| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:TNXB-VIM |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: TNXB-VIM | FusionPDB ID: 92962 | FusionGDB2.0 ID: 92962 | Hgene | Tgene | Gene symbol | TNXB | VIM | Gene ID | 7148 | 7431 |
| Gene name | tenascin XB | vimentin | |
| Synonyms | EDS3|EDSCLL|EDSCLL1|HXBL|TENX|TN-X|TNX|TNXB1|TNXB2|TNXBS|VUR8|XB|XBS | - | |
| Cytomap | 6p21.33-p21.32 | 10p13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | tenascin-Xgrowth-inhibiting protein 45hexabrachion-like proteintenascin XB1tenascin XB2 | vimentinepididymis secretory sperm binding protein | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | . | VMAC | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000375244, ENST00000375247, ENST00000383159, ENST00000424718, ENST00000433037, ENST00000440248, ENST00000451343, ENST00000464376, ENST00000465958, ENST00000469250, ENST00000471059, ENST00000475986, ENST00000479795, ENST00000489006, ENST00000546684, ENST00000548628, ENST00000548649, ENST00000549232, ENST00000549652, ENST00000550212, ENST00000550539, ENST00000551201, ENST00000551808, ENST00000552149, ENST00000552665, ENST00000553092, | ENST00000485947, ENST00000224237, ENST00000544301, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 4 X 4 X 4=64 | 42 X 25 X 11=11550 |
| # samples | 4 | 41 | |
| ** MAII score | log2(4/64*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(41/11550*10)=-4.81612513168534 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: TNXB [Title/Abstract] AND VIM [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | TNXB(32016140)-VIM(17278293), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | TNXB-VIM seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. TNXB-VIM seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. TNXB-VIM seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. TNXB-VIM seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across TNXB (5'-gene) Fusion gene breakpoints across TNXB (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across VIM (3'-gene) Fusion gene breakpoints across VIM (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
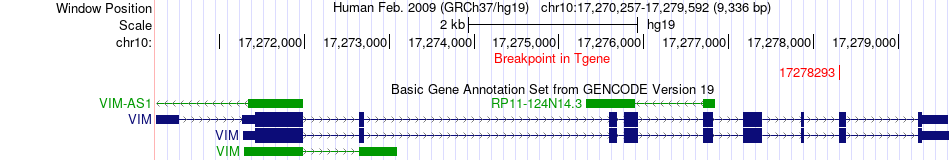 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-AB2T-01A | TNXB | chr6 | 32016140 | - | VIM | chr10 | 17278293 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000375244 | TNXB | chr6 | 32016140 | - | ENST00000544301 | VIM | chr10 | 17278293 | + | 10689 | 10247 | 202 | 10374 | 3390 |
| ENST00000375244 | TNXB | chr6 | 32016140 | - | ENST00000224237 | VIM | chr10 | 17278293 | + | 10697 | 10247 | 202 | 10374 | 3390 |
| ENST00000375247 | TNXB | chr6 | 32016140 | - | ENST00000544301 | VIM | chr10 | 17278293 | + | 10683 | 10241 | 202 | 10368 | 3388 |
| ENST00000375247 | TNXB | chr6 | 32016140 | - | ENST00000224237 | VIM | chr10 | 17278293 | + | 10691 | 10241 | 202 | 10368 | 3388 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000375244 | ENST00000544301 | TNXB | chr6 | 32016140 | - | VIM | chr10 | 17278293 | + | 0.000806098 | 0.99919397 |
| ENST00000375244 | ENST00000224237 | TNXB | chr6 | 32016140 | - | VIM | chr10 | 17278293 | + | 0.000805 | 0.99919504 |
| ENST00000375247 | ENST00000544301 | TNXB | chr6 | 32016140 | - | VIM | chr10 | 17278293 | + | 0.000647724 | 0.9993523 |
| ENST00000375247 | ENST00000224237 | TNXB | chr6 | 32016140 | - | VIM | chr10 | 17278293 | + | 0.000646794 | 0.99935323 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >92962_92962_1_TNXB-VIM_TNXB_chr6_32016140_ENST00000375244_VIM_chr10_17278293_ENST00000224237_length(amino acids)=3390AA_BP=3327 MMPAQYALTSSLVLLVLLSTARAGPFSSRSNVTLPAPRPPPQPGGHTVGAGVGSPSSQLYEHTVEGGEKQVVFTHRINLPPSTGCGCPPG TEPPVLASEVQALRVRLEILEELVKGLKEQCTGGCCPASAQAGTGQTDVRTLCSLHGVFDLSRCTCSCEPGWGGPTCSDPTDAEIPPSSP PSASGSCPDDCNDQGRCVRGRCVCFPGYTGPSCGWPSCPGDCQGRGRCVQGVCVCRAGFSGPDCSQRSCPRGCSQRGRCEGGRCVCDPGY TGDDCGMRSCPRGCSQRGRCENGRCVCNPGYTGEDCGVRSCPRGCSQRGRCKDGRCVCDPGYTGEDCGTRSCPWDCGEGGRCVDGRCVCW PGYTGEDCSTRTCPRDCRGRGRCEDGECICDTGYSGDDCGVRSCPGDCNQRGRCEDGRCVCWPGYTGTDCGSRACPRDCRGRGRCENGVC VCNAGYSGEDCGVRSCPGDCRGRGRCESGRCMCWPGYTGRDCGTRACPGDCRGRGRCVDGRCVCNPGFTGEDCGSRRCPGDCRGHGLCED GVCVCDAGYSGEDCSTRSCPGGCRGRGQCLDGRCVCEDGYSGEDCGVRQCPNDCSQHGVCQDGVCICWEGYVSEDCSIRTCPSNCHGRGR CEEGRCLCDPGYTGPTCATRMCPADCRGRGRCVQGVCLCHVGYGGEDCGQEEPPASACPGGCGPRELCRAGQCVCVEGFRGPDCAIQTCP GDCRGRGECHDGSCVCKDGYAGEDCGEEVPTIEGMRMHLLEETTVRTEWTPAPGPVDAYEIQFIPTTEGASPPFTARVPSSASAYDQRGL APGQEYQVTVRALRGTSWGLPASKTITTMIDGPQDLRVVAVTPTTLELGWLRPQAEVDRFVVSYVSAGNQRVRLEVPPEADGTLLTDLMP GVEYVVTVTAERGRAVSYPASVRANTGSSPLGLLGTTDEPPPSGPSTTQGAQAPLLQQRPQELGELRVLGRDETGRLRVVWTAQPDTFAY FQLRMRVPEGPGAHEEVLPGDVRQALVPPPPPGTPYELSLHGVPPGGKPSDPIIYQGIMDKDEEKPGKSSGPPRLGELTVTDRTSDSLLL RWTVPEGEFDSFVIQYKDRDGQPQVVPVEGPQRSAVITSLDPGRKYKFVLYGFVGKKRHGPLVAEAKILPQSDPSPGTPPHLGNLWVTDP TPDSLHLSWTVPEGQFDTFMVQYRDRDGRPQVVPVEGPERSFVVSSLDPDHKYRFTLFGIANKKRYGPLTADGTTAPERKEEPPRPEFLE QPLLGELTVTGVTPDSLRLSWTVAQGPFDSFMVQYKDAQGQPQAVPVAGDENEVTVPGLDPDRKYKMNLYGLRGRQRVGPESVVAKTAPQ EDVDETPSPTELGTEAPESPEEPLLGELTVTGSSPDSLSLFWTVPQGSFDSFTVQYKDRDGRPRAVRVGGKESEVTVGGLEPGHKYKMHL YGLHEGQRVGPVSAVGVTAPQQEETPPATESPLEPRLGELTVTDVTPNSVGLSWTVPEGQFDSFIVQYKDKDGQPQVVPVAADQREVTVY NLEPERKYKMNMYGLHDGQRMGPLSVVIVTAPLPPAPATEASKPPLEPRLGELTVTDITPDSVGLSWTVPEGEFDSFVVQYKDRDGQPQV VPVAADQREVTIPDLEPSRKYKFLLFGIQDGKRRSPVSVEAKTVARGDASPGAPPRLGELWVTDPTPDSLRLSWTVPEGQFDSFVVQFKD KDGPQVVPVEGHERSVTVTPLDAGRKYRFLLYGLLGKKRHGPLTADGTTEARSAMDDTGTKRPPKPRLGEELQVTTVTQNSVGLSWTVPE