| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:BAP1-PBRM1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: BAP1-PBRM1 | FusionPDB ID: 8972 | FusionGDB2.0 ID: 8972 | Hgene | Tgene | Gene symbol | BAP1 | PBRM1 | Gene ID | 9223 | 55193 |
| Gene name | membrane associated guanylate kinase, WW and PDZ domain containing 1 | polybromo 1 | |
| Synonyms | AIP-3|AIP3|BAIAP1|BAP-1|BAP1|MAGI-1|MAGI-1b|Magi1d|TNRC19|WWP3 | BAF180|PB1 | |
| Cytomap | 3p14.1 | 3p21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1BAI1-associated protein 1WW domain-containing protein 3atrophin-1-interacting protein 3membrane-associated guanylate kinase inverted 1trinucleotide repeat-containing gene 19 | protein polybromo-1BRG1-associated factor 180polybromo-1D | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q8IXM2 | . | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000296288, ENST00000460680, | ENST00000296302, ENST00000337303, ENST00000356770, ENST00000394830, ENST00000409057, ENST00000409114, ENST00000409767, ENST00000410007, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 12 X 13 X 7=1092 | 10 X 10 X 7=700 |
| # samples | 13 | 10 | |
| ** MAII score | log2(13/1092*10)=-3.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(10/700*10)=-2.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: BAP1 [Title/Abstract] AND PBRM1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | BAP1(52438469)-PBRM1(52663051), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | BAP1-PBRM1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BAP1-PBRM1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. BAP1-PBRM1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. BAP1-PBRM1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | BAP1 | GO:0022409 | positive regulation of cell-cell adhesion | 20298433 |
 Fusion gene breakpoints across BAP1 (5'-gene) Fusion gene breakpoints across BAP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
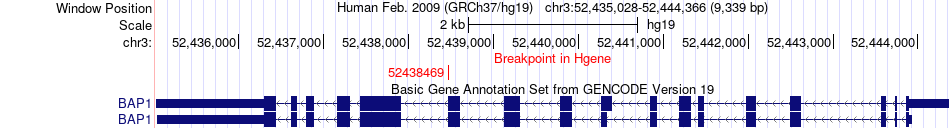 |
 Fusion gene breakpoints across PBRM1 (3'-gene) Fusion gene breakpoints across PBRM1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
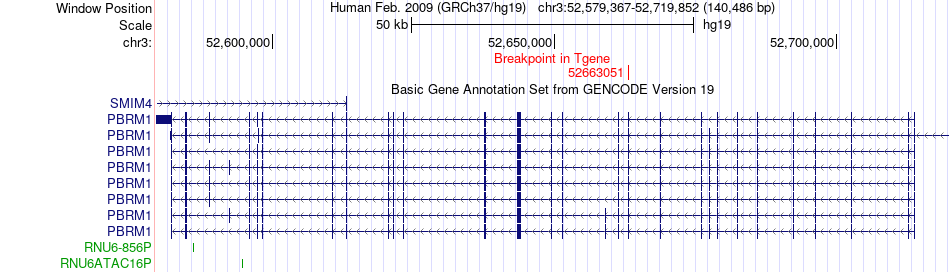 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-ER-A3ES-06A | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000460680 | BAP1 | chr3 | 52438469 | - | ENST00000356770 | PBRM1 | chr3 | 52663051 | - | 8037 | 1722 | 472 | 5325 | 1617 |
| ENST00000460680 | BAP1 | chr3 | 52438469 | - | ENST00000394830 | PBRM1 | chr3 | 52663051 | - | 5392 | 1722 | 472 | 5169 | 1565 |
| ENST00000460680 | BAP1 | chr3 | 52438469 | - | ENST00000296302 | PBRM1 | chr3 | 52663051 | - | 5564 | 1722 | 472 | 5490 | 1672 |
| ENST00000460680 | BAP1 | chr3 | 52438469 | - | ENST00000337303 | PBRM1 | chr3 | 52663051 | - | 5243 | 1722 | 472 | 5169 | 1565 |
| ENST00000460680 | BAP1 | chr3 | 52438469 | - | ENST00000410007 | PBRM1 | chr3 | 52663051 | - | 5323 | 1722 | 472 | 5250 | 1592 |
| ENST00000460680 | BAP1 | chr3 | 52438469 | - | ENST00000409057 | PBRM1 | chr3 | 52663051 | - | 5398 | 1722 | 472 | 5325 | 1617 |
| ENST00000460680 | BAP1 | chr3 | 52438469 | - | ENST00000409114 | PBRM1 | chr3 | 52663051 | - | 5433 | 1722 | 472 | 5379 | 1635 |
| ENST00000460680 | BAP1 | chr3 | 52438469 | - | ENST00000409767 | PBRM1 | chr3 | 52663051 | - | 5267 | 1722 | 472 | 5214 | 1580 |
| ENST00000296288 | BAP1 | chr3 | 52438469 | - | ENST00000356770 | PBRM1 | chr3 | 52663051 | - | 7547 | 1232 | 36 | 4835 | 1599 |
| ENST00000296288 | BAP1 | chr3 | 52438469 | - | ENST00000394830 | PBRM1 | chr3 | 52663051 | - | 4902 | 1232 | 36 | 4679 | 1547 |
| ENST00000296288 | BAP1 | chr3 | 52438469 | - | ENST00000296302 | PBRM1 | chr3 | 52663051 | - | 5074 | 1232 | 36 | 5000 | 1654 |
| ENST00000296288 | BAP1 | chr3 | 52438469 | - | ENST00000337303 | PBRM1 | chr3 | 52663051 | - | 4753 | 1232 | 36 | 4679 | 1547 |
| ENST00000296288 | BAP1 | chr3 | 52438469 | - | ENST00000410007 | PBRM1 | chr3 | 52663051 | - | 4833 | 1232 | 36 | 4760 | 1574 |
| ENST00000296288 | BAP1 | chr3 | 52438469 | - | ENST00000409057 | PBRM1 | chr3 | 52663051 | - | 4908 | 1232 | 36 | 4835 | 1599 |
| ENST00000296288 | BAP1 | chr3 | 52438469 | - | ENST00000409114 | PBRM1 | chr3 | 52663051 | - | 4943 | 1232 | 36 | 4889 | 1617 |
| ENST00000296288 | BAP1 | chr3 | 52438469 | - | ENST00000409767 | PBRM1 | chr3 | 52663051 | - | 4777 | 1232 | 36 | 4724 | 1562 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000460680 | ENST00000356770 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.000166417 | 0.9998336 |
| ENST00000460680 | ENST00000394830 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.001588807 | 0.9984112 |
| ENST00000460680 | ENST00000296302 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.002013928 | 0.9979861 |
| ENST00000460680 | ENST00000337303 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.001646103 | 0.99835396 |
| ENST00000460680 | ENST00000410007 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.002133053 | 0.9978669 |
| ENST00000460680 | ENST00000409057 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.000879415 | 0.99912053 |
| ENST00000460680 | ENST00000409114 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.002763297 | 0.9972367 |
| ENST00000460680 | ENST00000409767 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.001720536 | 0.99827945 |
