| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:TCF4-MAML3 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: TCF4-MAML3 | FusionPDB ID: 89788 | FusionGDB2.0 ID: 89820 | Hgene | Tgene | Gene symbol | TCF4 | MAML3 | Gene ID | 6934 | 55534 |
| Gene name | transcription factor 7 like 2 | mastermind like transcriptional coactivator 3 | |
| Synonyms | TCF-4|TCF4 | CAGH3|ERDA3|GDN|MAM-2|MAM2|TNRC3|mam-3 | |
| Cytomap | 10q25.2-q25.3 | 4q31.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transcription factor 7-like 2HMG box transcription factor 4T-cell factor 4T-cell-specific transcription factor 4hTCF-4transcription factor 7-like 2 (T-cell specific, HMG-box) | mastermind-like protein 3CAG repeat containing (glia-derived nexin I alpha)expanded repeat domain, CAG/CTG 3polyglutamine richtrinucleotide repeat containing 3 | |
| Modification date | 20200315 | 20200313 | |
| UniProtAcc | . | Q96JK9 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000354452, ENST00000356073, ENST00000398339, ENST00000457482, ENST00000537578, ENST00000537856, ENST00000540999, ENST00000543082, ENST00000544241, ENST00000561831, ENST00000561992, ENST00000563760, ENST00000564228, ENST00000564403, ENST00000564999, ENST00000565018, ENST00000566279, ENST00000566286, ENST00000567880, ENST00000568673, ENST00000568740, ENST00000570177, ENST00000570287, | ENST00000398940, ENST00000327122, ENST00000509479, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 13 X 13 X 4=676 | 13 X 13 X 8=1352 |
| # samples | 13 | 16 | |
| ** MAII score | log2(13/676*10)=-2.37851162325373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(16/1352*10)=-3.07895134139482 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: TCF4 [Title/Abstract] AND MAML3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | TCF4(52921728)-MAML3(140812121), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | TCF4-MAML3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. TCF4-MAML3 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. TCF4-MAML3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. TCF4-MAML3 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. TCF4-MAML3 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. TCF4-MAML3 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TCF4 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 12799378 |
| Hgene | TCF4 | GO:0006357 | regulation of transcription by RNA polymerase II | 9727977 |
| Hgene | TCF4 | GO:0032092 | positive regulation of protein binding | 12799378 |
| Hgene | TCF4 | GO:0032350 | regulation of hormone metabolic process | 15525634 |
| Hgene | TCF4 | GO:0042593 | glucose homeostasis | 15525634 |
| Hgene | TCF4 | GO:0043433 | negative regulation of DNA-binding transcription factor activity | 12799378 |
| Hgene | TCF4 | GO:0045444 | fat cell differentiation | 10937998 |
| Hgene | TCF4 | GO:0045892 | negative regulation of transcription, DNA-templated | 12799378|15525634 |
| Hgene | TCF4 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 9065401|19168596 |
| Hgene | TCF4 | GO:0048625 | myoblast fate commitment | 10937998 |
| Tgene | MAML3 | GO:0007219 | Notch signaling pathway | 12370315 |
| Tgene | MAML3 | GO:0045944 | positive regulation of transcription by RNA polymerase II | 12370315 |
 Fusion gene breakpoints across TCF4 (5'-gene) Fusion gene breakpoints across TCF4 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across MAML3 (3'-gene) Fusion gene breakpoints across MAML3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
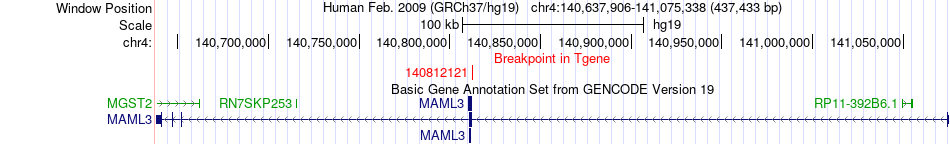 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PCPG | TCGA-WB-A80Y-01A | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000457482 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 6945 | 1426 | 556 | 4374 | 1272 |
| ENST00000457482 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 3055 | 1426 | 556 | 3054 | 833 |
| ENST00000543082 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 6915 | 1396 | 172 | 4344 | 1390 |
| ENST00000543082 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 3025 | 1396 | 172 | 3024 | 951 |
| ENST00000544241 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 6937 | 1418 | 206 | 4366 | 1386 |
| ENST00000544241 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 3047 | 1418 | 206 | 3046 | 947 |
| ENST00000561831 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 6531 | 1012 | 142 | 3960 | 1272 |
| ENST00000561831 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 2641 | 1012 | 142 | 2640 | 833 |
| ENST00000565018 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 7113 | 1594 | 226 | 4542 | 1438 |
| ENST00000565018 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 3223 | 1594 | 226 | 3222 | 999 |
| ENST00000570287 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 6637 | 1118 | 248 | 4066 | 1272 |
| ENST00000570287 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 2747 | 1118 | 248 | 2746 | 833 |
| ENST00000564228 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 6656 | 1137 | 0 | 4085 | 1361 |
| ENST00000564228 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 2766 | 1137 | 0 | 2765 | 922 |
| ENST00000567880 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 6689 | 1170 | 0 | 4118 | 1372 |
| ENST00000567880 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 2799 | 1170 | 0 | 2798 | 933 |
| ENST00000566286 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 6860 | 1341 | 0 | 4289 | 1429 |
| ENST00000566286 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 2970 | 1341 | 0 | 2969 | 990 |
| ENST00000564403 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 7414 | 1895 | 509 | 4843 | 1444 |
| ENST00000564403 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 3524 | 1895 | 509 | 3523 | 1005 |
| ENST00000398339 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 7232 | 1713 | 57 | 4661 | 1534 |
| ENST00000398339 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 3342 | 1713 | 57 | 3341 | 1095 |
| ENST00000570177 | TCF4 | chr18 | 52921728 | - | ENST00000509479 | MAML3 | chr4 | 140812121 | - | 6686 | 1167 | 78 | 4115 | 1345 |
| ENST00000570177 | TCF4 | chr18 | 52921728 | - | ENST00000327122 | MAML3 | chr4 | 140812121 | - | 2796 | 1167 | 78 | 2795 | 906 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000457482 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.007250679 | 0.99274933 |
| ENST00000457482 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.16395956 | 0.83604044 |
| ENST00000543082 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.006022517 | 0.9939774 |