GQFDSFVVQYKDRDGQPQVVPVEGSLREVSVPGLDPAHRYKLLLYGLHHGKRVGPISAVAITAGREETETETTAPTPPAPEPHLGELTVE EATSHTLHLSWMVTEGEFDSFEIQYTDRDGQLQMVRIGGDRNDITLSGLESDHRYLVTLYGFSDGKHVGPVHVEALTVPEEEKPSEPPTA TPEPPIKPRLGELTVTDATPDSLSLSWTVPEGQFDHFLVQYRNGDGQPKAVRVPGHEEGVTISGLEPDHKYKMNLYGFHGGQRMGPVSVV GVTAAEEETPSPTEPSMEAPEPAEEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGRPQVVRVGGEESEVTVGGLEPGRKYK MHLYGLHEGRRVGPVSAVGVTAPEEESPDAPLAKLRLGQMTVRDITSDSLSLSWTVPEGQFDHFLVQFKNGDGQPKAVRVPGHEDGVTIS GLEPDHKYKMNLYGFHGGQRVGPVSAVGLTAPGKDEEMAPASTEPPTPEPPIKPRLEELTVTDATPDSLSLSWTVPEGQFDHFLVQYKNG DGQPKATRVPGHEDRVTISGLEPDNKYKMNLYGFHGGQRVGPVSAIGVTAAEEETPSPTEPSMEAPEPPEEPLLGELTVTGSSPDSLSLS WTVPQGRFDSFTVQYKDRDGRPQVVRVGGEESEVTVGGLEPGRKYKMHLYGLHEGRRVGPVSTVGVTAPQEDVDETPSPTEPGTEAPGPP EEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGRPQAVRVGGQESKVTVRGLEPGRKYKMHLYGLHEGRRLGPVSAVGVTED EAETTQAVPTMTPEPPIKPRLGELTMTDATPDSLSLSWTVPEGQFDHFLVQYRNGDGQPKAVRVPGHEDGVTISGLEPDHKYKMNLYGFH GGQRVGPISVIGVTAAEEETPSPTELSTEAPEPPEEPLLGELTVTGSSPDSLSLSWTIPQGHFDSFTVQYKDRDGRPQVMRVRGEESEVT VGGLEPGRKYKMHLYGLHEGRRVGPVSTVGVTAPEDEAETTQAVPTTTPEPPNKPRLGELTVTDATPDSLSLSWMVPEGQFDHFLVQYRN GDGQPKVVRVPGHEDGVTISGLEPDHKYKMNLYGFHGGQRVGPISVIGVTAAEEETPAPTEPSTEAPEPPEEPLLGELTVTGSSPDSLSL SWTIPQGRFDSFTVQYKDRDGRPQVVRVRGEESEVTVGGLEPGCKYKMHLYGLHEGQRVGPVSAVGVTAPKDEAETTQAVPTMTPEPPIK PRLGELTVTDATPDSLSLSWMVPEGQFDHFLVQYRNGDGQPKAVRVPGHEDGVTISGLEPDHKYKMNLYGFHGGQRVGPVSAIGVTEEET PSPTEPSTEAPEAPEEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGQPQVVRVRGEESEVTVGGLEPGRKYKMHLYGLHEG QRVGPVSTVGITAPLPTPLPVEPRLGELAVAAVTSDSVGLSWTVAQGPFDSFLVQYRDAQGQPQAVPVSGDLRAVAVSGLDPARKYKFLL -------------------------------------------------------------- >92962_92962_2_TNXB-VIM_TNXB_chr6_32016140_ENST00000375244_VIM_chr10_17278293_ENST00000544301_length(amino acids)=3390AA_BP=3327 MMPAQYALTSSLVLLVLLSTARAGPFSSRSNVTLPAPRPPPQPGGHTVGAGVGSPSSQLYEHTVEGGEKQVVFTHRINLPPSTGCGCPPG TEPPVLASEVQALRVRLEILEELVKGLKEQCTGGCCPASAQAGTGQTDVRTLCSLHGVFDLSRCTCSCEPGWGGPTCSDPTDAEIPPSSP PSASGSCPDDCNDQGRCVRGRCVCFPGYTGPSCGWPSCPGDCQGRGRCVQGVCVCRAGFSGPDCSQRSCPRGCSQRGRCEGGRCVCDPGY TGDDCGMRSCPRGCSQRGRCENGRCVCNPGYTGEDCGVRSCPRGCSQRGRCKDGRCVCDPGYTGEDCGTRSCPWDCGEGGRCVDGRCVCW PGYTGEDCSTRTCPRDCRGRGRCEDGECICDTGYSGDDCGVRSCPGDCNQRGRCEDGRCVCWPGYTGTDCGSRACPRDCRGRGRCENGVC VCNAGYSGEDCGVRSCPGDCRGRGRCESGRCMCWPGYTGRDCGTRACPGDCRGRGRCVDGRCVCNPGFTGEDCGSRRCPGDCRGHGLCED GVCVCDAGYSGEDCSTRSCPGGCRGRGQCLDGRCVCEDGYSGEDCGVRQCPNDCSQHGVCQDGVCICWEGYVSEDCSIRTCPSNCHGRGR CEEGRCLCDPGYTGPTCATRMCPADCRGRGRCVQGVCLCHVGYGGEDCGQEEPPASACPGGCGPRELCRAGQCVCVEGFRGPDCAIQTCP GDCRGRGECHDGSCVCKDGYAGEDCGEEVPTIEGMRMHLLEETTVRTEWTPAPGPVDAYEIQFIPTTEGASPPFTARVPSSASAYDQRGL APGQEYQVTVRALRGTSWGLPASKTITTMIDGPQDLRVVAVTPTTLELGWLRPQAEVDRFVVSYVSAGNQRVRLEVPPEADGTLLTDLMP GVEYVVTVTAERGRAVSYPASVRANTGSSPLGLLGTTDEPPPSGPSTTQGAQAPLLQQRPQELGELRVLGRDETGRLRVVWTAQPDTFAY FQLRMRVPEGPGAHEEVLPGDVRQALVPPPPPGTPYELSLHGVPPGGKPSDPIIYQGIMDKDEEKPGKSSGPPRLGELTVTDRTSDSLLL RWTVPEGEFDSFVIQYKDRDGQPQVVPVEGPQRSAVITSLDPGRKYKFVLYGFVGKKRHGPLVAEAKILPQSDPSPGTPPHLGNLWVTDP TPDSLHLSWTVPEGQFDTFMVQYRDRDGRPQVVPVEGPERSFVVSSLDPDHKYRFTLFGIANKKRYGPLTADGTTAPERKEEPPRPEFLE QPLLGELTVTGVTPDSLRLSWTVAQGPFDSFMVQYKDAQGQPQAVPVAGDENEVTVPGLDPDRKYKMNLYGLRGRQRVGPESVVAKTAPQ EDVDETPSPTELGTEAPESPEEPLLGELTVTGSSPDSLSLFWTVPQGSFDSFTVQYKDRDGRPRAVRVGGKESEVTVGGLEPGHKYKMHL YGLHEGQRVGPVSAVGVTAPQQEETPPATESPLEPRLGELTVTDVTPNSVGLSWTVPEGQFDSFIVQYKDKDGQPQVVPVAADQREVTVY NLEPERKYKMNMYGLHDGQRMGPLSVVIVTAPLPPAPATEASKPPLEPRLGELTVTDITPDSVGLSWTVPEGEFDSFVVQYKDRDGQPQV VPVAADQREVTIPDLEPSRKYKFLLFGIQDGKRRSPVSVEAKTVARGDASPGAPPRLGELWVTDPTPDSLRLSWTVPEGQFDSFVVQFKD KDGPQVVPVEGHERSVTVTPLDAGRKYRFLLYGLLGKKRHGPLTADGTTEARSAMDDTGTKRPPKPRLGEELQVTTVTQNSVGLSWTVPE GQFDSFVVQYKDRDGQPQVVPVEGSLREVSVPGLDPAHRYKLLLYGLHHGKRVGPISAVAITAGREETETETTAPTPPAPEPHLGELTVE EATSHTLHLSWMVTEGEFDSFEIQYTDRDGQLQMVRIGGDRNDITLSGLESDHRYLVTLYGFSDGKHVGPVHVEALTVPEEEKPSEPPTA TPEPPIKPRLGELTVTDATPDSLSLSWTVPEGQFDHFLVQYRNGDGQPKAVRVPGHEEGVTISGLEPDHKYKMNLYGFHGGQRMGPVSVV GVTAAEEETPSPTEPSMEAPEPAEEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGRPQVVRVGGEESEVTVGGLEPGRKYK MHLYGLHEGRRVGPVSAVGVTAPEEESPDAPLAKLRLGQMTVRDITSDSLSLSWTVPEGQFDHFLVQFKNGDGQPKAVRVPGHEDGVTIS GLEPDHKYKMNLYGFHGGQRVGPVSAVGLTAPGKDEEMAPASTEPPTPEPPIKPRLEELTVTDATPDSLSLSWTVPEGQFDHFLVQYKNG DGQPKATRVPGHEDRVTISGLEPDNKYKMNLYGFHGGQRVGPVSAIGVTAAEEETPSPTEPSMEAPEPPEEPLLGELTVTGSSPDSLSLS WTVPQGRFDSFTVQYKDRDGRPQVVRVGGEESEVTVGGLEPGRKYKMHLYGLHEGRRVGPVSTVGVTAPQEDVDETPSPTEPGTEAPGPP EEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGRPQAVRVGGQESKVTVRGLEPGRKYKMHLYGLHEGRRLGPVSAVGVTED EAETTQAVPTMTPEPPIKPRLGELTMTDATPDSLSLSWTVPEGQFDHFLVQYRNGDGQPKAVRVPGHEDGVTISGLEPDHKYKMNLYGFH GGQRVGPISVIGVTAAEEETPSPTELSTEAPEPPEEPLLGELTVTGSSPDSLSLSWTIPQGHFDSFTVQYKDRDGRPQVMRVRGEESEVT VGGLEPGRKYKMHLYGLHEGRRVGPVSTVGVTAPEDEAETTQAVPTTTPEPPNKPRLGELTVTDATPDSLSLSWMVPEGQFDHFLVQYRN GDGQPKVVRVPGHEDGVTISGLEPDHKYKMNLYGFHGGQRVGPISVIGVTAAEEETPAPTEPSTEAPEPPEEPLLGELTVTGSSPDSLSL SWTIPQGRFDSFTVQYKDRDGRPQVVRVRGEESEVTVGGLEPGCKYKMHLYGLHEGQRVGPVSAVGVTAPKDEAETTQAVPTMTPEPPIK PRLGELTVTDATPDSLSLSWMVPEGQFDHFLVQYRNGDGQPKAVRVPGHEDGVTISGLEPDHKYKMNLYGFHGGQRVGPVSAIGVTEEET PSPTEPSTEAPEAPEEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGQPQVVRVRGEESEVTVGGLEPGRKYKMHLYGLHEG QRVGPVSTVGITAPLPTPLPVEPRLGELAVAAVTSDSVGLSWTVAQGPFDSFLVQYRDAQGQPQAVPVSGDLRAVAVSGLDPARKYKFLL -------------------------------------------------------------- >92962_92962_3_TNXB-VIM_TNXB_chr6_32016140_ENST00000375247_VIM_chr10_17278293_ENST00000224237_length(amino