| ENST00000296288 | ENST00000356770 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.000102246 | 0.9998977 |
| ENST00000296288 | ENST00000394830 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.000969003 | 0.99903107 |
| ENST00000296288 | ENST00000296302 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.001240788 | 0.99875927 |
| ENST00000296288 | ENST00000337303 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.000955329 | 0.9990446 |
| ENST00000296288 | ENST00000410007 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.001283769 | 0.99871624 |
| ENST00000296288 | ENST00000409057 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.000522561 | 0.99947745 |
| ENST00000296288 | ENST00000409114 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.001705387 | 0.99829465 |
| ENST00000296288 | ENST00000409767 | BAP1 | chr3 | 52438469 | - | PBRM1 | chr3 | 52663051 | - | 0.001007273 | 0.99899274 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >8972_8972_1_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000296288_PBRM1_chr3_52663051_ENST00000296302_length(amino acids)=1654AA_BP=398 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSA SNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLA AGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKK ELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNI RNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEV YEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKV LLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMF EVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSS GAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHLATRKFLEKEVFKSDYYNK VPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDN SEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHE PTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPI VPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSG YILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEERAAKVAEQQERERAAQQQQPSASPRAGTPVGALMGVVPPPTPMGM LNQQLTPVAGMMGGYPPGLPPLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGVPGLPGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVLG PPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAARRRDVHLSKEQESRLPSHWLKS -------------------------------------------------------------- >8972_8972_2_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000296288_PBRM1_chr3_52663051_ENST00000337303_length(amino acids)=1547AA_BP=398 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSA SNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLA AGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKK ELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNI RNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEV YEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKV LLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMF EVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSS GAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHLATRKFLEKEVFKSDYYNK VPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDN SEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHE PTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPI VPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSG YILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAG PPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAARRRDVHLSKEQESRLPSHWLKSKGAHTTMADALWRLRDL -------------------------------------------------------------- >8972_8972_3_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000296288_PBRM1_chr3_52663051_ENST00000356770_length(amino acids)=1599AA_BP=398 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSA SNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLA AGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKK ELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNI RNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEV YEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKV LLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMF EVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSS GAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHLATRKFLEKEVFKSDYYNK VPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDN SEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHE PTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPI VPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSG YILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGMMGGYPPGLPPLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGVPGL PGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAE -------------------------------------------------------------- >8972_8972_4_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000296288_PBRM1_chr3_52663051_ENST00000394830_length(amino acids)=1547AA_BP=398 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSA SNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLA AGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKK ELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNI RNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEV YEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKV LLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMF EVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSS GAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPEN FRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSN GEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCIL GKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELE GGDDDIEEMGEEDSESTPKSAKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGMMGGYPP GLPPLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGVPGLPGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAG PPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAARRRDVHLSKEQESRLPSHWLKSKGAHTTMADALWRLRDL -------------------------------------------------------------- >8972_8972_5_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000296288_PBRM1_chr3_52663051_ENST00000409057_length(amino acids)=1599AA_BP=398 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSA SNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLA AGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKK ELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNI RNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEV YEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKV LLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMF EVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSS GAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHLATRKFLEKEVFKSDYYNK VPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDN SEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHE PTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPI VPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSG YILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGMMGGYPPGLPPLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGVPGL PGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAE -------------------------------------------------------------- >8972_8972_6_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000296288_PBRM1_chr3_52663051_ENST00000409114_length(amino acids)=1617AA_BP=398 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSA SNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLA AGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKK ELARRDDIEDGDSMISSATSDTGSAKRKRNTHDSEMLGLRRLSSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYK IILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPK KSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNE PESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVE NNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLK REEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHLATR KFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASV FANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAY FYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLS AKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKG SAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEERAAKVAEQQERERAAQQQQPSASPRAGTPV GALMGVVPPPTPMGMLNQQLTPVAGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQ -------------------------------------------------------------- >8972_8972_7_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000296288_PBRM1_chr3_52663051_ENST00000409767_length(amino acids)=1562AA_BP=398 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSA SNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLA AGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKK ELARRDDIEDGDSMISSATSDTGSAKRKRNTHDSEMLGLRRLSSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYK IILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPK KSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNE PESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVE NNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLK REEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHLATR KFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASV FANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAY FYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLS AKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKG SAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPP GQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAARRRDVHLSKEQESRLPSHWLKSKG -------------------------------------------------------------- >8972_8972_8_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000296288_PBRM1_chr3_52663051_ENST00000410007_length(amino acids)=1574AA_BP=398 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGIKYEARLHVLKVNRQTVLEALQQLIRVTQPELIQTHKSQESQLPEESKSA SNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAFLDNHNYAKSPMQEEEDLA AGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNSAIYKRVLKLQQVMQAKKK ELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNI RNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGISPKKSKYMTPMQQKLNEV YEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACTYNEPESLIYKDALVLHKV LLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMF EVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEEDKLKREEEKREAEKSEDSS GAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPEN FRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSN GEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCIL GKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELE GGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELS RLVGTEWRNLETAKKAEYEGMMGGYPPGLPPLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGVPGLPGIPPPGVMNQGVAPMVGTPAPGGS PYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAARRRDVHLSKEQE -------------------------------------------------------------- >8972_8972_9_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000460680_PBRM1_chr3_52663051_ENST00000296302_length(amino acids)=1672AA_BP=416 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQPEL IQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAF LDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNS AIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPD YYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGI SPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACT YNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRK NVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEED KLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHL ATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRV ASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDG AAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRF SLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKS AKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEERAAKVAEQQERERAAQQQQPSASPRAG TPVGALMGVVPPPTPMGMLNQQLTPVAGMMGGYPPGLPPLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGVPGLPGIPPPGVMNQGVAPMV GTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAARRRD -------------------------------------------------------------- >8972_8972_10_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000460680_PBRM1_chr3_52663051_ENST00000337303_length(amino acids)=1565AA_BP=416 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQPEL IQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAF LDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNS AIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPD YYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGI SPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACT YNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRK NVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEED KLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHL ATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRV ASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDG AAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRF SLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKS AKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGVMNQGVAPMVGTPAPGGSPYGQQVGVL GPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAARRRDVHLSKEQESRLPSHWLK -------------------------------------------------------------- >8972_8972_11_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000460680_PBRM1_chr3_52663051_ENST00000356770_length(amino acids)=1617AA_BP=416 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQPEL IQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAF LDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNS AIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPD YYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGI SPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACT YNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRK NVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEED KLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHL ATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRV ASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDG AAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRF SLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKS AKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGMMGGYPPGLPPLQGPVDGLVSMGSMQP LHPGGPPPHHLPPGVPGLPGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQ -------------------------------------------------------------- >8972_8972_12_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000460680_PBRM1_chr3_52663051_ENST00000394830_length(amino acids)=1565AA_BP=416 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQPEL IQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAF LDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNS AIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPD YYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGI SPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACT YNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRK NVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEED KLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAEKEVFKSDYYNKVPVSKIL GKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAE DNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYK KEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPS PLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSESTPKSAKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRN LETAKKAEYEGMMGGYPPGLPPLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGVPGLPGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVL GPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAARRRDVHLSKEQESRLPSHWLK -------------------------------------------------------------- >8972_8972_13_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000460680_PBRM1_chr3_52663051_ENST00000409057_length(amino acids)=1617AA_BP=416 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQPEL IQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAF LDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNS AIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPD YYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGI SPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACT YNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRK NVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEED KLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKWLYGCWFYRPNETFHL ATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRV ASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDG AAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRF SLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKS AKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGMMGGYPPGLPPLQGPVDGLVSMGSMQP LHPGGPPPHHLPPGVPGLPGIPPPGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQ -------------------------------------------------------------- >8972_8972_14_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000460680_PBRM1_chr3_52663051_ENST00000409114_length(amino acids)=1635AA_BP=416 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQPEL IQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAF LDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNS AIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKRNTHDSEMLGLRRLSSKKNIRKQRMKILFNVVLEAREPGSGRRL CDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDD DMASPKLKLSRKSGISPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDID SMVEDFVMMFNNACTYNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDP NFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDV EKERKEKLPKEIEEDKLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKW LYGCWFYRPNETFHLATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISS VRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLV RPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYN ESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASE LDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEERAAKVAEQQERE RAAQQQQPSASPRAGTPVGALMGVVPPPTPMGMLNQQLTPVAGVMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPP VIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAARRRDVHLSKEQESRLPSHWLKSKGAHTTMADALWRLRDLML -------------------------------------------------------------- >8972_8972_15_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000460680_PBRM1_chr3_52663051_ENST00000409767_length(amino acids)=1580AA_BP=416 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQPEL IQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAF LDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNS AIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKRNTHDSEMLGLRRLSSKKNIRKQRMKILFNVVLEAREPGSGRRL CDLFMVKPSKKDYPDYYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDD DMASPKLKLSRKSGISPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDID SMVEDFVMMFNNACTYNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDP NFPNKPPLTFDIIRKNVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDV EKERKEKLPKEIEEDKLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAGEKW LYGCWFYRPNETFHLATRKFLEKEVFKSDYYNKVPVSKILGKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISS VRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAEDNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLV RPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYKKEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYN ESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPSPLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASE LDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSGYILFSSEMRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGVMNQGVAPMVGT PAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKWDQTLAARRRDVH -------------------------------------------------------------- >8972_8972_16_BAP1-PBRM1_BAP1_chr3_52438469_ENST00000460680_PBRM1_chr3_52663051_ENST00000410007_length(amino acids)=1592AA_BP=416 MNKGWLELESDPGLFTLLVEDFGVKGVQVEEIYDLQSKCQGPVYGFIFLFKWIEERRSRRKVSTLVDDTSVIDDDIVNNMFFAHQLIPNS CATHALLSVLLNCSSVDLGPTLSRMKDFTKGFSPESKGYAIGNAPELAKAHNSHARPEPRHLPEKQNGLSAVRTMEAFHFVSYVPITGRL FELDGLKVYPIDHGPWGEDEEWTDKARRVIMERIGLATAGEPYHDIRFNLMAVVPDRRIKYEARLHVLKVNRQTVLEALQQLIRVTQPEL IQTHKSQESQLPEESKSASNKSPLVLEANRAPAASEGNHTDGAEEAAGSCAQAPSHSPPNKPKLVVKPPGSSLNGVHPNPTPIVQRLPAF LDNHNYAKSPMQEEEDLAAGVGRSRVPVRPPQQYSDDEDDYEDDEEDDVQNTNSALRTKLKNQEYETLDHLECDLNLMFENAKRYNVPNS AIYKRVLKLQQVMQAKKKELARRDDIEDGDSMISSATSDTGSAKRKSKKNIRKQRMKILFNVVLEAREPGSGRRLCDLFMVKPSKKDYPD YYKIILEPMDLKIIEHNIRNDKYAGEEGMIEDMKLMFRNARHYNEEGSQVYNDAHILEKLLKEKRKELGPLPDDDDMASPKLKLSRKSGI SPKKSKYMTPMQQKLNEVYEAVKNYTDKRGRRLSAIFLRLPSRSELPDYYLTIKKPMDMEKIRSHMMANKYQDIDSMVEDFVMMFNNACT YNEPESLIYKDALVLHKVLLETRRDLEGDEDSHVPNVTLLIQELIHNLFVSVMSHQDDEGRCYSDSLAEIPAVDPNFPNKPPLTFDIIRK NVENNRYRRLDLFQEHMFEVLERARRMNRTDSEIYEDAVELQQFFIKIRDELCKNGEILLSPALSYTTKHLHNDVEKERKEKLPKEIEED KLKREEEKREAEKSEDSSGAAGLSGLHRTYSQDCSFKNSMYHVGDYVYVEPAEANLQPHIVCIERLWEDSAEKEVFKSDYYNKVPVSKIL GKCVVMFVKEYFKLCPENFRDEDVFVCESRYSAKTKSFKKIKLWTMPISSVRFVPRDVPLPVVRVASVFANADKGDDEKNTDNSEDSRAE DNFNLEKEKEDVPVEMSNGEPGCHYFEQLHYNDMWLKVGDCVFIKSHGLVRPRVGRIEKVWVRDGAAYFYGPIFIHPEETEHEPTKMFYK KEVFLSNLEETCPMTCILGKCAVLSFKDFLSCRPTEIPENDILLCESRYNESDKQMKKFKGLKRFSLSAKVVDDEIYYFRKPIVPQKEPS PLLEKKIQLLEAKFAELEGGDDDIEEMGEEDSEVIEPPSLPQLQTPLASELDLMPYTPPQSTPKSAKGSAKKEGSKRKINMSGYILFSSE MRAVIKAQHPDYSFGELSRLVGTEWRNLETAKKAEYEGMMGGYPPGLPPLQGPVDGLVSMGSMQPLHPGGPPPHHLPPGVPGLPGIPPPG VMNQGVAPMVGTPAPGGSPYGQQVGVLGPPGQQAPPPYPGPHPAGPPVIQQPTTPMFVAPPPKTQRLLHSEAYLKYIEGLSAESNSISKW -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:52438469/chr3:52663051) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| BAP1 | . |
