| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:TPR-NTRK1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: TPR-NTRK1 | FusionPDB ID: 93453 | FusionGDB2.0 ID: 93453 | Hgene | Tgene | Gene symbol | TPR | NTRK1 | Gene ID | 7175 | 4914 |
| Gene name | translocated promoter region, nuclear basket protein | neurotrophic receptor tyrosine kinase 1 | |
| Synonyms | - | MTC|TRK|TRK1|TRKA|Trk-A|p140-TrkA | |
| Cytomap | 1q31.1 | 1q23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | nucleoprotein TPRNPC-associated intranuclear proteinmegatornuclear pore complex-associated protein TPRtranslocated promoter region (to activated MET oncogene)translocated promoter region proteintumor potentiating region | high affinity nerve growth factor receptorOncogene TRKTRK1-transforming tyrosine kinase proteingp140trkneurotrophic tyrosine kinase, receptor, type 1tropomyosin-related kinase Atyrosine kinase receptor A | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | TRANK1 | P04629 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000367478, ENST00000474852, | ENST00000531606, ENST00000358660, ENST00000368196, ENST00000392302, ENST00000524377, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 8 X 11 X 4=352 | 25 X 24 X 13=7800 |
| # samples | 8 | 33 | |
| ** MAII score | log2(8/352*10)=-2.13750352374993 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(33/7800*10)=-4.56293619439116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: TPR [Title/Abstract] AND NTRK1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | TPR(186332090)-NTRK1(156845411), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TPR | GO:0006404 | RNA import into nucleus | 9864356 |
| Hgene | TPR | GO:0006606 | protein import into nucleus | 9864356 |
| Hgene | TPR | GO:0031990 | mRNA export from nucleus in response to heat stress | 17897941 |
| Hgene | TPR | GO:0034605 | cellular response to heat | 17897941 |
| Hgene | TPR | GO:0046832 | negative regulation of RNA export from nucleus | 22253824 |
| Hgene | TPR | GO:0070849 | response to epidermal growth factor | 18794356 |
| Tgene | NTRK1 | GO:0006468 | protein phosphorylation | 15488758 |
| Tgene | NTRK1 | GO:0008285 | negative regulation of cell proliferation | 15488758 |
| Tgene | NTRK1 | GO:0010976 | positive regulation of neuron projection development | 15488758 |
| Tgene | NTRK1 | GO:0018108 | peptidyl-tyrosine phosphorylation | 2927393 |
| Tgene | NTRK1 | GO:0043547 | positive regulation of GTPase activity | 15488758 |
| Tgene | NTRK1 | GO:0046579 | positive regulation of Ras protein signal transduction | 15488758 |
| Tgene | NTRK1 | GO:0046777 | protein autophosphorylation | 15488758 |
| Tgene | NTRK1 | GO:0048011 | neurotrophin TRK receptor signaling pathway | 15488758 |
| Tgene | NTRK1 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity | 15488758 |
| Tgene | NTRK1 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | 15488758 |
| Tgene | NTRK1 | GO:1904646 | cellular response to amyloid-beta | 11927634 |
 Fusion gene breakpoints across TPR (5'-gene) Fusion gene breakpoints across TPR (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 Fusion gene breakpoints across NTRK1 (3'-gene) Fusion gene breakpoints across NTRK1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerKB3 | . | . | TPR | chr1 | 184610797 | - | NTRK1 | chr1 | 155112034 | + |
| ChimerKB3 | . | . | TPR | chr1 | 186319354 | - | NTRK1 | chr1 | 156844362 | + |
| ChiTaRS5.0 | N/A | X62947 | TPR | chr1 | 186332090 | - | NTRK1 | chr1 | 156845411 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000367478 | TPR | chr1 | 186319354 | - | ENST00000392302 | NTRK1 | chr1 | 156844362 | + | 4421 | 3073 | 297 | 4268 | 1323 |
| ENST00000367478 | TPR | chr1 | 186319354 | - | ENST00000368196 | NTRK1 | chr1 | 156844362 | + | 4477 | 3073 | 297 | 4268 | 1323 |
| ENST00000367478 | TPR | chr1 | 186319354 | - | ENST00000524377 | NTRK1 | chr1 | 156844362 | + | 4269 | 3073 | 297 | 4268 | 1323 |