acids)=3388AA_BP=3325 MMPAQYALTSSLVLLVLLSTARAGPFSSRSNVTLPAPRPPPQPGGHTVGAGVGSPSSQLYEHTVEGGEKQVVFTHRINLPPSTGCGCPPG TEPPVLASEVQALRVRLEILEELVKGLKEQCTGGCCPASAQAGTGQTDVRTLCSLHGVFDLSRCTCSCEPGWGGPTCSDPTDAEIPPSSP PSASGSCPDDCNDQGRCVRGRCVCFPGYTGPSCGWPSCPGDCQGRGRCVQGVCVCRAGFSGPDCSQRSCPRGCSQRGRCEGGRCVCDPGY TGDDCGMRSCPRGCSQRGRCENGRCVCNPGYTGEDCGVRSCPRGCSQRGRCKDGRCVCDPGYTGEDCGTRSCPWDCGEGGRCVDGRCVCW PGYTGEDCSTRTCPRDCRGRGRCEDGECICDTGYSGDDCGVRSCPGDCNQRGRCEDGRCVCWPGYTGTDCGSRACPRDCRGRGRCENGVC VCNAGYSGEDCGVRSCPGDCRGRGRCESGRCMCWPGYTGRDCGTRACPGDCRGRGRCVDGRCVCNPGFTGEDCGSRRCPGDCRGHGLCED GVCVCDAGYSGEDCSTRSCPGGCRGRGQCLDGRCVCEDGYSGEDCGVRQCPNDCSQHGVCQDGVCICWEGYVSEDCSIRTCPSNCHGRGR CEEGRCLCDPGYTGPTCATRMCPADCRGRGRCVQGVCLCHVGYGGEDCGQEEPPASACPGGCGPRELCRAGQCVCVEGFRGPDCAIQTCP GDCRGRGECHDGSCVCKDGYAGEDCGEEVPTIEGMRMHLLEETTVRTEWTPAPGPVDAYEIQFIPTTEGASPPFTARVPSSASAYDQRGL APGQEYQVTVRALRGTSWGLPASKTITTMIDGPQDLRVVAVTPTTLELGWLRPQAEVDRFVVSYVSAGNQRVRLEVPPEADGTLLTDLMP GVEYVVTVTAERGRAVSYPASVRANTGSSPLGLLGTTDEPPPSGPSTTQGAQAPLLQQRPQELGELRVLGRDETGRLRVVWTAQPDTFAY FQLRMRVPEGPGAHEEVLPGDVRQALVPPPPPGTPYELSLHGVPPGGKPSDPIIYQGIMDKDEEKPGKSSGPPRLGELTVTDRTSDSLLL RWTVPEGEFDSFVIQYKDRDGQPQVVPVEGPQRSAVITSLDPGRKYKFVLYGFVGKKRHGPLVAEAKILPQSDPSPGTPPHLGNLWVTDP TPDSLHLSWTVPEGQFDTFMVQYRDRDGRPQVVPVEGPERSFVVSSLDPDHKYRFTLFGIANKKRYGPLTADGTTAPERKEEPPRPEFLE QPLLGELTVTGVTPDSLRLSWTVAQGPFDSFMVQYKDAQGQPQAVPVAGDENEVTVPGLDPDRKYKMNLYGLRGRQRVGPESVVAKTAPQ EDVDETPSPTELGTEAPESPEEPLLGELTVTGSSPDSLSLFWTVPQGSFDSFTVQYKDRDGRPRAVRVGGKESEVTVGGLEPGHKYKMHL YGLHEGQRVGPVSAVGVTAPQQEETPPATESPLEPRLGELTVTDVTPNSVGLSWTVPEGQFDSFIVQYKDKDGQPQVVPVAADQREVTVY NLEPERKYKMNMYGLHDGQRMGPLSVVIVTAPLPPAPATEASKPPLEPRLGELTVTDITPDSVGLSWTVPEGEFDSFVVQYKDRDGQPQV VPVAADQREVTIPDLEPSRKYKFLLFGIQDGKRRSPVSVEAKTVARGDASPGAPPRLGELWVTDPTPDSLRLSWTVPEGQFDSFVVQFKD KDGPQVVPVEGHERSVTVTPLDAGRKYRFLLYGLLGKKRHGPLTADGTTEARSAMDDTGTKRPPKPRLGEELQVTTVTQNSVGLSWTVPE GQFDSFVVQYKDRDGQPQVVPVEGSLREVSVPGLDPAHRYKLLLYGLHHGKRVGPISAVAITAGREETETETTAPTPPAPEPHLGELTVE EATSHTLHLSWMVTEGEFDSFEIQYTDRDGQLQMVRIGGDRNDITLSGLESDHRYLVTLYGFSDGKHVGPVHVEALTVPEEEKPSEPPTA TPEPPIKPRLGELTVTDATPDSLSLSWTVPEGQFDHFLVQYRNGDGQPKAVRVPGHEEGVTISGLEPDHKYKMNLYGFHGGQRMGPVSVV GVTAAEEETPSPTEPSMEAPEPAEEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGRPQVVRVGGEESEVTVGGLEPGRKYK MHLYGLHEGRRVGPVSAVGVTAPEEESPDAPLAKLRLGQMTVRDITSDSLSLSWTVPEGQFDHFLVQFKNGDGQPKAVRVPGHEDGVTIS GLEPDHKYKMNLYGFHGGQRVGPVSAVGLTAPGKDEEMAPASTEPPTPEPPIKPRLEELTVTDATPDSLSLSWTVPEGQFDHFLVQYKNG DGQPKATRVPGHEDRVTISGLEPDNKYKMNLYGFHGGQRVGPVSAIGVTAAEEETPSPTEPSMEAPEPPEEPLLGELTVTGSSPDSLSLS WTVPQGRFDSFTVQYKDRDGRPQVVRVGGEESEVTVGGLEPGRKYKMHLYGLHEGRRVGPVSTVGVTAPQEDVDETPSPTEPGTEAPGPP EEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGRPQAVRVGGQESKVTVRGLEPGRKYKMHLYGLHEGRRLGPVSAVGVTED EAETTQAVPTMTPEPPIKPRLGELTMTDATPDSLSLSWTVPEGQFDHFLVQYRNGDGQPKAVRVPGHEDGVTISGLEPDHKYKMNLYGFH GGQRVGPISVIGVTAAEEETPSPTELSTEAPEPPEEPLLGELTVTGSSPDSLSLSWTIPQGHFDSFTVQYKDRDGRPQVMRVRGEESEVT VGGLEPGRKYKMHLYGLHEGRRVGPVSTVGVTEDEAETTQAVPTTTPEPPNKPRLGELTVTDATPDSLSLSWMVPEGQFDHFLVQYRNGD GQPKVVRVPGHEDGVTISGLEPDHKYKMNLYGFHGGQRVGPISVIGVTAAEEETPAPTEPSTEAPEPPEEPLLGELTVTGSSPDSLSLSW TIPQGRFDSFTVQYKDRDGRPQVVRVRGEESEVTVGGLEPGCKYKMHLYGLHEGQRVGPVSAVGVTAPKDEAETTQAVPTMTPEPPIKPR LGELTVTDATPDSLSLSWMVPEGQFDHFLVQYRNGDGQPKAVRVPGHEDGVTISGLEPDHKYKMNLYGFHGGQRVGPVSAIGVTEEETPS PTEPSTEAPEAPEEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGQPQVVRVRGEESEVTVGGLEPGRKYKMHLYGLHEGQR VGPVSTVGITAPLPTPLPVEPRLGELAVAAVTSDSVGLSWTVAQGPFDSFLVQYRDAQGQPQAVPVSGDLRAVAVSGLDPARKYKFLLFG -------------------------------------------------------------- >92962_92962_4_TNXB-VIM_TNXB_chr6_32016140_ENST00000375247_VIM_chr10_17278293_ENST00000544301_length(amino acids)=3388AA_BP=3325 MMPAQYALTSSLVLLVLLSTARAGPFSSRSNVTLPAPRPPPQPGGHTVGAGVGSPSSQLYEHTVEGGEKQVVFTHRINLPPSTGCGCPPG TEPPVLASEVQALRVRLEILEELVKGLKEQCTGGCCPASAQAGTGQTDVRTLCSLHGVFDLSRCTCSCEPGWGGPTCSDPTDAEIPPSSP PSASGSCPDDCNDQGRCVRGRCVCFPGYTGPSCGWPSCPGDCQGRGRCVQGVCVCRAGFSGPDCSQRSCPRGCSQRGRCEGGRCVCDPGY TGDDCGMRSCPRGCSQRGRCENGRCVCNPGYTGEDCGVRSCPRGCSQRGRCKDGRCVCDPGYTGEDCGTRSCPWDCGEGGRCVDGRCVCW PGYTGEDCSTRTCPRDCRGRGRCEDGECICDTGYSGDDCGVRSCPGDCNQRGRCEDGRCVCWPGYTGTDCGSRACPRDCRGRGRCENGVC VCNAGYSGEDCGVRSCPGDCRGRGRCESGRCMCWPGYTGRDCGTRACPGDCRGRGRCVDGRCVCNPGFTGEDCGSRRCPGDCRGHGLCED GVCVCDAGYSGEDCSTRSCPGGCRGRGQCLDGRCVCEDGYSGEDCGVRQCPNDCSQHGVCQDGVCICWEGYVSEDCSIRTCPSNCHGRGR CEEGRCLCDPGYTGPTCATRMCPADCRGRGRCVQGVCLCHVGYGGEDCGQEEPPASACPGGCGPRELCRAGQCVCVEGFRGPDCAIQTCP GDCRGRGECHDGSCVCKDGYAGEDCGEEVPTIEGMRMHLLEETTVRTEWTPAPGPVDAYEIQFIPTTEGASPPFTARVPSSASAYDQRGL APGQEYQVTVRALRGTSWGLPASKTITTMIDGPQDLRVVAVTPTTLELGWLRPQAEVDRFVVSYVSAGNQRVRLEVPPEADGTLLTDLMP GVEYVVTVTAERGRAVSYPASVRANTGSSPLGLLGTTDEPPPSGPSTTQGAQAPLLQQRPQELGELRVLGRDETGRLRVVWTAQPDTFAY FQLRMRVPEGPGAHEEVLPGDVRQALVPPPPPGTPYELSLHGVPPGGKPSDPIIYQGIMDKDEEKPGKSSGPPRLGELTVTDRTSDSLLL RWTVPEGEFDSFVIQYKDRDGQPQVVPVEGPQRSAVITSLDPGRKYKFVLYGFVGKKRHGPLVAEAKILPQSDPSPGTPPHLGNLWVTDP TPDSLHLSWTVPEGQFDTFMVQYRDRDGRPQVVPVEGPERSFVVSSLDPDHKYRFTLFGIANKKRYGPLTADGTTAPERKEEPPRPEFLE QPLLGELTVTGVTPDSLRLSWTVAQGPFDSFMVQYKDAQGQPQAVPVAGDENEVTVPGLDPDRKYKMNLYGLRGRQRVGPESVVAKTAPQ EDVDETPSPTELGTEAPESPEEPLLGELTVTGSSPDSLSLFWTVPQGSFDSFTVQYKDRDGRPRAVRVGGKESEVTVGGLEPGHKYKMHL YGLHEGQRVGPVSAVGVTAPQQEETPPATESPLEPRLGELTVTDVTPNSVGLSWTVPEGQFDSFIVQYKDKDGQPQVVPVAADQREVTVY NLEPERKYKMNMYGLHDGQRMGPLSVVIVTAPLPPAPATEASKPPLEPRLGELTVTDITPDSVGLSWTVPEGEFDSFVVQYKDRDGQPQV VPVAADQREVTIPDLEPSRKYKFLLFGIQDGKRRSPVSVEAKTVARGDASPGAPPRLGELWVTDPTPDSLRLSWTVPEGQFDSFVVQFKD KDGPQVVPVEGHERSVTVTPLDAGRKYRFLLYGLLGKKRHGPLTADGTTEARSAMDDTGTKRPPKPRLGEELQVTTVTQNSVGLSWTVPE