 1 | CGI;CTD_human |
| Tgene | NF1 | C0020796 | Profound Mental Retardation | 1 | CTD_human |
| Tgene | NF1 | C0023186 | Learning Disorders | 1 | CTD_human |
| Tgene | NF1 | C0023827 | liposarcoma | 1 | CTD_human |
| Tgene | NF1 | C0025363 | Mental Retardation, Psychosocial | 1 | CTD_human |
| Tgene | NF1 | C0026654 | Moyamoya Disease | 1 | GENOMICS_ENGLAND |
| Tgene | NF1 | C0027809 | Neurilemmoma | 1 | CTD_human |
| Tgene | NF1 | C0027830 | neurofibroma | 1 | CTD_human |
| Tgene | NF1 | C0027962 | Melanocytic nevus | 1 | CTD_human |
| Tgene | NF1 | C0028326 | Noonan Syndrome | 1 | GENOMICS_ENGLAND |
| Tgene | NF1 | C0031511 | Pheochromocytoma | 1 | CTD_human |
| Tgene | NF1 | C0035320 | Retinal Neovascularization | 1 | CTD_human |
| Tgene | NF1 | C0205646 | Adenoma, Basal Cell | 1 | CTD_human |
| Tgene | NF1 | C0205647 | Follicular adenoma | 1 | CTD_human |
| Tgene | NF1 | C0205648 | Adenoma, Microcystic | 1 | CTD_human |
| Tgene | NF1 | C0205649 | Adenoma, Monomorphic | 1 | CTD_human |
| Tgene | NF1 | C0205650 | Papillary adenoma | 1 | CTD_human |
| Tgene | NF1 | C0205651 | Adenoma, Trabecular | 1 | CTD_human |
| Tgene | NF1 | C0205824 | Liposarcoma, Dedifferentiated | 1 | CTD_human |
| Tgene | NF1 | C0205825 | Liposarcoma, Pleomorphic | 1 | CTD_human |
| Tgene | NF1 | C0205944 | Sarcoma, Epithelioid | 1 | CTD_human |
| Tgene | NF1 | C0205945 | Sarcoma, Spindle Cell | 1 | CTD_human |
| Tgene | NF1 | C0259783 | mixed gliomas | 1 | CTD_human |
| Tgene | NF1 | C0334588 | Giant Cell Glioblastoma | 1 | CTD_human |
| Tgene | NF1 | C0555198 | Malignant Glioma | 1 | CTD_human |
| Tgene | NF1 | C0751262 | Adult Learning Disorders | 1 | CTD_human |
| Tgene | NF1 | C0751263 | Learning Disturbance | 1 | CTD_human |
| Tgene | NF1 | C0751265 | Learning Disabilities | 1 | CTD_human |
| Tgene | NF1 | C0751374 | Schwannomatosis, Plexiform | 1 | CTD_human |
| Tgene | NF1 | C0917816 | Mental deficiency | 1 | CTD_human |
| Tgene | NF1 | C0917817 | Neurofibromatosis 3 | 1 | CTD_human |
| Tgene | NF1 | C1257877 | Pheochromocytoma, Extra-Adrenal | 1 | CTD_human |
| Tgene | NF1 | C1261473 | Sarcoma | 1 | CTD_human |
| Tgene | NF1 | C1330966 | Developmental Academic Disorder | 1 | CTD_human |
| Tgene | NF1 | C1370889 | Liposarcoma, well differentiated | 1 | CTD_human |
| Tgene | NF1 | C1621958 | Glioblastoma Multiforme | 1 | CTD_human |
| Tgene | NF1 | C3150928 | NF1 Microdeletion Syndrome | 1 | ORPHANET |
| Tgene | NF1 | C3714756 | Intellectual Disability | 1 | CTD_human |