| ENST00000367478 | TPR | chr1 | 186319354 | - | ENST00000358660 | NTRK1 | chr1 | 156844362 | + | 4347 | 3073 | 297 | 4277 | 1326 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >93453_93453_1_TPR-NTRK1_TPR_chr1_186319354_ENST00000367478_NTRK1_chr1_156844362_ENST00000358660_length(amino acids)=1326AA_BP=925 MAAVLQQVLERTELNKLPKSVQNKLEKFLADQQSEIDGLKGRHEKFKVESEQQYFEIEKRLSHSQERLVNETRECQSLRLELEKLNNQLK ALTEKNKELEIAQDRNIAIQSQFTRTKEELEAEKRDLIRTNERLSQELEYLTEDVKRLNEKLKESNTTKGELQLKLDELQASDVSVKYRE KRLEQEKELLHSQNTWLNTELKTKTDELLALGREKGNEILELKCNLENKKEEVSRLEEQMNGLKTSNEHLQKHVEDLLTKLKEAKEQQAS MEEKFHNELNAHIKLSNLYKSAADDSEAKSNELTRAVEELHKLLKEAGEANKAIQDHLLEVEQSKDQMEKEMLEKIGRLEKELENANDLL SATKRKGAILSEEELAAMSPTAAAVAKIVKPGMKLTELYNAYVETQDQLLLEKLENKRINKYLDEIVKEVEAKAPILKRQREEYERAQKA VASLSVKLEQAMKEIQRLQEDTDKANKQSSVLERDNRRMEIQVKDLSQQIRVLLMELEEARGNHVIRDEEVSSADISSSSEVISQHLVSY RNIEELQQQNQRLLVALRELGETREREEQETTSSKITELQLKLESALTELEQLRKSRQHQMQLVDSIVRQRDMYRILLSQTTGVAIPLHA SSLDDVSLASTPKRPSTSQTVSTPAPVPVIESTEAIEAKAALKQLQEIFENYKKEKAENEKIQNEQLEKLQEQVTDLRSQNTKISTQLDF ASKRYEMLQDNVEGYRREITSLHERNQKLTATTQKQEQIINTMTQDLRGANEKLAVAEVRAENLKKEKEMLKLSEVRLSQQRESLLAEQR GQNLLLTNLQTIQGILERSETETKQRLSSQIEKLEHEISHLKKKLENEVEQRHTLTRNLDVQLLDTKRQLDTETNLHLNTKELLKNAQKE IATLKQHLSNMEVQVASQSSQRTGKDTNSTSGDPVEKKDETPFGVSVAVGLAVFACLFLSTLLLVLNKCGRRNKFGINRPAVLAPEDGLA MSLHFMTLGGSSLSPTEGKGSGLQGHIIENPQYFSDASPSGVHHIKRRDIVLKWELGEGAFGKVFLAECHNLLPEQDKMLVAVKALKEAS ESARQDFQREAELLTMLQHQHIVRFFGVCTEGRPLLMVFEYMRHGDLNRFLRSHGPDAKLLAGGEDVAPGPLGLGQLLAVASQVAAGMVY LAGLHFVHRDLATRNCLVGQGLVVKIGDFGMSRDIYSTDYYRVGGRTMLPIRWMPPESILYRKFTTESDVWSFGVVLWEIFTYGKQPWYQ -------------------------------------------------------------- >93453_93453_2_TPR-NTRK1_TPR_chr1_186319354_ENST00000367478_NTRK1_chr1_156844362_ENST00000368196_length(amino acids)=1323AA_BP=925 MAAVLQQVLERTELNKLPKSVQNKLEKFLADQQSEIDGLKGRHEKFKVESEQQYFEIEKRLSHSQERLVNETRECQSLRLELEKLNNQLK ALTEKNKELEIAQDRNIAIQSQFTRTKEELEAEKRDLIRTNERLSQELEYLTEDVKRLNEKLKESNTTKGELQLKLDELQASDVSVKYRE KRLEQEKELLHSQNTWLNTELKTKTDELLALGREKGNEILELKCNLENKKEEVSRLEEQMNGLKTSNEHLQKHVEDLLTKLKEAKEQQAS MEEKFHNELNAHIKLSNLYKSAADDSEAKSNELTRAVEELHKLLKEAGEANKAIQDHLLEVEQSKDQMEKEMLEKIGRLEKELENANDLL SATKRKGAILSEEELAAMSPTAAAVAKIVKPGMKLTELYNAYVETQDQLLLEKLENKRINKYLDEIVKEVEAKAPILKRQREEYERAQKA VASLSVKLEQAMKEIQRLQEDTDKANKQSSVLERDNRRMEIQVKDLSQQIRVLLMELEEARGNHVIRDEEVSSADISSSSEVISQHLVSY RNIEELQQQNQRLLVALRELGETREREEQETTSSKITELQLKLESALTELEQLRKSRQHQMQLVDSIVRQRDMYRILLSQTTGVAIPLHA SSLDDVSLASTPKRPSTSQTVSTPAPVPVIESTEAIEAKAALKQLQEIFENYKKEKAENEKIQNEQLEKLQEQVTDLRSQNTKISTQLDF ASKRYEMLQDNVEGYRREITSLHERNQKLTATTQKQEQIINTMTQDLRGANEKLAVAEVRAENLKKEKEMLKLSEVRLSQQRESLLAEQR GQNLLLTNLQTIQGILERSETETKQRLSSQIEKLEHEISHLKKKLENEVEQRHTLTRNLDVQLLDTKRQLDTETNLHLNTKELLKNAQKE IATLKQHLSNMEVQVASQSSQRTGKDTNSTSGDPVEKKDETPFGVSVAVGLAVFACLFLSTLLLVLNKCGRRNKFGINRPAVLAPEDGLA MSLHFMTLGGSSLSPTEGKGSGLQGHIIENPQYFSDACVHHIKRRDIVLKWELGEGAFGKVFLAECHNLLPEQDKMLVAVKALKEASESA RQDFQREAELLTMLQHQHIVRFFGVCTEGRPLLMVFEYMRHGDLNRFLRSHGPDAKLLAGGEDVAPGPLGLGQLLAVASQVAAGMVYLAG LHFVHRDLATRNCLVGQGLVVKIGDFGMSRDIYSTDYYRVGGRTMLPIRWMPPESILYRKFTTESDVWSFGVVLWEIFTYGKQPWYQLSN -------------------------------------------------------------- >93453_93453_3_TPR-NTRK1_TPR_chr1_186319354_ENST00000367478_NTRK1_chr1_156844362_ENST00000392302_length(amino acids)=1323AA_BP=925 MAAVLQQVLERTELNKLPKSVQNKLEKFLADQQSEIDGLKGRHEKFKVESEQQYFEIEKRLSHSQERLVNETRECQSLRLELEKLNNQLK ALTEKNKELEIAQDRNIAIQSQFTRTKEELEAEKRDLIRTNERLSQELEYLTEDVKRLNEKLKESNTTKGELQLKLDELQASDVSVKYRE KRLEQEKELLHSQNTWLNTELKTKTDELLALGREKGNEILELKCNLENKKEEVSRLEEQMNGLKTSNEHLQKHVEDLLTKLKEAKEQQAS MEEKFHNELNAHIKLSNLYKSAADDSEAKSNELTRAVEELHKLLKEAGEANKAIQDHLLEVEQSKDQMEKEMLEKIGRLEKELENANDLL SATKRKGAILSEEELAAMSPTAAAVAKIVKPGMKLTELYNAYVETQDQLLLEKLENKRINKYLDEIVKEVEAKAPILKRQREEYERAQKA VASLSVKLEQAMKEIQRLQEDTDKANKQSSVLERDNRRMEIQVKDLSQQIRVLLMELEEARGNHVIRDEEVSSADISSSSEVISQHLVSY RNIEELQQQNQRLLVALRELGETREREEQETTSSKITELQLKLESALTELEQLRKSRQHQMQLVDSIVRQRDMYRILLSQTTGVAIPLHA SSLDDVSLASTPKRPSTSQTVSTPAPVPVIESTEAIEAKAALKQLQEIFENYKKEKAENEKIQNEQLEKLQEQVTDLRSQNTKISTQLDF