GQFDSFVVQYKDRDGQPQVVPVEGSLREVSVPGLDPAHRYKLLLYGLHHGKRVGPISAVAITAGREETETETTAPTPPAPEPHLGELTVE EATSHTLHLSWMVTEGEFDSFEIQYTDRDGQLQMVRIGGDRNDITLSGLESDHRYLVTLYGFSDGKHVGPVHVEALTVPEEEKPSEPPTA TPEPPIKPRLGELTVTDATPDSLSLSWTVPEGQFDHFLVQYRNGDGQPKAVRVPGHEEGVTISGLEPDHKYKMNLYGFHGGQRMGPVSVV GVTAAEEETPSPTEPSMEAPEPAEEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGRPQVVRVGGEESEVTVGGLEPGRKYK MHLYGLHEGRRVGPVSAVGVTAPEEESPDAPLAKLRLGQMTVRDITSDSLSLSWTVPEGQFDHFLVQFKNGDGQPKAVRVPGHEDGVTIS GLEPDHKYKMNLYGFHGGQRVGPVSAVGLTAPGKDEEMAPASTEPPTPEPPIKPRLEELTVTDATPDSLSLSWTVPEGQFDHFLVQYKNG DGQPKATRVPGHEDRVTISGLEPDNKYKMNLYGFHGGQRVGPVSAIGVTAAEEETPSPTEPSMEAPEPPEEPLLGELTVTGSSPDSLSLS WTVPQGRFDSFTVQYKDRDGRPQVVRVGGEESEVTVGGLEPGRKYKMHLYGLHEGRRVGPVSTVGVTAPQEDVDETPSPTEPGTEAPGPP EEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGRPQAVRVGGQESKVTVRGLEPGRKYKMHLYGLHEGRRLGPVSAVGVTED EAETTQAVPTMTPEPPIKPRLGELTMTDATPDSLSLSWTVPEGQFDHFLVQYRNGDGQPKAVRVPGHEDGVTISGLEPDHKYKMNLYGFH GGQRVGPISVIGVTAAEEETPSPTELSTEAPEPPEEPLLGELTVTGSSPDSLSLSWTIPQGHFDSFTVQYKDRDGRPQVMRVRGEESEVT VGGLEPGRKYKMHLYGLHEGRRVGPVSTVGVTEDEAETTQAVPTTTPEPPNKPRLGELTVTDATPDSLSLSWMVPEGQFDHFLVQYRNGD GQPKVVRVPGHEDGVTISGLEPDHKYKMNLYGFHGGQRVGPISVIGVTAAEEETPAPTEPSTEAPEPPEEPLLGELTVTGSSPDSLSLSW TIPQGRFDSFTVQYKDRDGRPQVVRVRGEESEVTVGGLEPGCKYKMHLYGLHEGQRVGPVSAVGVTAPKDEAETTQAVPTMTPEPPIKPR LGELTVTDATPDSLSLSWMVPEGQFDHFLVQYRNGDGQPKAVRVPGHEDGVTISGLEPDHKYKMNLYGFHGGQRVGPVSAIGVTEEETPS PTEPSTEAPEAPEEPLLGELTVTGSSPDSLSLSWTVPQGRFDSFTVQYKDRDGQPQVVRVRGEESEVTVGGLEPGRKYKMHLYGLHEGQR VGPVSTVGITAPLPTPLPVEPRLGELAVAAVTSDSVGLSWTVAQGPFDSFLVQYRDAQGQPQAVPVSGDLRAVAVSGLDPARKYKFLLFG -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr6:32016140/chr10:17278293) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | VIM |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | 169 |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1064_1153 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 2 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1161_1249 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 3 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1263_1352 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 4 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1374_1468 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 5 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1476_1572 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 6 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 156_168 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 1%3B incomplete |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1574_1669 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 7 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1674_1764 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 8 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1778_1868 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 9 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 183_213 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 2 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1883_1971 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 10 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1989_2089 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 11 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 2097_2185 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 12 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 214_244 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 3 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 2196_2296 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 13 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 2305_2398 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 14 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 2408_2502 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 15 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 245_275 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 4 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 2519_2617 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 16 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 2625_2723 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 17 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 2733_2840 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 18 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 276_306 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 5 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 2841_2939 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 19 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 2949_3042 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 20 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 3062_3153 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 21 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 307_337 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 6 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 3168_3260 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 22 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 338_368 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 7 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 369_399 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 8 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 400_430 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 