| ENST00000543082 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.019252932 | 0.98074704 |
| ENST00000544241 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.005739211 | 0.9942608 |
| ENST00000544241 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.06099865 | 0.9390013 |
| ENST00000561831 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.007213175 | 0.9927868 |
| ENST00000561831 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.16509603 | 0.83490396 |
| ENST00000565018 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.005643476 | 0.99435645 |
| ENST00000565018 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.030478628 | 0.96952134 |
| ENST00000570287 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.006973014 | 0.9930269 |
| ENST00000570287 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.15038846 | 0.8496115 |
| ENST00000564228 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.005642818 | 0.99435717 |
| ENST00000564228 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.024299286 | 0.9757007 |
| ENST00000567880 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.005977881 | 0.9940221 |
| ENST00000567880 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.06629192 | 0.9337081 |
| ENST00000566286 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.005559165 | 0.99444085 |
| ENST00000566286 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.006172056 | 0.9938279 |
| ENST00000564403 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.006392723 | 0.9936073 |
| ENST00000564403 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.04562081 | 0.95437914 |
| ENST00000398339 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.005128549 | 0.9948715 |
| ENST00000398339 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.025570443 | 0.9744296 |
| ENST00000570177 | ENST00000509479 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.006894576 | 0.9931055 |
| ENST00000570177 | ENST00000327122 | TCF4 | chr18 | 52921728 | - | MAML3 | chr4 | 140812121 | - | 0.010713556 | 0.9892865 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >89788_89788_1_TCF4-MAML3_TCF4_chr18_52921728_ENST00000398339_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=1095AA_BP=552 MFSKRLEKIAQVPLLFPFIFIILTNYSKMEGAVESQPSFFKTSQDIVTCTWVENCYSSFSRRPLEQMFCKHQSKNIISWTGMVAHTCNPS TLGGQGLCDFAKMHHQQRMAALGTDKELSDLLDFSAMFSPPVSSGKNGPTSLASGHFTGSNVEDRSSSGSWGNGGHPSPSRNYGDGTPYD HMTSRDLGSHDNLSPPFVNSRIQSKTERGSYSSYGRESNLQGCHQQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEV QTKKVRKVPPGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLH PHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTP VGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYG TGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGT HTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDD IQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCA VQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQ QQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQH NILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMA -------------------------------------------------------------- >89788_89788_2_TCF4-MAML3_TCF4_chr18_52921728_ENST00000398339_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1534AA_BP=552 MFSKRLEKIAQVPLLFPFIFIILTNYSKMEGAVESQPSFFKTSQDIVTCTWVENCYSSFSRRPLEQMFCKHQSKNIISWTGMVAHTCNPS TLGGQGLCDFAKMHHQQRMAALGTDKELSDLLDFSAMFSPPVSSGKNGPTSLASGHFTGSNVEDRSSSGSWGNGGHPSPSRNYGDGTPYD HMTSRDLGSHDNLSPPFVNSRIQSKTERGSYSSYGRESNLQGCHQQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEV QTKKVRKVPPGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLH PHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTP VGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYG TGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGT HTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDD IQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCA VQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQ QQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQH NILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMA YAALPSHGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQMLIDQRAQLIEQQKQQFLREQRQQQQQQQQQIL AEQQLQQSHLPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQNAGMMGIGPSQNPGTMATAAAQSEMGLAPYSTT PTSQPGMYNMSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVNSAITQQHPQMKGPVGQALPRPQAPPRLQSLMG TVQQGAQSWQQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVDANTGTVRTLNPAAMGRQMMPSLPGQQGTSQAR PMVMSGLSQGVPGMPAFSQPPAQQQIPSGSFAPSSQSQAYERNAPQDVSYNYSGDGAGGSFPGLPDGADLVDSIIKGGPGDEWMQELDEL -------------------------------------------------------------- >89788_89788_3_TCF4-MAML3_TCF4_chr18_52921728_ENST00000457482_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=833AA_BP=290 MYCAYTIPGMGGNSLMYYYNGKAVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQ SSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNS FSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAM GGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPS LPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDEL ANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNP ASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQ QQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLG TNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQK -------------------------------------------------------------- >89788_89788_4_TCF4-MAML3_TCF4_chr18_52921728_ENST00000457482_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1272AA_BP=290 MYCAYTIPGMGGNSLMYYYNGKAVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQ SSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNS FSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAM GGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPS LPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDEL ANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNP ASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQ QQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLG TNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQK GVMNQPMAYAALPSHGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQMLIDQRAQLIEQQKQQFLREQRQQQ QQQQQQILAEQQLQQSHLPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQNAGMMGIGPSQNPGTMATAAAQSEM GLAPYSTTPTSQPGMYNMSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVNSAITQQHPQMKGPVGQALPRPQAP PRLQSLMGTVQQGAQSWQQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVDANTGTVRTLNPAAMGRQMMPSLPG QQGTSQARPMVMSGLSQGVPGMPAFSQPPAQQQIPSGSFAPSSQSQAYERNAPQDVSYNYSGDGAGGSFPGLPDGADLVDSIIKGGPGDE -------------------------------------------------------------- >89788_89788_5_TCF4-MAML3_TCF4_chr18_52921728_ENST00000543082_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=951AA_BP=408 MEEDSRDVEDRSSSGSWGNGGHPSPSRNYGDGTPYDHMTSRDLGSHDNLSPPFVNSRIQSKTERGSYSSYGRESNLQGCHQQSLLGGDMD MGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVRKVPPGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHH SSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGS GAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRN HAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISA GMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTS ETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARP SSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQL KQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMA NNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQ -------------------------------------------------------------- >89788_89788_6_TCF4-MAML3_TCF4_chr18_52921728_ENST00000543082_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1390AA_BP=408 MEEDSRDVEDRSSSGSWGNGGHPSPSRNYGDGTPYDHMTSRDLGSHDNLSPPFVNSRIQSKTERGSYSSYGRESNLQGCHQQSLLGGDMD MGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVRKVPPGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHH SSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGS GAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRN HAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISA GMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTS ETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARP SSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQL KQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMA NNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQ QQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMAYAALPSHGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAI MKQMLIDQRAQLIEQQKQQFLREQRQQQQQQQQQILAEQQLQQSHLPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRL MAQNAGMMGIGPSQNPGTMATAAAQSEMGLAPYSTTPTSQPGMYNMSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQS MLVNSAITQQHPQMKGPVGQALPRPQAPPRLQSLMGTVQQGAQSWQQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQ SVVDANTGTVRTLNPAAMGRQMMPSLPGQQGTSQARPMVMSGLSQGVPGMPAFSQPPAQQQIPSGSFAPSSQSQAYERNAPQDVSYNYSG -------------------------------------------------------------- >89788_89788_7_TCF4-MAML3_TCF4_chr18_52921728_ENST00000544241_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=947AA_BP=404 MFVRESGGHWNDSFWKCGAVWLRIGMKDIFFQFIIARVRKCYSLSCLHTLPVVPTLRKTERGSYSSYGRESNLQGCHQSLLGGDMDMGNP GTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVRKVPPGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDP WSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAG SSQTGDALGKALASIYSPDHTNNSFSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVG PSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEA INNLPSNMPLPSASPLHQLDLKPSLPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSL SNQNKLFSDINLNDQEWQELIDELANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSG PPFSTVSTATSLPSVASTPAAPNPASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMA AQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQ KTTMNNYLPQNHMNMINQQPNNLGTNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPP -------------------------------------------------------------- >89788_89788_8_TCF4-MAML3_TCF4_chr18_52921728_ENST00000544241_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1386AA_BP=404 MFVRESGGHWNDSFWKCGAVWLRIGMKDIFFQFIIARVRKCYSLSCLHTLPVVPTLRKTERGSYSSYGRESNLQGCHQSLLGGDMDMGNP GTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVRKVPPGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDP WSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAG SSQTGDALGKALASIYSPDHTNNSFSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVG PSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEA INNLPSNMPLPSASPLHQLDLKPSLPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSL SNQNKLFSDINLNDQEWQELIDELANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSG PPFSTVSTATSLPSVASTPAAPNPASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMA AQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQ KTTMNNYLPQNHMNMINQQPNNLGTNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPP PPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMAYAALPSHGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQM LIDQRAQLIEQQKQQFLREQRQQQQQQQQQILAEQQLQQSHLPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQN AGMMGIGPSQNPGTMATAAAQSEMGLAPYSTTPTSQPGMYNMSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVN SAITQQHPQMKGPVGQALPRPQAPPRLQSLMGTVQQGAQSWQQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVD ANTGTVRTLNPAAMGRQMMPSLPGQQGTSQARPMVMSGLSQGVPGMPAFSQPPAQQQIPSGSFAPSSQSQAYERNAPQDVSYNYSGDGAG -------------------------------------------------------------- >89788_89788_9_TCF4-MAML3_TCF4_chr18_52921728_ENST00000561831_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=833AA_BP=290 MKFKQCRCSDTGLCCLDHEGKAEVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQ SSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNS FSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAM GGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPS LPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDEL ANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNP ASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQ QQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLG TNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQK -------------------------------------------------------------- >89788_89788_10_TCF4-MAML3_TCF4_chr18_52921728_ENST00000561831_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1272AA_BP=290 MKFKQCRCSDTGLCCLDHEGKAEVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQ SSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNS FSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAM GGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPS LPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDEL ANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNP ASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQ QQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLG TNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQK GVMNQPMAYAALPSHGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQMLIDQRAQLIEQQKQQFLREQRQQQ QQQQQQILAEQQLQQSHLPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQNAGMMGIGPSQNPGTMATAAAQSEM GLAPYSTTPTSQPGMYNMSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVNSAITQQHPQMKGPVGQALPRPQAP PRLQSLMGTVQQGAQSWQQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVDANTGTVRTLNPAAMGRQMMPSLPG