ASKRYEMLQDNVEGYRREITSLHERNQKLTATTQKQEQIINTMTQDLRGANEKLAVAEVRAENLKKEKEMLKLSEVRLSQQRESLLAEQR GQNLLLTNLQTIQGILERSETETKQRLSSQIEKLEHEISHLKKKLENEVEQRHTLTRNLDVQLLDTKRQLDTETNLHLNTKELLKNAQKE IATLKQHLSNMEVQVASQSSQRTGKDTNSTSGDPVEKKDETPFGVSVAVGLAVFACLFLSTLLLVLNKCGRRNKFGINRPAVLAPEDGLA MSLHFMTLGGSSLSPTEGKGSGLQGHIIENPQYFSDACVHHIKRRDIVLKWELGEGAFGKVFLAECHNLLPEQDKMLVAVKALKEASESA RQDFQREAELLTMLQHQHIVRFFGVCTEGRPLLMVFEYMRHGDLNRFLRSHGPDAKLLAGGEDVAPGPLGLGQLLAVASQVAAGMVYLAG LHFVHRDLATRNCLVGQGLVVKIGDFGMSRDIYSTDYYRVGGRTMLPIRWMPPESILYRKFTTESDVWSFGVVLWEIFTYGKQPWYQLSN -------------------------------------------------------------- >93453_93453_4_TPR-NTRK1_TPR_chr1_186319354_ENST00000367478_NTRK1_chr1_156844362_ENST00000524377_length(amino acids)=1323AA_BP=925 MAAVLQQVLERTELNKLPKSVQNKLEKFLADQQSEIDGLKGRHEKFKVESEQQYFEIEKRLSHSQERLVNETRECQSLRLELEKLNNQLK ALTEKNKELEIAQDRNIAIQSQFTRTKEELEAEKRDLIRTNERLSQELEYLTEDVKRLNEKLKESNTTKGELQLKLDELQASDVSVKYRE KRLEQEKELLHSQNTWLNTELKTKTDELLALGREKGNEILELKCNLENKKEEVSRLEEQMNGLKTSNEHLQKHVEDLLTKLKEAKEQQAS MEEKFHNELNAHIKLSNLYKSAADDSEAKSNELTRAVEELHKLLKEAGEANKAIQDHLLEVEQSKDQMEKEMLEKIGRLEKELENANDLL SATKRKGAILSEEELAAMSPTAAAVAKIVKPGMKLTELYNAYVETQDQLLLEKLENKRINKYLDEIVKEVEAKAPILKRQREEYERAQKA VASLSVKLEQAMKEIQRLQEDTDKANKQSSVLERDNRRMEIQVKDLSQQIRVLLMELEEARGNHVIRDEEVSSADISSSSEVISQHLVSY RNIEELQQQNQRLLVALRELGETREREEQETTSSKITELQLKLESALTELEQLRKSRQHQMQLVDSIVRQRDMYRILLSQTTGVAIPLHA SSLDDVSLASTPKRPSTSQTVSTPAPVPVIESTEAIEAKAALKQLQEIFENYKKEKAENEKIQNEQLEKLQEQVTDLRSQNTKISTQLDF ASKRYEMLQDNVEGYRREITSLHERNQKLTATTQKQEQIINTMTQDLRGANEKLAVAEVRAENLKKEKEMLKLSEVRLSQQRESLLAEQR GQNLLLTNLQTIQGILERSETETKQRLSSQIEKLEHEISHLKKKLENEVEQRHTLTRNLDVQLLDTKRQLDTETNLHLNTKELLKNAQKE IATLKQHLSNMEVQVASQSSQRTGKDTNSTSGDPVEKKDETPFGVSVAVGLAVFACLFLSTLLLVLNKCGRRNKFGINRPAVLAPEDGLA MSLHFMTLGGSSLSPTEGKGSGLQGHIIENPQYFSDACVHHIKRRDIVLKWELGEGAFGKVFLAECHNLLPEQDKMLVAVKALKEASESA RQDFQREAELLTMLQHQHIVRFFGVCTEGRPLLMVFEYMRHGDLNRFLRSHGPDAKLLAGGEDVAPGPLGLGQLLAVASQVAAGMVYLAG LHFVHRDLATRNCLVGQGLVVKIGDFGMSRDIYSTDYYRVGGRTMLPIRWMPPESILYRKFTTESDVWSFGVVLWEIFTYGKQPWYQLSN -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:186332090/chr1:156845411) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| TPR | NTRK1 |
| 2925 | FUNCTION: Receptor tyrosine kinase involved in the development and the maturation of the central and peripheral nervous systems through regulation of proliferation, differentiation and survival of sympathetic and nervous neurons. High affinity receptor for NGF which is its primary ligand (PubMed:1850821, PubMed:1849459, PubMed:1281417, PubMed:8325889, PubMed:15488758, PubMed:22649032, PubMed:17196528, PubMed:27445338). Can also bind and be activated by NTF3/neurotrophin-3. However, NTF3 only supports axonal extension through NTRK1 but has no effect on neuron survival (By similarity). Upon dimeric NGF ligand-binding, undergoes homodimerization, autophosphorylation and activation (PubMed:1281417). Recruits, phosphorylates and/or activates several downstream effectors including SHC1, FRS2, SH2B1, SH2B2 and PLCG1 that regulate distinct overlapping signaling cascades driving cell survival and differentiation. Through SHC1 and FRS2 activates a GRB2-Ras-MAPK cascade that regulates cell differentiation and survival. Through PLCG1 controls NF-Kappa-B activation and the transcription of genes involved in cell survival. Through SHC1 and SH2B1 controls a Ras-PI3 kinase-AKT1 signaling cascade that is also regulating survival. In absence of ligand and activation, may promote cell death, making the survival of neurons dependent on trophic factors. {ECO:0000250|UniProtKB:P35739, ECO:0000250|UniProtKB:Q3UFB7, ECO:0000269|PubMed:11244088, ECO:0000269|PubMed:1281417, ECO:0000269|PubMed:15488758, ECO:0000269|PubMed:17196528, ECO:0000269|PubMed:1849459, ECO:0000269|PubMed:1850821, ECO:0000269|PubMed:22649032, ECO:0000269|PubMed:27445338, ECO:0000269|PubMed:27676246, ECO:0000269|PubMed:8155326, ECO:0000269|PubMed:8325889}.; FUNCTION: [Isoform TrkA-III]: Resistant to NGF, it constitutively activates AKT1 and NF-kappa-B and is unable to activate the Ras-MAPK signaling cascade. Antagonizes the anti-proliferative NGF-NTRK1 signaling that promotes neuronal precursors differentiation. Isoform TrkA-III promotes angiogenesis and has oncogenic activity when overexpressed. {ECO:0000269|PubMed:15488758}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Protein Structures |