9 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 431_461 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 10 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 462_492 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 11 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 493_523 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 12 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 524_554 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 13 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 555_585 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 14 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 586_616 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 15 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 617_647 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 16 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 648_679 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 17 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 684_714 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 18 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 715_746 | 3346.3333333333335 | 4243.0 | Domain | EGF-like 19 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 959_1051 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 1 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 1666_1668 | 3346.3333333333335 | 4243.0 | Motif | Cell attachment site |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 3264_3355 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 23 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 3357_3446 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 24 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 3451_3544 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 25 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 3553_3647 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 26 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 3657_3754 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 27 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 3758_3847 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 28 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 3848_3934 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 29 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 3935_4025 | 3346.3333333333335 | 4243.0 | Domain | Fibronectin type-III 30 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000375247 | - | 29 | 44 | 4021_4236 | 3346.3333333333335 | 4243.0 | Domain | Fibrinogen C-terminal |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1064_1153 | 0 | 674.0 | Domain | Fibronectin type-III 2 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1161_1249 | 0 | 674.0 | Domain | Fibronectin type-III 3 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1263_1352 | 0 | 674.0 | Domain | Fibronectin type-III 4 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1374_1468 | 0 | 674.0 | Domain | Fibronectin type-III 5 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1476_1572 | 0 | 674.0 | Domain | Fibronectin type-III 6 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 156_168 | 0 | 674.0 | Domain | EGF-like 1%3B incomplete |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1574_1669 | 0 | 674.0 | Domain | Fibronectin type-III 7 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1674_1764 | 0 | 674.0 | Domain | Fibronectin type-III 8 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1778_1868 | 0 | 674.0 | Domain | Fibronectin type-III 9 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 183_213 | 0 | 674.0 | Domain | EGF-like 2 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1883_1971 | 0 | 674.0 | Domain | Fibronectin type-III 10 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1989_2089 | 0 | 674.0 | Domain | Fibronectin type-III 11 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 2097_2185 | 0 | 674.0 | Domain | Fibronectin type-III 12 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 214_244 | 0 | 674.0 | Domain | EGF-like 3 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 2196_2296 | 0 | 674.0 | Domain | Fibronectin type-III 13 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 2305_2398 | 0 | 674.0 | Domain | Fibronectin type-III 14 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 2408_2502 | 0 | 674.0 | Domain | Fibronectin type-III 15 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 245_275 | 0 | 674.0 | Domain | EGF-like 4 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 2519_2617 | 0 | 674.0 | Domain | Fibronectin type-III 16 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 2625_2723 | 0 | 674.0 | Domain | Fibronectin type-III 17 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 2733_2840 | 0 | 674.0 | Domain | Fibronectin type-III 18 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 276_306 | 0 | 674.0 | Domain | EGF-like 5 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 2841_2939 | 0 | 674.0 | Domain | Fibronectin type-III 19 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 2949_3042 | 0 | 674.0 | Domain | Fibronectin type-III 20 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 3062_3153 | 0 | 674.0 | Domain | Fibronectin type-III 21 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 307_337 | 0 | 674.0 | Domain | EGF-like 6 