QQGTSQARPMVMSGLSQGVPGMPAFSQPPAQQQIPSGSFAPSSQSQAYERNAPQDVSYNYSGDGAGGSFPGLPDGADLVDSIIKGGPGDE -------------------------------------------------------------- >89788_89788_11_TCF4-MAML3_TCF4_chr18_52921728_ENST00000564228_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=922AA_BP=379 MKDIFFQFIIARVRKCYSLSCLHTLPVVPTLRKTERGSYSSYGRESNLQGCHQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPL HSSAMEVQTKKVRKVPPGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQS SSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSF SSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMG GLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSL PLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELA NTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPA SSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQ QQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGT NSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKG -------------------------------------------------------------- >89788_89788_12_TCF4-MAML3_TCF4_chr18_52921728_ENST00000564228_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1361AA_BP=379 MKDIFFQFIIARVRKCYSLSCLHTLPVVPTLRKTERGSYSSYGRESNLQGCHQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPL HSSAMEVQTKKVRKVPPGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQS SSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSF SSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMG GLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSL PLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELA NTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPA SSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQ QQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGT NSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKG VMNQPMAYAALPSHGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQMLIDQRAQLIEQQKQQFLREQRQQQQ QQQQQILAEQQLQQSHLPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQNAGMMGIGPSQNPGTMATAAAQSEMG LAPYSTTPTSQPGMYNMSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVNSAITQQHPQMKGPVGQALPRPQAPP RLQSLMGTVQQGAQSWQQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVDANTGTVRTLNPAAMGRQMMPSLPGQ QGTSQARPMVMSGLSQGVPGMPAFSQPPAQQQIPSGSFAPSSQSQAYERNAPQDVSYNYSGDGAGGSFPGLPDGADLVDSIIKGGPGDEW -------------------------------------------------------------- >89788_89788_13_TCF4-MAML3_TCF4_chr18_52921728_ENST00000564403_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=1005AA_BP=462 MCDFAKMHHQQRMAALGTDKELSDLLDFSAMFSPPVSSGKNGPTSLASGHFTGSNVEDRSSSGSWGNGGHPSPSRNYGDGTPYDHMTSRD LGSHDNLSPPFVNSRIQSKTERGSYSSYGRESNLQGCHQQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVR KVPPGLPSSLVLSPQVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLH PHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTP VGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYG TGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGT HTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDD IQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCA VQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQ QQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQH NILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMA -------------------------------------------------------------- >89788_89788_14_TCF4-MAML3_TCF4_chr18_52921728_ENST00000564403_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1444AA_BP=462 MCDFAKMHHQQRMAALGTDKELSDLLDFSAMFSPPVSSGKNGPTSLASGHFTGSNVEDRSSSGSWGNGGHPSPSRNYGDGTPYDHMTSRD LGSHDNLSPPFVNSRIQSKTERGSYSSYGRESNLQGCHQQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVR KVPPGLPSSLVLSPQVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLH PHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTP VGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYG TGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGT HTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDD IQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCA VQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQ QQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQH NILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMA YAALPSHGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQMLIDQRAQLIEQQKQQFLREQRQQQQQQQQQIL AEQQLQQSHLPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQNAGMMGIGPSQNPGTMATAAAQSEMGLAPYSTT PTSQPGMYNMSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVNSAITQQHPQMKGPVGQALPRPQAPPRLQSLMG TVQQGAQSWQQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVDANTGTVRTLNPAAMGRQMMPSLPGQQGTSQAR PMVMSGLSQGVPGMPAFSQPPAQQQIPSGSFAPSSQSQAYERNAPQDVSYNYSGDGAGGSFPGLPDGADLVDSIIKGGPGDEWMQELDEL -------------------------------------------------------------- >89788_89788_15_TCF4-MAML3_TCF4_chr18_52921728_ENST00000565018_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=999AA_BP=456 MCDFAKMHHQQRMAALGTDKELSDLLDFSAMFSPPVSSGKNGPTSLASGHFTGSNVEDRSSSGSWGNGGHPSPSRNYGDGTPYDHMTSRD LGSHDNLSPPFVNSRIQSKTERGSYSSYGRESNLQGCHQQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVR KVPPGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLHPHERLS YPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTPVGSPPS LSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYGTGLLSA NRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGTHTPGLL EDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDDIQDLFN EDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCAVQSPQT PNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQQQQQQQ HSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQHNILTYG NTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMAYAALPS -------------------------------------------------------------- >89788_89788_16_TCF4-MAML3_TCF4_chr18_52921728_ENST00000565018_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1438AA_BP=456 MCDFAKMHHQQRMAALGTDKELSDLLDFSAMFSPPVSSGKNGPTSLASGHFTGSNVEDRSSSGSWGNGGHPSPSRNYGDGTPYDHMTSRD LGSHDNLSPPFVNSRIQSKTERGSYSSYGRESNLQGCHQQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVR KVPPGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLHPHERLS YPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTPVGSPPS LSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYGTGLLSA NRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGTHTPGLL EDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDDIQDLFN EDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCAVQSPQT PNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQQQQQQQ HSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQHNILTYG NTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMAYAALPS HGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQMLIDQRAQLIEQQKQQFLREQRQQQQQQQQQILAEQQLQ QSHLPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQNAGMMGIGPSQNPGTMATAAAQSEMGLAPYSTTPTSQPG MYNMSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVNSAITQQHPQMKGPVGQALPRPQAPPRLQSLMGTVQQGA QSWQQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVDANTGTVRTLNPAAMGRQMMPSLPGQQGTSQARPMVMSG -------------------------------------------------------------- >89788_89788_17_TCF4-MAML3_TCF4_chr18_52921728_ENST00000566286_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=990AA_BP=447 MQRAKTELFRLQIVTDDLRKNEMFSPPVSSGKNGPTSLASGHFTGSNVEDRSSSGSWGNGGHPSPSRNYGDGTPYDHMTSRDLGSHDNLS PPFVNSRIQSKTERGSYSSYGRESNLQGCHQQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVRKVPPGLPS SVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLHPHERLSYPSHSSAD INSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTPVGSPPSLSGTAVWS RNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYGTGLLSANRHSLMLQE TVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGTHTPGLLEDLSKNGRL PEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDDIQDLFNEDFEEKKEP EFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCAVQSPQTPNQAHTPGQ APPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQQQQQQQHSNQTSNWS PLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQHNILTYGNTKPLTHFN ADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMAYAALPSHGQEQQPVG -------------------------------------------------------------- >89788_89788_18_TCF4-MAML3_TCF4_chr18_52921728_ENST00000566286_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1429AA_BP=447 MQRAKTELFRLQIVTDDLRKNEMFSPPVSSGKNGPTSLASGHFTGSNVEDRSSSGSWGNGGHPSPSRNYGDGTPYDHMTSRDLGSHDNLS PPFVNSRIQSKTERGSYSSYGRESNLQGCHQQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVRKVPPGLPS SVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLHPHERLSYPSHSSAD INSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTPVGSPPSLSGTAVWS RNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYGTGLLSANRHSLMLQE TVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGTHTPGLLEDLSKNGRL PEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDDIQDLFNEDFEEKKEP EFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCAVQSPQTPNQAHTPGQ APPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQQQQQQQHSNQTSNWS PLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQHNILTYGNTKPLTHFN ADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMAYAALPSHGQEQHPVG LPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQMLIDQRAQLIEQQKQQFLREQRQQQQQQQQQILAEQQLQQSHLPRQHL QPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQNAGMMGIGPSQNPGTMATAAAQSEMGLAPYSTTPTSQPGMYNMSTGMT QMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVNSAITQQHPQMKGPVGQALPRPQAPPRLQSLMGTVQQGAQSWQQRSLQ GMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVDANTGTVRTLNPAAMGRQMMPSLPGQQGTSQARPMVMSGLSQGVPGMP -------------------------------------------------------------- >89788_89788_19_TCF4-MAML3_TCF4_chr18_52921728_ENST00000567880_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=933AA_BP=390 MHHQQRMAALGTDKELSDLLDFSAMFSPPVSSGKNGPTSLASGHFTGSNVEDRSSSGSWGNGGHPSPSRNYGDGTPYDHMTSRDLGSHDN LSPPFVNSRIQSKTERGSYSSYGRESNLQGCHQVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAG MLGNSSHIPQSSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALAS IYSPDHTNNSFSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHG IIGPSHNGAMGGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSAS PLHQLDLKPSLPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLND QEWQELIDELANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPS VASTPAAPNPASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQ QQQQQQQQQQQQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMN MINQQPNNLGTNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSED -------------------------------------------------------------- >89788_89788_20_TCF4-MAML3_TCF4_chr18_52921728_ENST00000567880_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1372AA_BP=390 MHHQQRMAALGTDKELSDLLDFSAMFSPPVSSGKNGPTSLASGHFTGSNVEDRSSSGSWGNGGHPSPSRNYGDGTPYDHMTSRDLGSHDN LSPPFVNSRIQSKTERGSYSSYGRESNLQGCHQVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAG MLGNSSHIPQSSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALAS IYSPDHTNNSFSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHG IIGPSHNGAMGGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSAS PLHQLDLKPSLPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLND QEWQELIDELANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPS VASTPAAPNPASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQ QQQQQQQQQQQQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMN MINQQPNNLGTNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSED QKRLLLMKQKGVMNQPMAYAALPSHGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQMLIDQRAQLIEQQKQ QFLREQRQQQQQQQQQILAEQQLQQSHLPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQNAGMMGIGPSQNPGT MATAAAQSEMGLAPYSTTPTSQPGMYNMSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVNSAITQQHPQMKGPV GQALPRPQAPPRLQSLMGTVQQGAQSWQQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVDANTGTVRTLNPAAM GRQMMPSLPGQQGTSQARPMVMSGLSQGVPGMPAFSQPPAQQQIPSGSFAPSSQSQAYERNAPQDVSYNYSGDGAGGSFPGLPDGADLVD -------------------------------------------------------------- >89788_89788_21_TCF4-MAML3_TCF4_chr18_52921728_ENST00000570177_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=906AA_BP=363 MVKSSCRSAVHGSLFGKNSAGKKFTAIPAQRKHLWNQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVRKVP PGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLHPHERLSYPS HSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTPVGSPPSLSA GTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYGTGLLSANRH SLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGTHTPGLLEDL SKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDDIQDLFNEDF EEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCAVQSPQTPNQ AHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQQQQQQQHSN QTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQHNILTYGNTK PLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMAYAALPSHGQ -------------------------------------------------------------- >89788_89788_22_TCF4-MAML3_TCF4_chr18_52921728_ENST00000570177_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1345AA_BP=363 MVKSSCRSAVHGSLFGKNSAGKKFTAIPAQRKHLWNQSLLGGDMDMGNPGTLSPTKPGSQYYQYSSNNPRRRPLHSSAMEVQTKKVRKVP PGLPSSVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQSSSYCSLHPHERLSYPS HSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNSFSSNPSTPVGSPPSLSA GTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAMGGLGSGYGTGLLSANRH SLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPSLPLQNSGTHTPGLLEDL SKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDELANTVPEDDIQDLFNEDF EEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNPASSPANCAVQSPQTPNQ AHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQQQQQQQQQQQQQQQHSN QTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLGTNSLNKQHNILTYGNTK PLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQKGVMNQPMAYAALPSHGQ EQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQMLIDQRAQLIEQQKQQFLREQRQQQQQQQQQILAEQQLQQSH LPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQNAGMMGIGPSQNPGTMATAAAQSEMGLAPYSTTPTSQPGMYN MSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVNSAITQQHPQMKGPVGQALPRPQAPPRLQSLMGTVQQGAQSW QQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVDANTGTVRTLNPAAMGRQMMPSLPGQQGTSQARPMVMSGLSQ -------------------------------------------------------------- >89788_89788_23_TCF4-MAML3_TCF4_chr18_52921728_ENST00000570287_MAML3_chr4_140812121_ENST00000327122_length(amino acids)=833AA_BP=290 MYCAYTIPGMGGNSLMYYYNGKAVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQ SSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNS FSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAM GGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPS LPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDEL ANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNP ASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQ QQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLG TNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQK -------------------------------------------------------------- >89788_89788_24_TCF4-MAML3_TCF4_chr18_52921728_ENST00000570287_MAML3_chr4_140812121_ENST00000509479_length(amino acids)=1272AA_BP=290 MYCAYTIPGMGGNSLMYYYNGKAVYAPSASTADYNRDSPGYPSSKPATSTFPSSFFMQDGHHSSDPWSSSSGMNQPGYAGMLGNSSHIPQ SSSYCSLHPHERLSYPSHSSADINSSLPPMSTFHRSGTNHYSTSSCTPPANGTDSIMANRGSGAAGSSQTGDALGKALASIYSPDHTNNS FSSNPSTPVGSPPSLSAGTAVWSRNGGQASSSPNYEGPLHSLQSRIEDRLERLDDAIHVLRNHAVGPSTAMPGGHGDMHGIIGPSHNGAM GGLGSGYGTGLLSANRHSLMLQETVKRKLEGARSPLNGDQQNGACDGNFSPTSKRIRKDISAGMEAINNLPSNMPLPSASPLHQLDLKPS LPLQNSGTHTPGLLEDLSKNGRLPEIKLPVNGCSDLEDSFTILQSKDLKQEPLDDPTCIDTSETSLSNQNKLFSDINLNDQEWQELIDEL ANTVPEDDIQDLFNEDFEEKKEPEFSQPATETPLSQESASVKSDPSHSPFAHVSMGSPQARPSSSGPPFSTVSTATSLPSVASTPAAPNP ASSPANCAVQSPQTPNQAHTPGQAPPRPGNGYLLNPAAVTVAGSASGPVAVPSSDMSPAEQLKQMAAQQQQRAKLMQQKQQQQQQQQQQQ QQQQQQQQQQQQQQHSNQTSNWSPLGPPSSPYGAAFTAEKPNSPMMYPQAFNNQNPIVPPMANNLQKTTMNNYLPQNHMNMINQQPNNLG TNSLNKQHNILTYGNTKPLTHFNADLSQRMTPPVANPNKNPLMPYIQQQQQQQQQQQQQQQQQQPPPPQLQAPRAHLSEDQKRLLLMKQK GVMNQPMAYAALPSHGQEQHPVGLPRTTGPMQSSVPPGSGGMVSGASPAGPGFLGSQPQAAIMKQMLIDQRAQLIEQQKQQFLREQRQQQ QQQQQQILAEQQLQQSHLPRQHLQPQRNPYPVQQVNQFQGSPQDIAAVRSQAALQSMRTSRLMAQNAGMMGIGPSQNPGTMATAAAQSEM GLAPYSTTPTSQPGMYNMSTGMTQMLQHPNQSGMSITHNQAQGPRQPASGQGVGMVSGFGQSMLVNSAITQQHPQMKGPVGQALPRPQAP PRLQSLMGTVQQGAQSWQQRSLQGMPGRTSGELGPFNNGASYPLQAGQPRLTKQHFPQGLSQSVVDANTGTVRTLNPAAMGRQMMPSLPG QQGTSQARPMVMSGLSQGVPGMPAFSQPPAQQQIPSGSFAPSSQSQAYERNAPQDVSYNYSGDGAGGSFPGLPDGADLVDSIIKGGPGDE -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr18:52921728/chr4:140812121) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MAML3 |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | FUNCTION: Acts as a transcriptional coactivator for NOTCH proteins. Has been shown to amplify NOTCH-induced transcription of HES1. {ECO:0000269|PubMed:12370315, ECO:0000269|PubMed:12386158}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000354452 | - | 15 | 20 | 228_231 | 450.0 | 2391.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000356073 | - | 15 | 20 | 228_231 | 450.0 | 2386.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000457482 | - | 8 | 13 | 228_231 | 290.0 | 429.3333333333333 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537578 | - | 14 | 19 | 228_231 | 426.0 | 591.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537856 | - | 10 | 15 | 228_231 | 320.0 | 566.3333333333334 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000540999 | - | 14 | 19 | 228_231 | 426.0 | 682.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000543082 | - | 13 | 18 | 228_231 | 408.0 | 702.6666666666666 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000544241 | - | 11 | 16 | 228_231 | 379.0 | 560.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561831 | - | 8 | 13 | 228_231 | 290.0 | 500.3333333333333 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561992 | - | 11 | 16 | 228_231 | 320.0 | 514.6666666666666 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564228 | - | 11 | 16 | 228_231 | 379.0 | 631.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564999 | - | 15 | 20 | 228_231 | 450.0 | 700.6666666666666 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000565018 | - | 15 | 20 | 228_231 | 450.0 | 662.6666666666666 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566279 | - | 13 | 18 | 228_231 | 390.0 | 619.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566286 | - | 14 | 19 | 228_231 | 447.0 | 705.6666666666666 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000567880 | - | 12 | 17 | 228_231 | 390.0 | 646.6666666666666 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000568673 | - | 14 | 19 | 228_231 | 426.0 | 757.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570177 | - | 10 | 15 | 228_231 | 320.0 | 495.3333333333333 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570287 | - | 8 | 13 | 228_231 | 290.0 | 454.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000354452 | - | 15 | 20 | 18_26 | 450.0 | 2391.0 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000356073 | - | 15 | 20 | 18_26 | 450.0 | 2386.0 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000457482 | - | 8 | 13 | 18_26 | 290.0 | 429.3333333333333 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537578 | - | 14 | 19 | 18_26 | 426.0 | 591.0 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537856 | - | 10 | 15 | 18_26 | 320.0 | 566.3333333333334 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000540999 | - | 14 | 19 | 18_26 | 426.0 | 682.0 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000543082 | - | 13 | 18 | 18_26 | 408.0 | 702.6666666666666 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000544241 | - | 11 | 16 | 18_26 | 379.0 | 560.0 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561831 | - | 8 | 13 | 18_26 | 290.0 | 500.3333333333333 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561992 | - | 11 | 16 | 18_26 | 320.0 | 