 PDB and CIF files of the predicted fusion proteins PDB and CIF files of the predicted fusion proteins * Here we show the 3D structure of the fusion proteins using Mol*. AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. Model confidence is shown from the pLDDT values per residue. pLDDT corresponds to the model’s prediction of its score on the local Distance Difference Test. It is a measure of local accuracy (from AlphfaFold website). To color code individual residues, we transformed individual PDB files into CIF format. |
| Fusion protein PDB link (fusion AA seq ID in FusionPDB) | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | AA seq | Len(AA seq) |
| PDB file (885)CIF file (885) >>>885.cif | TPR | chr1 | 186319354 | - | NTRK1 | chr1 | 156844362 | + | MAAVLQQVLERTELNKLPKSVQNKLEKFLADQQSEIDGLKGRHEKFKVES EQQYFEIEKRLSHSQERLVNETRECQSLRLELEKLNNQLKALTEKNKELE IAQDRNIAIQSQFTRTKEELEAEKRDLIRTNERLSQELEYLTEDVKRLNE KLKESNTTKGELQLKLDELQASDVSVKYREKRLEQEKELLHSQNTWLNTE LKTKTDELLALGREKGNEILELKCNLENKKEEVSRLEEQMNGLKTSNEHL QKHVEDLLTKLKEAKEQQASMEEKFHNELNAHIKLSNLYKSAADDSEAKS NELTRAVEELHKLLKEAGEANKAIQDHLLEVEQSKDQMEKEMLEKIGRLE KELENANDLLSATKRKGAILSEEELAAMSPTAAAVAKIVKPGMKLTELYN AYVETQDQLLLEKLENKRINKYLDEIVKEVEAKAPILKRQREEYERAQKA VASLSVKLEQAMKEIQRLQEDTDKANKQSSVLERDNRRMEIQVKDLSQQI RVLLMELEEARGNHVIRDEEVSSADISSSSEVISQHLVSYRNIEELQQQN QRLLVALRELGETREREEQETTSSKITELQLKLESALTELEQLRKSRQHQ MQLVDSIVRQRDMYRILLSQTTGVAIPLHASSLDDVSLASTPKRPSTSQT VSTPAPVPVIESTEAIEAKAALKQLQEIFENYKKEKAENEKIQNEQLEKL QEQVTDLRSQNTKISTQLDFASKRYEMLQDNVEGYRREITSLHERNQKLT ATTQKQEQIINTMTQDLRGANEKLAVAEVRAENLKKEKEMLKLSEVRLSQ QRESLLAEQRGQNLLLTNLQTIQGILERSETETKQRLSSQIEKLEHEISH LKKKLENEVEQRHTLTRNLDVQLLDTKRQLDTETNLHLNTKELLKNAQKE IATLKQHLSNMEVQVASQSSQRTGKDTNSTSGDPVEKKDETPFGVSVAVG LAVFACLFLSTLLLVLNKCGRRNKFGINRPAVLAPEDGLAMSLHFMTLGG SSLSPTEGKGSGLQGHIIENPQYFSDACVHHIKRRDIVLKWELGEGAFGK VFLAECHNLLPEQDKMLVAVKALKEASESARQDFQREAELLTMLQHQHIV RFFGVCTEGRPLLMVFEYMRHGDLNRFLRSHGPDAKLLAGGEDVAPGPLG LGQLLAVASQVAAGMVYLAGLHFVHRDLATRNCLVGQGLVVKIGDFGMSR DIYSTDYYRVGGRTMLPIRWMPPESILYRKFTTESDVWSFGVVLWEIFTY GKQPWYQLSNTEAIDCITQGRELERPRACPPEVYAIMRGCWQREPQQRHS | 1323 |
| 3D view using mol* of 885 (AA BP:) | ||||||||||
 | ||||||||||
| PDB file (889)CIF file (889) >>>889.cif | TPR | chr1 | 186319354 | - | NTRK1 | chr1 | 156844362 | + | MAAVLQQVLERTELNKLPKSVQNKLEKFLADQQSEIDGLKGRHEKFKVES EQQYFEIEKRLSHSQERLVNETRECQSLRLELEKLNNQLKALTEKNKELE IAQDRNIAIQSQFTRTKEELEAEKRDLIRTNERLSQELEYLTEDVKRLNE KLKESNTTKGELQLKLDELQASDVSVKYREKRLEQEKELLHSQNTWLNTE LKTKTDELLALGREKGNEILELKCNLENKKEEVSRLEEQMNGLKTSNEHL QKHVEDLLTKLKEAKEQQASMEEKFHNELNAHIKLSNLYKSAADDSEAKS NELTRAVEELHKLLKEAGEANKAIQDHLLEVEQSKDQMEKEMLEKIGRLE KELENANDLLSATKRKGAILSEEELAAMSPTAAAVAKIVKPGMKLTELYN AYVETQDQLLLEKLENKRINKYLDEIVKEVEAKAPILKRQREEYERAQKA VASLSVKLEQAMKEIQRLQEDTDKANKQSSVLERDNRRMEIQVKDLSQQI RVLLMELEEARGNHVIRDEEVSSADISSSSEVISQHLVSYRNIEELQQQN QRLLVALRELGETREREEQETTSSKITELQLKLESALTELEQLRKSRQHQ MQLVDSIVRQRDMYRILLSQTTGVAIPLHASSLDDVSLASTPKRPSTSQT VSTPAPVPVIESTEAIEAKAALKQLQEIFENYKKEKAENEKIQNEQLEKL QEQVTDLRSQNTKISTQLDFASKRYEMLQDNVEGYRREITSLHERNQKLT ATTQKQEQIINTMTQDLRGANEKLAVAEVRAENLKKEKEMLKLSEVRLSQ QRESLLAEQRGQNLLLTNLQTIQGILERSETETKQRLSSQIEKLEHEISH LKKKLENEVEQRHTLTRNLDVQLLDTKRQLDTETNLHLNTKELLKNAQKE IATLKQHLSNMEVQVASQSSQRTGKDTNSTSGDPVEKKDETPFGVSVAVG LAVFACLFLSTLLLVLNKCGRRNKFGINRPAVLAPEDGLAMSLHFMTLGG SSLSPTEGKGSGLQGHIIENPQYFSDASPSGVHHIKRRDIVLKWELGEGA FGKVFLAECHNLLPEQDKMLVAVKALKEASESARQDFQREAELLTMLQHQ HIVRFFGVCTEGRPLLMVFEYMRHGDLNRFLRSHGPDAKLLAGGEDVAPG PLGLGQLLAVASQVAAGMVYLAGLHFVHRDLATRNCLVGQGLVVKIGDFG MSRDIYSTDYYRVGGRTMLPIRWMPPESILYRKFTTESDVWSFGVVLWEI FTYGKQPWYQLSNTEAIDCITQGRELERPRACPPEVYAIMRGCWQREPQQ | 1326 |