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 3168_3260 | 0 | 674.0 | Domain | Fibronectin type-III 22 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 3264_3355 | 0 | 674.0 | Domain | Fibronectin type-III 23 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 3357_3446 | 0 | 674.0 | Domain | Fibronectin type-III 24 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 338_368 | 0 | 674.0 | Domain | EGF-like 7 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 3451_3544 | 0 | 674.0 | Domain | Fibronectin type-III 25 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 3553_3647 | 0 | 674.0 | Domain | Fibronectin type-III 26 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 3657_3754 | 0 | 674.0 | Domain | Fibronectin type-III 27 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 369_399 | 0 | 674.0 | Domain | EGF-like 8 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 3758_3847 | 0 | 674.0 | Domain | Fibronectin type-III 28 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 3848_3934 | 0 | 674.0 | Domain | Fibronectin type-III 29 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 3935_4025 | 0 | 674.0 | Domain | Fibronectin type-III 30 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 400_430 | 0 | 674.0 | Domain | EGF-like 9 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 4021_4236 | 0 | 674.0 | Domain | Fibrinogen C-terminal |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 431_461 | 0 | 674.0 | Domain | EGF-like 10 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 462_492 | 0 | 674.0 | Domain | EGF-like 11 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 493_523 | 0 | 674.0 | Domain | EGF-like 12 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 524_554 | 0 | 674.0 | Domain | EGF-like 13 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 555_585 | 0 | 674.0 | Domain | EGF-like 14 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 586_616 | 0 | 674.0 | Domain | EGF-like 15 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 617_647 | 0 | 674.0 | Domain | EGF-like 16 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 648_679 | 0 | 674.0 | Domain | EGF-like 17 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 684_714 | 0 | 674.0 | Domain | EGF-like 18 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 715_746 | 0 | 674.0 | Domain | EGF-like 19 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 959_1051 | 0 | 674.0 | Domain | Fibronectin type-III 1 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1064_1153 | 0 | 674.0 | Domain | Fibronectin type-III 2 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1161_1249 | 0 | 674.0 | Domain | Fibronectin type-III 3 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1263_1352 | 0 | 674.0 | Domain | Fibronectin type-III 4 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1374_1468 | 0 | 674.0 | Domain | Fibronectin type-III 5 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1476_1572 | 0 | 674.0 | Domain | Fibronectin type-III 6 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 156_168 | 0 | 674.0 | Domain | EGF-like 1%3B incomplete |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1574_1669 | 0 | 674.0 | Domain | Fibronectin type-III 7 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1674_1764 | 0 | 674.0 | Domain | Fibronectin type-III 8 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1778_1868 | 0 | 674.0 | Domain | Fibronectin type-III 9 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 183_213 | 0 | 674.0 | Domain | EGF-like 2 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1883_1971 | 0 | 674.0 | Domain | Fibronectin type-III 10 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1989_2089 | 0 | 674.0 | Domain | Fibronectin type-III 11 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 2097_2185 | 0 | 674.0 | Domain | Fibronectin type-III 12 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 214_244 | 0 | 674.0 | Domain | EGF-like 3 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 2196_2296 | 0 | 674.0 | Domain | Fibronectin type-III 13 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 2305_2398 | 0 | 674.0 | Domain | Fibronectin type-III 14 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 2408_2502 | 0 | 674.0 | Domain | Fibronectin type-III 15 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 245_275 | 0 | 674.0 | Domain | EGF-like 4 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 2519_2617 | 0 | 674.0 | Domain | Fibronectin type-III 16 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 2625_2723 | 0 | 674.0 | Domain | Fibronectin type-III 17 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 2733_2840 | 0 | 674.0 | Domain | Fibronectin type-III 18 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 276_306 | 0 | 674.0 | Domain | EGF-like 5 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 2841_2939 | 0 | 674.0 | Domain | Fibronectin type-III 19 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 2949_3042 | 0 | 674.0 | Domain | Fibronectin type-III 20 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 3062_3153 | 0 | 674.0 | Domain | Fibronectin type-III 21 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 307_337 | 0 | 674.0 | Domain | EGF-like 6 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 3168_3260 | 0 | 674.0 | Domain | Fibronectin type-III 22 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 