| FUNCTION: Component of chromatin complexes such as the MLL1/MLL and NURF complexes. | FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 1468_1599 | 433.6666666666667 | 1690.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 630_633 | 433.6666666666667 | 1690.0 | Compositional bias | Note=Poly-Asp | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 1468_1599 | 433.6666666666667 | 1583.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 630_633 | 433.6666666666667 | 1583.0 | Compositional bias | Note=Poly-Asp | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 1468_1599 | 401.6666666666667 | 1603.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 630_633 | 401.6666666666667 | 1603.0 | Compositional bias | Note=Poly-Asp | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 1468_1599 | 433.6666666666667 | 1583.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 630_633 | 433.6666666666667 | 1583.0 | Compositional bias | Note=Poly-Asp | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 1468_1599 | 433.6666666666667 | 1635.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 630_633 | 433.6666666666667 | 1635.0 | Compositional bias | Note=Poly-Asp | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 1468_1599 | 433.6666666666667 | 1653.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 630_633 | 433.6666666666667 | 1653.0 | Compositional bias | Note=Poly-Asp | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 1468_1599 | 433.6666666666667 | 1598.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 630_633 | 433.6666666666667 | 1598.0 | Compositional bias | Note=Poly-Asp | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 1468_1599 | 433.6666666666667 | 1610.0 | Compositional bias | Note=Pro-rich | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 630_633 | 433.6666666666667 | 1610.0 | Compositional bias | Note=Poly-Asp | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 1379_1447 | 433.6666666666667 | 1690.0 | DNA binding | HMG box | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 1379_1447 | 433.6666666666667 | 1583.0 | DNA binding | HMG box | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 1379_1447 | 401.6666666666667 | 1603.0 | DNA binding | HMG box | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 1379_1447 | 433.6666666666667 | 1583.0 | DNA binding | HMG box | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 1379_1447 | 433.6666666666667 | 1635.0 | DNA binding | HMG box | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 1379_1447 | 433.6666666666667 | 1653.0 | DNA binding | HMG box | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 1379_1447 | 433.6666666666667 | 1598.0 | DNA binding | HMG box | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 1379_1447 | 433.6666666666667 | 1610.0 | DNA binding | HMG box | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 1156_1272 | 433.6666666666667 | 1690.0 | Domain | BAH 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 538_608 | 433.6666666666667 | 1690.0 | Domain | Bromo 4 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 676_746 | 433.6666666666667 | 1690.0 | Domain | Bromo 5 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 792_862 | 433.6666666666667 | 1690.0 | Domain | Bromo 6 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 956_1074 | 433.6666666666667 | 1690.0 | Domain | BAH 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 1156_1272 | 433.6666666666667 | 1583.0 | Domain | BAH 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 538_608 | 433.6666666666667 | 1583.0 | Domain | Bromo 4 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 676_746 | 433.6666666666667 | 1583.0 | Domain | Bromo 5 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 792_862 | 433.6666666666667 | 1583.0 | Domain | Bromo 6 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 956_1074 | 433.6666666666667 | 1583.0 | Domain | BAH 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 1156_1272 | 401.6666666666667 | 1603.0 | Domain | BAH 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 400_470 | 401.6666666666667 | 1603.0 | Domain | Bromo 3 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 538_608 | 401.6666666666667 | 1603.0 | Domain | Bromo 4 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 676_746 | 401.6666666666667 | 1603.0 | Domain | Bromo 5 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 792_862 | 401.6666666666667 | 1603.0 | Domain | Bromo 6 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 956_1074 | 401.6666666666667 | 1603.0 | Domain | BAH 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 1156_1272 | 433.6666666666667 | 1583.0 | Domain | BAH 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 538_608 | 433.6666666666667 | 1583.0 | Domain | Bromo 4 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 676_746 | 433.6666666666667 | 1583.0 | Domain | Bromo 5 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 792_862 | 433.6666666666667 | 1583.0 | Domain | Bromo 6 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 956_1074 | 433.6666666666667 | 1583.0 | Domain | BAH 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 1156_1272 | 433.6666666666667 | 1635.0 | Domain | BAH 