| 3D view using mol* of 889 (AA BP:) | ||||||||||
 | ||||||||||
Top |
pLDDT score distribution |
 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2 pLDDT score distribution of the predicted wild-type structures of two partner proteins from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
TPR_pLDDT.png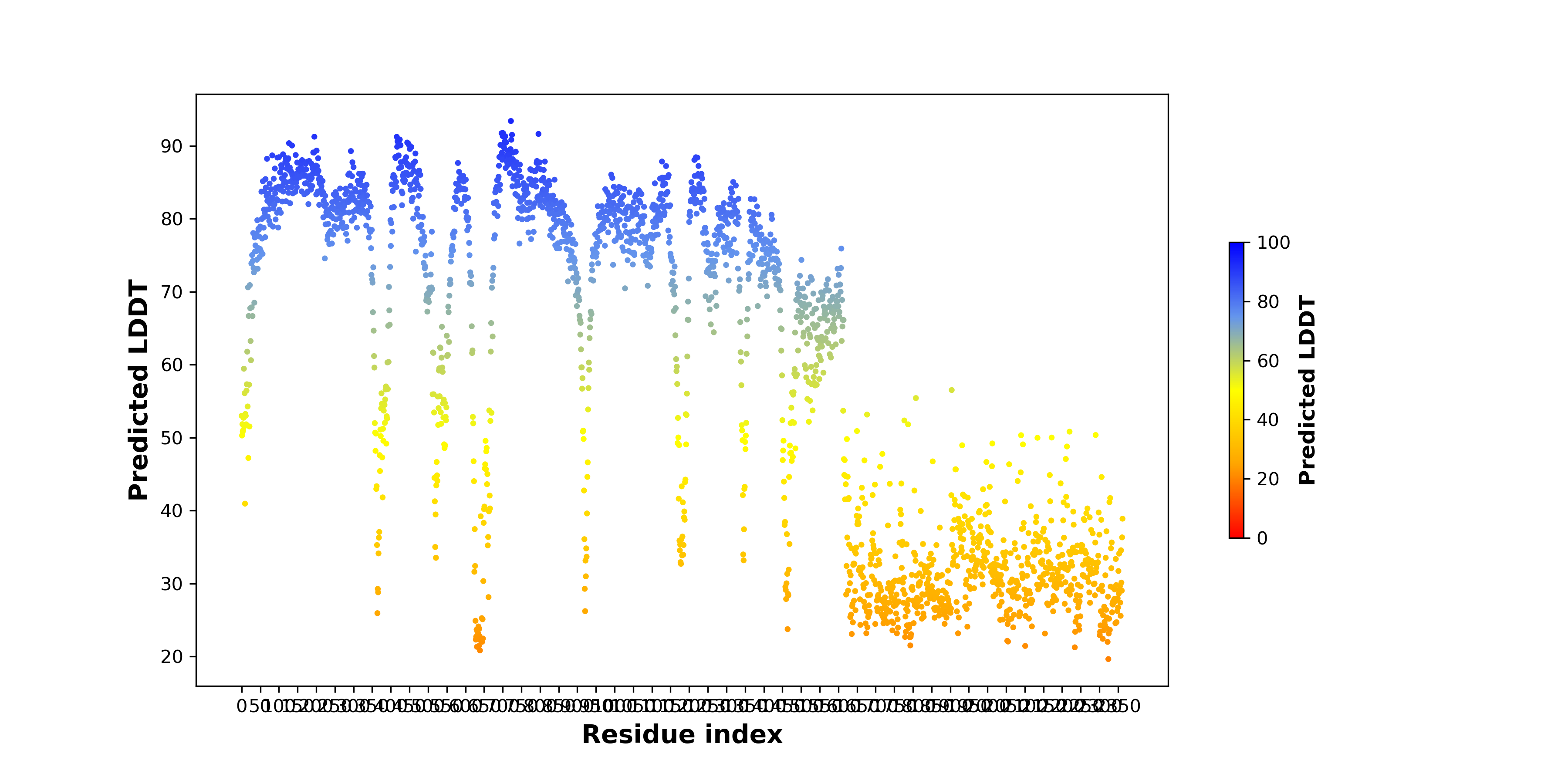 |
NTRK1_pLDDT.png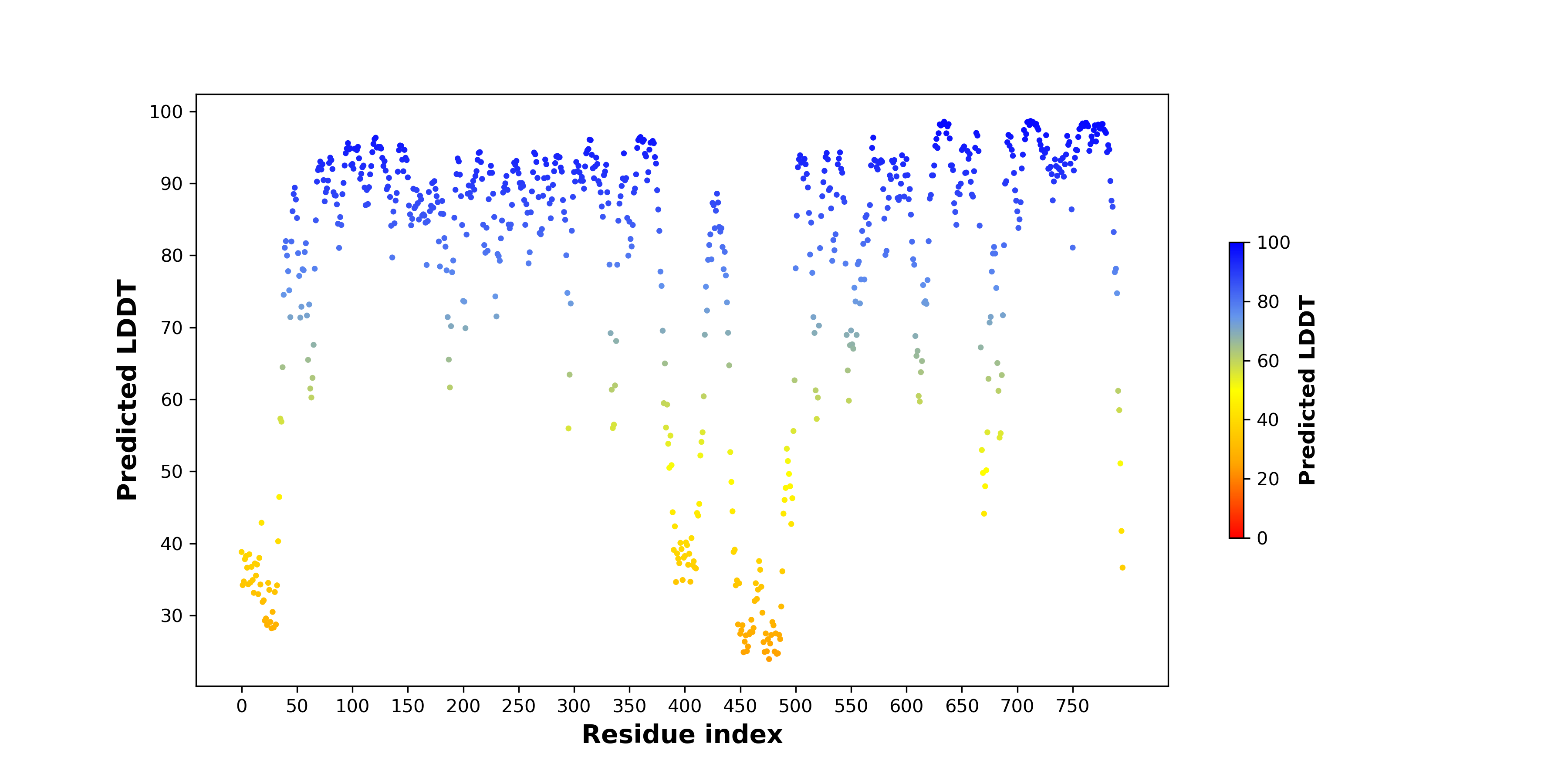 |
 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2 pLDDT score distribution of the predicted fusion protein structures from AlphaFold2* AlphaFold produces a per-residue confidence score (pLDDT) between 0 and 100. |
TPR_NTRK1_889_pLDDT_and_active_sites.png (AA BP:)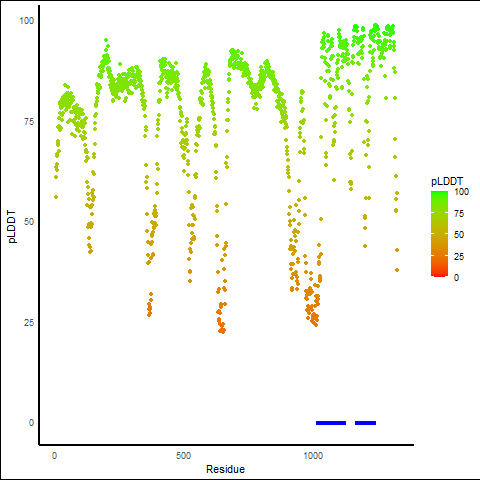 |
TPR_NTRK1_889_violinplot.png (AA BP:)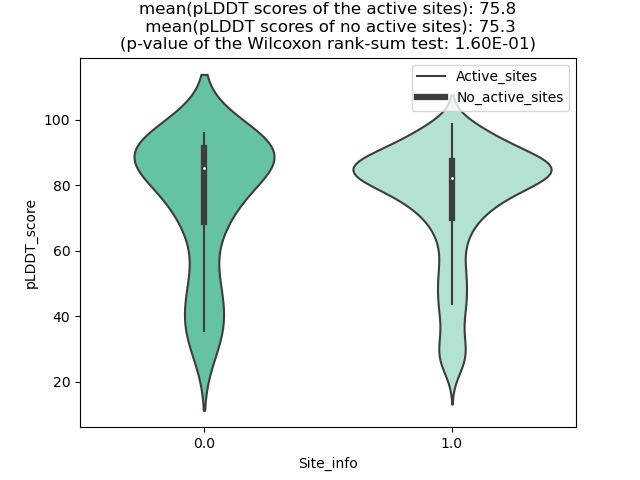 |
Top |
Ramachandran Plot of Fusion Protein Structure |
 Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. Ramachandran plot of the torsional angles - phi (φ)and psi (ψ) - of the residues (amino acids) contained in this fusion protein peptide. |
| Fusion AA seq ID in FusionPDB and their Ramachandran plots |
Top |
Potential Active Site Information |
 The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. The potential binding sites of these fusion proteins were identified using SiteMap, a module of the Schrodinger suite. |
| Fusion AA seq ID in FusionPDB | Site score | Size | D score | Volume | Exposure | Enclosure | Contact | Phobic | Philic | Balance | Don/Acc | Residues |
| 889 | 1.019 | 251 | 1.049 | 948.052 | 0.635 | 0.703 | 0.892 | 0.688 | 0.958 | 0.718 | 1.132 | Chain A: 1018,1019,1020,1023,1024,1025,1027,1028,1 029,1030,1031,1032,1033,1051,1062,1063,1074,1076,1 077,1078,1082,1083,1086,1087,1089,1090,1092,1093,1 094,1095,1096,1097,1098,1102,1103,1104,1105,1106,1 107,1119,1171,1175,1176,1177,1178,1179,1180,1184,1 185,1196,1197,1198,1199,1200,1202,1203,1210,1217,1 218,1219,1220,1234 |
Top |
Potentially Interacting Small Molecules through Virtual Screening |
 The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. The FDA-approved small molecule library molecules were subjected to virtual screening using the Glide. |
| Fusion AA seq ID in FusionPDB | ZINC ID | DrugBank ID | Drug name | Docking score | Glide gscore |
Top |
 Drug information from DrugBank of the top 20 interacting small molecules. Drug information from DrugBank of the top 20 interacting small molecules. |
| ZINC ID | DrugBank ID | Drug name | Drug type | SMILES | Drug group |
Top |
Biochemical Features of Small Molecules |
 ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) ADME (Absorption, Distribution, Metabolism, and Excretion) of drugs using QikProp(v3.9) |
| ZINC ID | mol_MW | dipole | SASA | FOSA | FISA | PISA | WPSA | volume | donorHB | accptHB | IP | Human Oral Absorption | Percent Human Oral Absorption | Rule Of Five | Rule Of Three |
Top |
Drug Toxicity Information |
 Toxicity information of individual drugs using eToxPred Toxicity information of individual drugs using eToxPred |
| ZINC ID | Smile | Surface Accessibility | Toxicity |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
| NTRK1 | ABL1, NGF, MATK, PLCG1, GRB2, CAV1, NTF3, SQSTM1, SH2B1, SHC1, SH2B2, ARHGAP32, FRS2, NTRK1, RAP1A, SHC3, GIPC1, RGS19, PTPN1, TRAF6, NEDD4, CBL, HSP90AA1, HSPA4, CDC37, NEDD4L, MYC, SGSM3, SORBS1, TNK2, SNX17, SNX27, GAREML, SOS2, PIK3CA, GAREM, PIK3C2B, ARHGEF40, PIK3R1, PIK3R2, SOS1, PIK3CB, PML, UBC, AMFR, CBLB, GAB1, EPN1, PIK3C2A, ANKRD13A, DOCK4, TRIO, LGR5, WRNIP1, RABEP1, PDGFRA, KANK2, RABGEF1, VAV2, TBC1D15, PTPN11, GAK, IRS2, PCDHGC3, EPS15, LRRN3, HUWE1, ACVR1B, MIB1, OFD1, PIBF1, ZNF497, TXN, CAT, FABP3, JUP, DSG1, TGM1, CFL1, LCN1, HIST2H4A, DSP, PFN1, LGALS3BP, HIST1H2BL, LTF, S100A9, IGLL5, TGM3, DSC1, LYZ, DCD, LEKR1, ENO2, PPIA, BTN1A1, XRCC5, XRCC6, HLA-A, GSTP1, ABCB7, PA2G4, PRDX1, HDGF, HSPA4L, TBC1D10B, HP1BP3, KIF26B, SYT4, RHOBTB3, NOTCH3, GAB2, ATAD