3264_3355 | 0 | 674.0 | Domain | Fibronectin type-III 23 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 3357_3446 | 0 | 674.0 | Domain | Fibronectin type-III 24 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 338_368 | 0 | 674.0 | Domain | EGF-like 7 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 3451_3544 | 0 | 674.0 | Domain | Fibronectin type-III 25 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 3553_3647 | 0 | 674.0 | Domain | Fibronectin type-III 26 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 3657_3754 | 0 | 674.0 | Domain | Fibronectin type-III 27 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 369_399 | 0 | 674.0 | Domain | EGF-like 8 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 3758_3847 | 0 | 674.0 | Domain | Fibronectin type-III 28 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 3848_3934 | 0 | 674.0 | Domain | Fibronectin type-III 29 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 3935_4025 | 0 | 674.0 | Domain | Fibronectin type-III 30 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 400_430 | 0 | 674.0 | Domain | EGF-like 9 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 4021_4236 | 0 | 674.0 | Domain | Fibrinogen C-terminal |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 431_461 | 0 | 674.0 | Domain | EGF-like 10 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 462_492 | 0 | 674.0 | Domain | EGF-like 11 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 493_523 | 0 | 674.0 | Domain | EGF-like 12 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 524_554 | 0 | 674.0 | Domain | EGF-like 13 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 555_585 | 0 | 674.0 | Domain | EGF-like 14 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 586_616 | 0 | 674.0 | Domain | EGF-like 15 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 617_647 | 0 | 674.0 | Domain | EGF-like 16 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 648_679 | 0 | 674.0 | Domain | EGF-like 17 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 684_714 | 0 | 674.0 | Domain | EGF-like 18 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 715_746 | 0 | 674.0 | Domain | EGF-like 19 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 959_1051 | 0 | 674.0 | Domain | Fibronectin type-III 1 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1064_1153 | 0 | 674.0 | Domain | Fibronectin type-III 2 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1161_1249 | 0 | 674.0 | Domain | Fibronectin type-III 3 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1263_1352 | 0 | 674.0 | Domain | Fibronectin type-III 4 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1374_1468 | 0 | 674.0 | Domain | Fibronectin type-III 5 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1476_1572 | 0 | 674.0 | Domain | Fibronectin type-III 6 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 156_168 | 0 | 674.0 | Domain | EGF-like 1%3B incomplete |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1574_1669 | 0 | 674.0 | Domain | Fibronectin type-III 7 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1674_1764 | 0 | 674.0 | Domain | Fibronectin type-III 8 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1778_1868 | 0 | 674.0 | Domain | Fibronectin type-III 9 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 183_213 | 0 | 674.0 | Domain | EGF-like 2 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1883_1971 | 0 | 674.0 | Domain | Fibronectin type-III 10 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1989_2089 | 0 | 674.0 | Domain | Fibronectin type-III 11 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 2097_2185 | 0 | 674.0 | Domain | Fibronectin type-III 12 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 214_244 | 0 | 674.0 | Domain | EGF-like 3 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 2196_2296 | 0 | 674.0 | Domain | Fibronectin type-III 13 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 2305_2398 | 0 | 674.0 | Domain | Fibronectin type-III 14 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 2408_2502 | 0 | 674.0 | Domain | Fibronectin type-III 15 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 245_275 | 0 | 674.0 | Domain | EGF-like 4 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 2519_2617 | 0 | 674.0 | Domain | Fibronectin type-III 16 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 2625_2723 | 0 | 674.0 | Domain | Fibronectin type-III 17 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 2733_2840 | 0 | 674.0 | Domain | Fibronectin type-III 18 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 276_306 | 0 | 674.0 | Domain | EGF-like 5 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 2841_2939 | 0 | 674.0 | Domain | Fibronectin type-III 19 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 2949_3042 | 0 | 674.0 | Domain | Fibronectin type-III 20 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 3062_3153 | 0 | 674.0 | Domain | Fibronectin type-III 21 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 307_337 | 0 | 674.0 | Domain | EGF-like 6 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 3168_3260 | 0 | 674.0 | Domain | Fibronectin type-III 22 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 3264_3355 | 0 | 674.0 | Domain | Fibronectin type-III 23 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 