514.6666666666666 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564228 | - | 11 | 16 | 18_26 | 379.0 | 631.0 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564999 | - | 15 | 20 | 18_26 | 450.0 | 700.6666666666666 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000565018 | - | 15 | 20 | 18_26 | 450.0 | 662.6666666666666 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566279 | - | 13 | 18 | 18_26 | 390.0 | 619.0 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566286 | - | 14 | 19 | 18_26 | 447.0 | 705.6666666666666 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000567880 | - | 12 | 17 | 18_26 | 390.0 | 646.6666666666666 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000568673 | - | 14 | 19 | 18_26 | 426.0 | 757.0 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570177 | - | 10 | 15 | 18_26 | 320.0 | 495.3333333333333 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570287 | - | 8 | 13 | 18_26 | 290.0 | 454.0 | Motif | Note=9aaTAD |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000354452 | - | 15 | 20 | 1_83 | 450.0 | 2391.0 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000354452 | - | 15 | 20 | 379_400 | 450.0 | 2391.0 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000356073 | - | 15 | 20 | 1_83 | 450.0 | 2386.0 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000356073 | - | 15 | 20 | 379_400 | 450.0 | 2386.0 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000457482 | - | 8 | 13 | 1_83 | 290.0 | 429.3333333333333 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537578 | - | 14 | 19 | 1_83 | 426.0 | 591.0 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537578 | - | 14 | 19 | 379_400 | 426.0 | 591.0 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537856 | - | 10 | 15 | 1_83 | 320.0 | 566.3333333333334 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000540999 | - | 14 | 19 | 1_83 | 426.0 | 682.0 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000540999 | - | 14 | 19 | 379_400 | 426.0 | 682.0 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000543082 | - | 13 | 18 | 1_83 | 408.0 | 702.6666666666666 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000543082 | - | 13 | 18 | 379_400 | 408.0 | 702.6666666666666 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000544241 | - | 11 | 16 | 1_83 | 379.0 | 560.0 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561831 | - | 8 | 13 | 1_83 | 290.0 | 500.3333333333333 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561992 | - | 11 | 16 | 1_83 | 320.0 | 514.6666666666666 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564228 | - | 11 | 16 | 1_83 | 379.0 | 631.0 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564999 | - | 15 | 20 | 1_83 | 450.0 | 700.6666666666666 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564999 | - | 15 | 20 | 379_400 | 450.0 | 700.6666666666666 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000565018 | - | 15 | 20 | 1_83 | 450.0 | 662.6666666666666 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000565018 | - | 15 | 20 | 379_400 | 450.0 | 662.6666666666666 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566279 | - | 13 | 18 | 1_83 | 390.0 | 619.0 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566286 | - | 14 | 19 | 1_83 | 447.0 | 705.6666666666666 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566286 | - | 14 | 19 | 379_400 | 447.0 | 705.6666666666666 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000567880 | - | 12 | 17 | 1_83 | 390.0 | 646.6666666666666 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000568673 | - | 14 | 19 | 1_83 | 426.0 | 757.0 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000568673 | - | 14 | 19 | 379_400 | 426.0 | 757.0 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570177 | - | 10 | 15 | 1_83 | 320.0 | 495.3333333333333 | Region | Essential for MYOD1 inhibition |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570287 | - | 8 | 13 | 1_83 | 290.0 | 454.0 | Region | Essential for MYOD1 inhibition |
| Tgene | MAML3 | chr18:52921728 | chr4:140812121 | ENST00000509479 | 0 | 5 | 467_968 | 156.0 | 1139.0 | Compositional bias | Note=Gln-rich | |
| Tgene | MAML3 | chr18:52921728 | chr4:140812121 | ENST00000509479 | 0 | 5 | 548_595 | 156.0 | 1139.0 | Compositional bias | Note=Asn-rich |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000354452 | - | 15 | 20 | 564_617 | 450.0 | 2391.0 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000356073 | - | 15 | 20 | 564_617 | 450.0 | 2386.0 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000457482 | - | 8 | 13 | 564_617 | 290.0 | 429.3333333333333 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537578 | - | 14 | 19 | 564_617 | 426.0 | 591.0 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537856 | - | 10 | 15 | 564_617 | 320.0 | 566.3333333333334 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000540999 | - | 14 | 19 | 564_617 | 426.0 | 682.0 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000543082 | - | 13 | 18 | 564_617 | 408.0 | 702.6666666666666 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000544241 | - | 11 | 16 | 564_617 | 379.0 | 560.0 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561831 | - | 8 | 13 | 564_617 | 290.0 | 500.3333333333333 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561992 | - | 11 | 16 | 564_617 | 320.0 | 514.6666666666666 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564228 | - | 11 | 16 | 564_617 | 379.0 | 631.0 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564999 | - | 15 | 20 | 564_617 | 450.0 | 700.6666666666666 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000565018 | - | 15 | 20 | 564_617 | 450.0 | 662.6666666666666 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566279 | - | 13 | 18 | 564_617 | 390.0 | 619.0 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566286 | - | 14 | 19 | 564_617 | 447.0 | 705.6666666666666 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000567880 | - | 12 | 17 | 564_617 | 390.0 | 646.6666666666666 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000568673 | - | 14 | 19 | 564_617 | 426.0 | 757.0 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570177 | - | 10 | 15 | 564_617 | 320.0 | 495.3333333333333 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570287 | - | 8 | 13 | 564_617 | 290.0 | 454.0 | Domain | bHLH |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000354452 | - | 15 | 20 | 619_642 | 450.0 | 2391.0 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000356073 | - | 15 | 20 | 619_642 | 450.0 | 2386.0 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000457482 | - | 8 | 13 | 379_400 | 290.0 | 429.3333333333333 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000457482 | - | 8 | 13 | 619_642 | 290.0 | 429.3333333333333 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537578 | - | 14 | 19 | 619_642 | 426.0 | 591.0 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537856 | - | 10 | 15 | 379_400 | 320.0 | 566.3333333333334 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000537856 | - | 10 | 15 | 619_642 | 320.0 | 566.3333333333334 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000540999 | - | 14 | 19 | 619_642 | 426.0 | 682.0 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000543082 | - | 13 | 18 | 619_642 | 408.0 | 702.6666666666666 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000544241 | - | 11 | 16 | 379_400 | 379.0 | 560.0 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000544241 | - | 11 | 16 | 619_642 | 379.0 | 560.0 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561831 | - | 8 | 13 | 379_400 | 290.0 | 500.3333333333333 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561831 | - | 8 | 13 | 619_642 | 290.0 | 500.3333333333333 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561992 | - | 11 | 16 | 379_400 | 320.0 | 514.6666666666666 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000561992 | - | 11 | 16 | 619_642 | 320.0 | 514.6666666666666 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564228 | - | 11 | 16 | 379_400 | 379.0 | 631.0 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564228 | - | 11 | 16 | 619_642 | 379.0 | 631.0 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000564999 | - | 15 | 20 | 619_642 | 450.0 | 700.6666666666666 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000565018 | - | 15 | 20 | 619_642 | 450.0 | 662.6666666666666 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566279 | - | 13 | 18 | 379_400 | 390.0 | 619.0 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566279 | - | 13 | 18 | 619_642 | 390.0 | 619.0 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000566286 | - | 14 | 19 | 619_642 | 447.0 | 705.6666666666666 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000567880 | - | 12 | 17 | 379_400 | 390.0 | 646.6666666666666 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000567880 | - | 12 | 17 | 619_642 | 390.0 | 646.6666666666666 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000568673 | - | 14 | 19 | 619_642 | 426.0 | 757.0 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570177 | - | 10 | 15 | 379_400 | 320.0 | 495.3333333333333 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570177 | - | 10 | 15 | 619_642 | 320.0 | 495.3333333333333 | Region | Note=Class A specific domain |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570287 | - | 8 | 13 | 379_400 | 290.0 | 454.0 | Region | Note=Leucine-zipper |
| Hgene | TCF4 | chr18:52921728 | chr4:140812121 | ENST00000570287 | - | 8 | 13 | 619_642 | 290.0 | 454.0 | Region | Note=Class A specific domain |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| TCF4 | |
| MAML3 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to TCF4-MAML3 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to TCF4-MAML3 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | TCF4 | C1970431 | PITT-HOPKINS SYNDROME | 14 | CLINGEN;CTD_human;GENOMICS_ENGLAND;ORPHANET;UNIPROT |
| Hgene | TCF4 | C0005586 | Bipolar Disorder | 5 | PSYGENET |
| Hgene | TCF4 | C0036341 | Schizophrenia | 4 | PSYGENET |
| Hgene | TCF4 | C0016781 | Fuchs Endothelial Dystrophy | 1 | ORPHANET |
| Hgene | TCF4 | C0018799 | Heart Diseases | 1 | CTD_human |
| Hgene | TCF4 | C0022333 | Jacksonian Seizure | 1 | CTD_human |
| Hgene | TCF4 | C0023893 | Liver Cirrhosis, Experimental | 1 | CTD_human |
| Hgene | TCF4 | C0023903 | Liver neoplasms | 1 | CTD_human |
| Hgene | TCF4 | C0025958 | Microcephaly | 1 | CTD_human |
| Hgene | TCF4 | C0033975 | Psychotic Disorders | 1 | PSYGENET |
| Hgene | TCF4 | C0036572 | Seizures | 1 | CTD_human |
| Hgene | TCF4 | C0149958 | Complex partial seizures | 1 | CTD_human |
| Hgene | TCF4 | C0234533 | Generalized seizures | 1 | CTD_human |
| Hgene | TCF4 | C0234535 | Clonic Seizures | 1 | CTD_human |
| Hgene | TCF4 | C0270824 | Visual seizure | 1 | CTD_human |
| Hgene | TCF4 | C0270844 | Tonic Seizures | 1 | CTD_human |
| Hgene | TCF4 | C0270846 | Epileptic drop attack | 1 | CTD_human |
| Hgene | TCF4 | C0345904 | Malignant neoplasm of liver | 1 | CTD_human |
| Hgene | TCF4 | C0349204 | Nonorganic psychosis | 1 | PSYGENET |
| Hgene | TCF4 | C0376634 | Craniofacial Abnormalities | 1 | CTD_human |
| Hgene | TCF4 | C0422850 | Seizures, Somatosensory | 1 | CTD_human |
| Hgene | TCF4 | C0422852 | Seizures, Auditory | 1 | CTD_human |
| Hgene | TCF4 | C0422853 | Olfactory seizure | 1 | CTD_human |
| Hgene | TCF4 | C0422854 | Gustatory seizure | 1 | CTD_human |
| Hgene | TCF4 | C0422855 | Vertiginous seizure | 1 | CTD_human |
| Hgene | TCF4 | C0494475 | Tonic - clonic seizures | 1 | CTD_human |
| Hgene | TCF4 | C0566602 | Primary sclerosing cholangitis | 1 | ORPHANET |
| Hgene | TCF4 | C0751056 | Non-epileptic convulsion | 1 | CTD_human |
| Hgene | TCF4 | C0751110 | Single Seizure | 1 | CTD_human |
| Hgene | TCF4 | C0751123 | Atonic Absence Seizures | 1 | CTD_human |
| Hgene | TCF4 | C0751494 | Convulsive Seizures | 1 | CTD_human |
| Hgene | TCF4 | C0751495 | Seizures, Focal | 1 | CTD_human |
| Hgene | TCF4 | C0751496 | Seizures, Sensory | 1 | CTD_human |
| Hgene | TCF4 | C1456784 | Paranoia | 1 | PSYGENET |
| Hgene | TCF4 | C1535926 | Neurodevelopmental Disorders | 1 | CTD_human |
| Hgene | TCF4 | C1956147 | Microlissencephaly | 1 | CTD_human |
| Hgene | TCF4 | C2750451 | CORNEAL DYSTROPHY, FUCHS ENDOTHELIAL, 3 | 1 | CTD_human;GENOMICS_ENGLAND |
| Hgene | TCF4 | C2931456 | Prostate cancer, familial | 1 | CTD_human |
| Hgene | TCF4 | C3495874 | Nonepileptic Seizures | 1 | CTD_human |
| Hgene | TCF4 | C3853041 | Severe Congenital Microcephaly | 1 | CTD_human |
| Hgene | TCF4 | C4048158 | Convulsions | 1 | CTD_human |
| Hgene | TCF4 | C4316903 | Absence Seizures | 1 | CTD_human |
| Hgene | TCF4 | C4317109 | Epileptic Seizures | 1 | CTD_human |
| Hgene | TCF4 | C4317123 | Myoclonic Seizures | 1 | CTD_human |
| Hgene | TCF4 | C4505436 | Generalized Absence Seizures | 1 | CTD_human |
| Hgene | TCF4 | C4721453 | Peripheral Nervous System Diseases | 1 | CTD_human |
| Hgene | TCF4 | C4722327 | PROSTATE CANCER, HEREDITARY, 1 | 1 | CTD_human |