3A, RBM15B, KCNH2, USP15, PDCD11, RBFOX2, RBM14, RNF31, ADNP, ZC3HAV1, MAP7D1, USP4, JAK1, PTPRS, PDGFRB, HIST1H1C, USP11, PPFIBP1, INTS6, VGF, ARHGEF11, SMPD4, SYNPO2, HNRNPM, SLC27A4, DDX41, EGFR, H1FX, ASAP1, HIST1H1B, RRBP1, EPHA2, KIDINS220, CPT1A, UNC5B, IGF1R, DDR2, AUP1, XPR1, RP9, MARK2, REPS1, EMD, SMARCA4, DDX21, DAB2IP, C7, MSTO1, CDC27, ZNF629, ASAP2, NCAPH2, SRBD1, ATAD3B, TCF12, PLEKHA6, INPPL1, PRPF4B, DPM1, SMARCA1, FIP1L1, DLGAP5, PLEKHH3, TRA2A, ACIN1, DDX10, TAX1BP1, SMARCA5, RABEP2, DDX5, MYO1C, SRSF10, DNAJA2, RET, TRA2B, MYEF2, PNPLA6, SLC12A2, TAF6L, RSL1D1, ARAP3, DDX18, ABL2, KHDRBS1, CERK, SRSF7, SPTLC2, PBXIP1, DNAJA1, IQGAP3, DDX31, ZC3H11A, PLRG1, CNP, FMR1, HIP1R, KIAA1429, CDC5L, ARAP1, ZNF316, SMARCC2, EML5, VANGL2, SLC25A1, UNC5C, SLC16A1, RFC1, ZKSCAN1, CHD1, PTK7, SMARCD2, AP3B1, YTHDC2, AKAP8L, RPL15, CTNND1, PHLDB1, ZNF574, DDX27, TOM1, RASA1, MORC2, MTCH2, IMMT, SMARCC1, ROR2, CAPN2, DNAJC11, SYT11, AP2A1, LBR, MEPCE, NCAPD3, CEP97, QKI, LRRC41, LPCAT2, EPS15L1, LAS1L, TMEM131, CWF19L2, NCAPG2, MAP4K5, CCP110, ARHGEF2, ITGAV, ZC3H18, CWC22, VIM, FAM171A1, TMEM132A, SRSF9, SOGA1, NKRF, RBL1, FMNL3, TOP1, PIGT, RBM15, GBAS, EDC3, WDR5, NOP2, GNL2, SLC25A11, RRP1B, FAM115A, INTS5, SLC25A5, AAK1, HERC5, CCNT1, FERMT2, CDC42BPA, MRPL28, UBAP2, WWP2, SEC24B, HNRNPU, GPD2, FXR2, DHX15, SLC25A22, SSR1, PLXNB2, PROX1, TEX10, USP19, FTSJ3, TMEM59, GTF3C2, PGAM5, CPSF1, KLHL22, TMX3, SIN3A, NOP58, PTCD3, RPRD2, DHX9, PICALM, FARP1, SLC25A6, ERLIN2, SMARCD1, RHOT1, ALDH1B1, SGPL1, USP9X, HNRNPF, KIAA1211, MAPK7, DDX17, GSE1, DVL3, MRPL44, DNAJB6, GTPBP4, HNRNPA0, PLXNA4, SFXN3, NAT10, DDX28, SNW1, MYO6, MRPL24, PRRT4, FAM98A, PURA, DHX29, C1QBP, TRIP4, TBK1, RBM28, ZNF609, POLRMT, PCDHB3, XRN1, HSD17B11, DKC1, DHX30, CDK13, SENP3, LPHN2, GPATCH4, NDUFA9, RASA4, MCCC1, DDX24, RPL14, RPS8, ATG2B, MYO1B, PTPN12, CEBPZ, ILVBL, RALY, KIF14, ZC3H7A, MOV10, MAP4K4, DHX57, RBMX, FXR1, MRPS35, SART1, MARK3, DECR1, CCDC88A, ANKLE2, PRPF31, LARP7, RPL10, PACS1, RPL13, FAM120A, SLC25A13, ADAR, CDK1, HNRNPR, CSNK1E, DLG1, LEMD3, NOL9, IFT122, NCLN, STK11IP, GATAD2B, CHERP, LARP4, MAP3K1, HNRNPUL2, PPIL2, ESYT2, MYH10, RPL8, UQCRC2, NDUFA10, HAX1, HELLS, MED14, RBM42, CHCHD3, RPL7, ABCD3, TUBGCP6, MRPS9, SNRNP200, IGF2BP3, LRCH3, MRPL3, RSBN1L, SAFB, TUBG1, ALDH16A1, ARHGEF12, EPB41L5, AP2A2, SRSF6, HLTF, PTPLAD1, TMPO, AGO2, POP1, RPS9, HDAC6, ILF3, VWA8, ABCB1, SCRIB, GTF3C5, YBX1, RBM4, PTPN9, PRPF8, BRD3, TRIM24, CLASP2, MADD, TCF4, MAP4K3, TSC2, DDX23, RPL7A, RRP12, PRKD2, TRMT1L, EDC4, CSTF3, RPS2, NDUFS2, RPL13A, PTPRR, ATP7A, EXD2, IK, HIP1, MATR3, IGSF3, PTPRF, SPATA5L1, EFTUD2, RPS3A, TUBGCP5, TTN, GIT1, PATL1, NUP160, PDE4B, SUN2, PTPN23, RPS6, DCP1A, CDC73, MEX3A, EIF2AK2, RPL3, PPP1R10, SRP72, ELAVL3, NOM1, RPL18, CEP170, THRAP3, MED17, MRPS7, DDX20, PDPR, SH3PXD2A, DBN1, DAP3, OAS3, STT3B, COPE, MED12, AP3D1, CAPRIN1, PHB2, U2SURP, BSG, TECR, FARSA, HSD17B12, RPL4, HNRNPUL1, PPFIA1, MRPS22, TBL3, GNL3, MTA2, CPSF2, ATP2B1, AP3B2, PPIP5K2, DDX56, ASCC3, FMNL2, SMCHD1, MYBBP1A, SRPK2, RHOT2, SRP68, PUM1, SLC25A3, SRPK1, RPL19, BEND3, SEC61A1, LSG1, NOC3L, TNKS1BP1, GTF3C3, TUBGCP3, PRPF40A, TRMT10C, URGCP, XRN2, MSH3, XRCC1, GEMIN4, DIS3L2, BCLAF1, SBF2, DHTKD1, ATXN2, ARID4B, TOR1AIP1, RPL29, NAA16, PRPF6, KIFAP3, TMEM33, WASF2, NACAD, LIG3, POLE, CKAP4, TCEB3, MTA1, GNA13, DHX38, PHF6, ELAVL4, DDX3X, CTR9, PRPF19, CAMSAP2, PRMT3, SH3PXD2B, SCAF8, MAGED1, ILF2, TONSL, PPP1R12A, SRSF5, KIAA0020, PTBP1, CAD, ACSL3, CHD1L, STAU1, MRPL38, DVL2, GNAI3, RPL10A, RPL6, MRPS5, CDK11B, MRPS34, PES1, ATP5C1, STXBP3, HNRNPLL, CHTF18, VPS39, SART3, COPA, PRPF3, UBE2O, ABCF2, HNRNPH3, EARS2, DHX40, ARHGEF7, EMC1, CASK, PTK2, SORT1, OGT, ATP12A, LMNB1, SLC4A7, PFKP, VDAC2, RBM25, ATL3, DHX36, TFIP11, EXOSC10, GTF3C1, SRSF1, PPP6C, TRAPPC9, RAD21, DNAJC13, ACOT1, ZC3H4, SRRM1, NDUFS3, DYRK1A, INTS1, SF3B2, EHHADH, MYO9B, ASCC2, WDR11, RPN1, EHD4, WDR91, SMARCAL1, NBAS, PI4KA, PAF1