3357_3446 | 0 | 674.0 | Domain | Fibronectin type-III 24 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 338_368 | 0 | 674.0 | Domain | EGF-like 7 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 3451_3544 | 0 | 674.0 | Domain | Fibronectin type-III 25 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 3553_3647 | 0 | 674.0 | Domain | Fibronectin type-III 26 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 3657_3754 | 0 | 674.0 | Domain | Fibronectin type-III 27 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 369_399 | 0 | 674.0 | Domain | EGF-like 8 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 3758_3847 | 0 | 674.0 | Domain | Fibronectin type-III 28 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 3848_3934 | 0 | 674.0 | Domain | Fibronectin type-III 29 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 3935_4025 | 0 | 674.0 | Domain | Fibronectin type-III 30 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 400_430 | 0 | 674.0 | Domain | EGF-like 9 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 4021_4236 | 0 | 674.0 | Domain | Fibrinogen C-terminal |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 431_461 | 0 | 674.0 | Domain | EGF-like 10 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 462_492 | 0 | 674.0 | Domain | EGF-like 11 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 493_523 | 0 | 674.0 | Domain | EGF-like 12 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 524_554 | 0 | 674.0 | Domain | EGF-like 13 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 555_585 | 0 | 674.0 | Domain | EGF-like 14 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 586_616 | 0 | 674.0 | Domain | EGF-like 15 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 617_647 | 0 | 674.0 | Domain | EGF-like 16 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 648_679 | 0 | 674.0 | Domain | EGF-like 17 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 684_714 | 0 | 674.0 | Domain | EGF-like 18 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 715_746 | 0 | 674.0 | Domain | EGF-like 19 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 959_1051 | 0 | 674.0 | Domain | Fibronectin type-III 1 |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000451343 | - | 1 | 13 | 1666_1668 | 0 | 674.0 | Motif | Cell attachment site |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000546684 | - | 1 | 15 | 1666_1668 | 0 | 674.0 | Motif | Cell attachment site |
| Hgene | TNXB | chr6:32016140 | chr10:17278293 | ENST00000550539 | - | 1 | 15 | 1666_1668 | 0 | 674.0 | Motif | Cell attachment site |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000224237 | 6 | 9 | 154_245 | 424.3333333333333 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000224237 | 6 | 9 | 303_407 | 424.3333333333333 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000224237 | 6 | 9 | 96_131 | 424.3333333333333 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000544301 | 7 | 10 | 154_245 | 424.3333333333333 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000544301 | 7 | 10 | 303_407 | 424.3333333333333 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000544301 | 7 | 10 | 96_131 | 424.3333333333333 | 467.0 | Coiled coil | Ontology_term=ECO:0000269 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000224237 | 6 | 9 | 103_411 | 424.3333333333333 | 467.0 | Domain | IF rod | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000544301 | 7 | 10 | 103_411 | 424.3333333333333 | 467.0 | Domain | IF rod | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000224237 | 6 | 9 | 326_329 | 424.3333333333333 | 467.0 | Motif | [IL]-x-C-x-x-[DE] motif | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000544301 | 7 | 10 | 326_329 | 424.3333333333333 | 467.0 | Motif | [IL]-x-C-x-x-[DE] motif | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000224237 | 6 | 9 | 132_153 | 424.3333333333333 | 467.0 | Region | Note=Linker 1 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000224237 | 6 | 9 | 246_268 | 424.3333333333333 | 467.0 | Region | Note=Linker 12 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000224237 | 6 | 9 | 269_407 | 424.3333333333333 | 467.0 | Region | Note=Coil 2 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000224237 | 6 | 9 | 2_95 | 424.3333333333333 | 467.0 | Region | Note=Head | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000224237 | 6 | 9 | 408_466 | 424.3333333333333 | 467.0 | Region | Note=Tail | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000544301 | 7 | 10 | 132_153 | 424.3333333333333 | 467.0 | Region | Note=Linker 1 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000544301 | 7 | 10 | 246_268 | 424.3333333333333 | 467.0 | Region | Note=Linker 12 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000544301 | 7 | 10 | 269_407 | 424.3333333333333 | 467.0 | Region | Note=Coil 2 | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000544301 | 7 | 10 | 2_95 | 424.3333333333333 | 467.0 | Region | Note=Head | |
| Tgene | VIM | chr6:32016140 | chr10:17278293 | ENST00000544301 | 7 | 10 | 408_466 | 424.3333333333333 | 467.0 | Region | Note=Tail |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| TNXB | |
| VIM |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to TNXB-VIM |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to TNXB-VIM |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