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 538_608 | 433.6666666666667 | 1635.0 | Domain | Bromo 4 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 676_746 | 433.6666666666667 | 1635.0 | Domain | Bromo 5 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 792_862 | 433.6666666666667 | 1635.0 | Domain | Bromo 6 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 956_1074 | 433.6666666666667 | 1635.0 | Domain | BAH 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 1156_1272 | 433.6666666666667 | 1653.0 | Domain | BAH 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 538_608 | 433.6666666666667 | 1653.0 | Domain | Bromo 4 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 676_746 | 433.6666666666667 | 1653.0 | Domain | Bromo 5 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 792_862 | 433.6666666666667 | 1653.0 | Domain | Bromo 6 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 956_1074 | 433.6666666666667 | 1653.0 | Domain | BAH 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 1156_1272 | 433.6666666666667 | 1598.0 | Domain | BAH 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 538_608 | 433.6666666666667 | 1598.0 | Domain | Bromo 4 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 676_746 | 433.6666666666667 | 1598.0 | Domain | Bromo 5 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 792_862 | 433.6666666666667 | 1598.0 | Domain | Bromo 6 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 956_1074 | 433.6666666666667 | 1598.0 | Domain | BAH 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 1156_1272 | 433.6666666666667 | 1610.0 | Domain | BAH 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 538_608 | 433.6666666666667 | 1610.0 | Domain | Bromo 4 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 676_746 | 433.6666666666667 | 1610.0 | Domain | Bromo 5 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 792_862 | 433.6666666666667 | 1610.0 | Domain | Bromo 6 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 956_1074 | 433.6666666666667 | 1610.0 | Domain | BAH 1 |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | BAP1 | chr3:52438469 | chr3:52663051 | ENST00000460680 | - | 12 | 17 | 630_661 | 416.6666666666667 | 730.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 200_270 | 433.6666666666667 | 1690.0 | Domain | Bromo 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 400_470 | 433.6666666666667 | 1690.0 | Domain | Bromo 3 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000296302 | 10 | 30 | 64_134 | 433.6666666666667 | 1690.0 | Domain | Bromo 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 200_270 | 433.6666666666667 | 1583.0 | Domain | Bromo 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 400_470 | 433.6666666666667 | 1583.0 | Domain | Bromo 3 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000337303 | 10 | 28 | 64_134 | 433.6666666666667 | 1583.0 | Domain | Bromo 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 200_270 | 401.6666666666667 | 1603.0 | Domain | Bromo 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000356770 | 9 | 28 | 64_134 | 401.6666666666667 | 1603.0 | Domain | Bromo 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 200_270 | 433.6666666666667 | 1583.0 | Domain | Bromo 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 400_470 | 433.6666666666667 | 1583.0 | Domain | Bromo 3 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000394830 | 11 | 30 | 64_134 | 433.6666666666667 | 1583.0 | Domain | Bromo 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 200_270 | 433.6666666666667 | 1635.0 | Domain | Bromo 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 400_470 | 433.6666666666667 | 1635.0 | Domain | Bromo 3 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409057 | 10 | 29 | 64_134 | 433.6666666666667 | 1635.0 | Domain | Bromo 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 200_270 | 433.6666666666667 | 1653.0 | Domain | Bromo 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 400_470 | 433.6666666666667 | 1653.0 | Domain | Bromo 3 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409114 | 10 | 30 | 64_134 | 433.6666666666667 | 1653.0 | Domain | Bromo 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 200_270 | 433.6666666666667 | 1598.0 | Domain | Bromo 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 400_470 | 433.6666666666667 | 1598.0 | Domain | Bromo 3 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000409767 | 10 | 29 | 64_134 | 433.6666666666667 | 1598.0 | Domain | Bromo 1 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 200_270 | 433.6666666666667 | 1610.0 | Domain | Bromo 2 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 400_470 | 433.6666666666667 | 1610.0 | Domain | Bromo 3 | |
| Tgene | PBRM1 | chr3:52438469 | chr3:52663051 | ENST00000410007 | 10 | 29 | 64_134 | 433.6666666666667 | 1610.0 | Domain | Bromo 1 |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| BAP1 | |
| PBRM1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | BAP1 | chr3:52438469 | chr3:52663051 | ENST00000460680 | - | 12 | 17 | 596_721 | 416.6666666666667 | 730.0 | BRCA1 |
Top |
Related Drugs to BAP1-PBRM1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to BAP1-PBRM1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