, EIF2B4, NUP107, EMC2, SYNE1, KLHL13, TBL1XR1, DNAJC10, SEC16A, METTL13, C7orf50, CDC42BPB, CLCN5, PGM3, DOCK7, ALYREF, POLR3A, TOP3A, WDR48, WDR81, DOCK1, CSNK2A2, CYC1, TLE3, THOC2, EIF2S3, MYO5A, SNX9, TARDBP, NAP1L4, CRKL, YTHDF2, PYGB, PABPC1, LTV1, PSMD4, KTN1, UBTF, WDR6, FUBP3, UBXN7, PHB, PDXP, CNOT3, TBL2, LLGL1, PLOD1, AHSA1, AAAS, ERGIC1, LARP4B, CYFIP1, DNM1, TARS2, ACSL1, ATXN1L, WDR59, SUCLA2, MLH1, CSNK2B, SYNCRIP, WDR36, FAM91A1, MTDH, UBE3C, CAPN1, BRIX1, DNAJB11, EDEM3, PLOD3, CLASP1, DPYSL3, MED16, NONO, EPM2AIP1, NT5DC2, ADRBK2, RPTOR, TELO2, MRPL37, PRKACB, ELMO1, TRAPPC8, ANAPC1, NIPSNAP1, EWSR1, PIK3R4, POLD1, IQGAP1, PRRC2A, COG1, ANKRD44, TFRC, DIEXF, FHOD1, SERPINH1, SPATA5, MED1, ABCF3, ZSWIM8, LMNA, HAT1, PABPC4, SMC4, HADH, RNGTT, TUBGCP2, ZNF598, SKIV2L2, GNB2L1, LARP1, DIP2B, BCR, IDH3B, LSM14A, FBL, NDUFS1, APPL2, USP39, SMC2, HCFC1, LANCL1, POLR2B, UPF2, CCBL2, MRPL15, MAGED2, AGK, GNPAT, AIMP1, GTF3C4, KDM1A, SCAMP3, RDH11, SRGAP2, CRNKL1, RFC5, PFKL, SPTBN1, PHOX2A, VDAC3, HNRNPA1, WDR26, DNM2, HAUS5, LCMT2, MCM7, CLINT1, UGDH, ACO1, TRIM46, PHKB, USP10, PCF11, YTHDF3, AARS2, FIGNL1, SFPQ, ATP5A1, NUP133, CPSF3, ANKRD28, PNN, C9orf64, ABCF1, ATL1, NAP1L1, HSPA9, MCCC2, FKBP5, CNOT1, TAF6, NEMF, UHRF1, SDHA, PELP1, HADHB, OAT, NEK9, AP3M1, PRKCA, LPCAT1, MRPL39, AFG3L2, WASF1, HNRNPL, RALGAPB, HK1, GBA, PLD3, CPSF7, HADHA, PDE12, DCX, LIMA1, HNRNPA2B1, POLR2A, MYH9, ZNF207, DDX55, ELP3, EIF2B5, MICAL1, TTI1, NES, HSDL1, GPSM1, PPM1F, C4orf27, SRRM2, OSBPL9, STAT3, IRF2BPL, KLC2, P3H1, STAT5B, UQCRC1, PCK2, CCAR2, TBC1D5, SACM1L, QARS, XPO4, CHD4, KIF2A, TACC3, NCL, BTAF1, RARS2, NCKAP1, EIF3D, RPS3, YME1L1, EIF4A3, HAUS6, HNRNPDL, MON2, G3BP2, WDHD1, CSNK1A1, SDAD1, RPS25, LSM14B, AP1M1, CCDC47, HNRNPK, DNMT1, NUP85, CUL5, TANC1, STAT1, COG5, STOML2, CDC45, OSBPL11, CIC, ABCD2, ARFGEF1, TROVE2, GBF1, SRPRB, UBE3A, GTPBP1, SCCPDH, RPS4X, PRPF4, ACTL6A, CCAR1, KIAA1033, TRAPPC12, HNRNPA3, FASTKD5, LYN, VPS18, PGRMC2, NUP214, AIFM1, TSG101, PPP6R3, OCRL, GANAB, IARS, SARS2, ATP2A2, EXOC1, NCAPH, TTC27, GNE, SRC, MIOS, ZCCHC8, GOLGA2, CARM1, ABCC1, PRMT5, LEO1, LMNB2, USP8, DARS, GYS1, APEH, ZYG11B, AKAP1, ELAVL1, MCM5, LRWD1, PSMC1, MOGS, TRIM28, EXOC6, LUC7L3, ANKFY1, NSUN2, HDAC2, G3BP1, QRICH1, INTS3, UBR1, XAB2, DYNC1LI1, TUFM, GAA, EXOC8, DHX8, LIMCH1, CLTC, SRPR, RNH1, ABI2, AQR, PREP, PPM1E, COG2, SEPT9, IDH2, MFN2, EXOC4, VPS33B, INTS4, GNB1, MSH6, HNRNPC, ERLIN1, EPRS, LRRC49, RPAP1, MCM3, PSMC4, PSMD3, GNL1, TRIP13, CUX1, PFAS, MAP1S, LRCH1, MRPL19, ABCD1, EIF3F, PDS5B, SMC3, KIF4A, FANCD2, IRF2BP1, SMC1A, RNF20, CLPX, NOC2L, DLAT, CTBP1, APPL1, NAA35, VPS45, PUM2, RELA, NUP210, RANBP9, DNAJC9, WRN, PRKAA1, AIMP2, VPS8, MAD1L1, MRE11A, ATP13A1, ATG7, CEP131, TRIM33, SLC25A10, HERC4, LRRFIP1, POLR1C, IPO9, GEMIN5, METAP2, SMG8, WASL, PPP1CB, SCFD1, RANBP3, RB1, CLTA, RNF40, SNRNP70, NCSTN, MGEA5, NARS, EIF4E, INPP4A, TFB2M, SEL1L, ARGLU1, PPP4R1, ANKRD17, TTK, RAD50, KIF2C, GOLGA3, TRAPPC11, GIGYF2, MSI2, CYFIP2, CDK5, HBS1L, IFT81, UFL1, PFKM, SMARCAD1, WDR35, PRIM2, NAMPT, PSMD2, MSH2, SOGA3, HEATR6, SFXN1, ARCN1, OGDH, FDPS, PSMC5, INSR, IRF2BP2, DDOST, ATXN2L, CSNK2A1, EIF2S2, MAP4, LTN1, COPB1, BRAT1, SMC6, SNRPN, RIC8A, SKIV2L, EIF4A1, KIF15, CLPTM1L, KPNA1, VPS16, VPS26B, RNPEP, RBM39, PMS1, FAM129B, RFC3, NUP155, RBM26, KIF21A, GAPVD1, C2CD5, WAPAL, RPS13, ARHGEF1, ACTR1A, KLC4, ISLR2, DCK, LRRC8A, STT3A, BOP1, RPN2, AFTPH, ORC3, FOXK1, CD2BP2, PDHA1, PPP6R1, PANK4, KIAA0196, WDR77, CHUK, DDX1, COLGALT1, VAPA, PPME1, CAMSAP1, RABL6, RTCB, SRRT, PIK3C3, NUMA1, STXBP1, OTUB1, UBAP2L, EXOC5, LRRC59, EXOC3, RQCD1, FLII, COPG2, USP5, XPOT, TIMM44, ATP1A1, SBF1, ARPC1B, PLK1, OSBPL8, BAG6, PPP6R2, FAM98B, VPS52, FADS2, EIF4G2, EIF4G3, HSPA8, POLR1B, PSMC6, SFSWAP, VPS33A, MCM10, AVL9, RINT1, PPIB, FBXL18, MFAP1, KIF13B, ATP2C1, ERCC2, CTTNBP2NL, WDR82, VARS, KPNA6, MIA3, HEATR1, SEC23B, AASDHPPT, POLR3B, ALDH18A1, PIKFYVE, MPP6, MRI1, CAPN7, RAB3GAP2, NCAPD2, UGGT1, IKBKAP, KDM3B, CLNS1A, FEN1, ECH1, AP2B1, HNRNPH1, RARS, AGPS, FANCI, PYCR2, EHD1, BUB3, KIF5B, SYMPK, PC, ACAA2, NDC80, PRRC2C, MTHFD1L, FASN, RANBP6, VPS11, NUP93, STRN, SCYL1, DDX19A, GTF2I, SEC23A, CTNNB1, STAM, VPRBP, VCL, PDHB, SEC24D, ERCC4, GRN, ACADM, HSPA6, ERCC6L, RPL5, ARHGAP17, DDX6, MAPK8IP3, TJP1, RIOK1, ERP29, PARG, NUP205, PALLD, COG4, TPP2, EPB41, VAC14, FUS, ZW10, KIF11, GET4, MCM6, EIF2S1, CSDE1, DHX16, GFM1, RABGAP1, TTC28, XPO1, EXOC2, DCTN1, ACBD3, RFC4, PARP1, COPG1, GPRIN1, EIF3C, CUL1, C14orf166, RTL1, STRIP1, MRPS2, PPP2R5D, EIF4G1, EXOSC4, MCM4, SEC63, WDR1, CUL4B, CRMP1, NSF, RNMT, BLVRA, UBE4A, PCBP1, MLLT4, RNF214, ARVCF, CKAP5, NUP188, SMCR8, CTNNA1, SYT1, PCM1, KNTC1, GFPT1, THYN1, SARM1, LPIN1, TIA1, NUP54, PKN1, SUMO4, DICER1, KIF3A, IPO8, CLIC4, PSMD14, GLG1, ABCE1, SMEK1, NCAPG, PPP5C, RTF1, IARS2, PDS5A, PHGDH, RPS6KA3, LONP1, ROCK1, CDK6, LARS, KPNA3, HNRNPAB, MCM2, PPP2R2A, HSP90AB2P, VCPIP1, PCBP2, RPA1, SEC24A, HECTD3, RCC2, DIS3, SMS, KHSRP, CLGN, GNAS, FOCAD, SEC23IP, ACOT7, HK2, CCDC6, MFN1, WDR7, CNN3, KIAA1524, RAB3GAP1, OXCT1, SBNO1, PRPSAP2, ACTR3, CTPS2, TMEM43, FAM21A, LRRC47, VAPB, SCYL2, STRN4, NAE1, SREK1, TFG, SF3A1, CDK2, GPKOW, GPHN, PDCD4, U2AF2, UNC45A, VPS35, NAA15, ASNS, TOP2B, ACO2, AGAP3, ROCK2, NUDT21, NPEPPS, ADD1, AP1G1, CPSF6, DNAJC2, RPS5, CTPS1, IDH1, SEC24C, KIF3B, ACTR2, CPD, EEF1D, SAMHD1, ETFB, DNM1L, LRPPRC, MCMBP, RBM12, OPA1, FARSB, UPF1, PCMT1, USO1, COG3, CLPTM1, TLK2, PRUNE, ASPSCR1, MARS, PYCR1, TCERG1, MED23, PITPNB, ARPC2, UBE4B, DBR1, ETFA, BICD1, KLC1, WDR70, PXDN, DPYSL4, USP7, CLUH, TBRG4, OGFR, SV2A, PDCD6IP, PDLIM5, PSMC2, TRAP1, DRG1, SMEK2, TCP1, TNPO2, CALD1, ARIH2, POR, FKBP3, DCTN2, NPM1, USP48, IPO13, NUP88, NNT, TAOK1, PPWD1, PSPC1, PABPN1, POLA1, MTR, BCCIP, RUVBL2, C16orf62, SORD, PPP2CA, STRN3, RUVBL1, PPP1CA, CACTIN, RPAP3, VPS51, TUBB3, PRKDC, HSPA5, CCDC132, MRPS27, RBBP4, CAPZA1, SLC4A1AP, THUMPD3, FAF1, CDK4, GSTO1, NCBP1, CAPZB, SNRPA, TXLNG, NELFE, MUT, DYNC1LI2, CTTN, DYNC1I2, TIMELESS, CBS, TTLL12, ESYT1, LAMB1, KARS, PPIL4, LRRC40, TXLNA, RAD23B, TBCE, ACAD9, THADA, GCN1L1, SLK, PNKP, ACLY, ATP6V0A1, VTA1, TXNDC5, OXSR1, ACACA, TNPO3, CCDC93, TIPRL, RAN, HOOK3, PSME4, YWHAQ, PPM1G, PSMC3, SCARB2, EXOC7, DFNA5, TRMT6, CPNE3, SLC3A2, G6PD, EIF2B1, SF3B1, EXOC6B, HGS, DYNC1H1, DNAJC7, CANX, TSR1, TTC37, DARS2, ZPR1, SERBP1, HSP90AB1, TBCB, UCHL5, IMPDH2, PUF60, RANGAP1, PDXDC1, YWHAH, LARS2, ZC3H15, GMPS, YWHAG, SUPV3L1, SPAG9, IMPDH1, HSD17B10, GTF2F1, EXOSC9, CUL3, ARHGAP35, TUBB4B, EIF5B, KIF5C, PDIA6, LDHA, ATL2, SNX1, SF3B3, SRM, MAP2, DIAPH1, DBH, NHLRC2, ELP2, GRPEL1, TWF1, WBP11, ERP44, SERPINB6, VCP, HSPH1, CMTR1, KIF1A, USP47, EML4, PRMT1, ATP6V1B2, GPS1, CAND1, CLPB, PCNA, RPLP0, CORO7, TUBB, ANXA2, API5, SUPT16H, RUFY1, KIAA1279, PDIA3, ACTA1, SF3A3, DCTN4, CUL2, WARS, PGM2, NELFB, XPO5, UGP2, HSP90B1, GSR, SHMT2, IPO11, EIF3E, RDX, DNAJB1, KIAA1598, AARS, EFTUD1, HEATR5B, ENAH, TNPO1, LIG1, NRD1, DLD, STRA6, PAFAH1B1, NT5C2, SSRP1, NTRK3, HDGFRP2, EIF3G, CSE1L, ACAT1, TBCD, NAA10, MTHFD1, RALA, EIF3A, PRPS1, ISOC1, IPO7, HNRNPD, GAPDH, U2AF1, SUPT5H, PSMD8, RPS6KA1, NUF2, GDAP1, VPS26A, TUBB2B, CDH2, TMOD1, CBR1, HDLBP, CS, HSPD1, LMAN1, EIF3B, DDB1, PSMD1, DPYSL2, STRAP, RTN4, FLNA, AHCY, HSD17B4, GOLIM4, SPTAN1, IPO4, PKM, EIF3I, SF1, BZW2, XPO7, ACADVL, STAG2, DPP9, RECQL, GDI1, GARS, MMS19, EIF2A, IPO5, TCOF1, RAB5C, SAP30BP, VDAC1, TUBA1A, SEPT2, DBNL, PLAA, TOMM70A, NOMO1, ISYNA1, CTNNA2, EEF1B2, PAK4, NELFCD, GSPT1, KPNB1, MAP1A, COPS4, YWHAZ, KPNA2, EIF4B, PLS3, COPS3, HSPBP1, AP1B1, NASP, DDX39A, LUC7L2, APMAP, SEC31A, SRSF2, TXNRD1, ATP1A3, RRM2, COG7, FSD1, PRDX4, DCPS, NCAM1, NUDCD1, NAA25, RPL9, NPLOC4, YWHAB, ATP1B1, LSS, DNAAF5, METTL16, COPS6, XPNPEP1, ELAC2, PUS7, CCT3, SFN, AGL, PAPOLA, SRSF11, ALDH2, PSME2, CKMT1B, LDHB, PSMA7, RAB11A, PPP2R1A, GNAO1, LUC7L, COPS5, IMPA1, RRM1, PPID, DPP3, KIAA0368, SLC1A5, YARS, NUP50, TPM4, ITGB1, SARS, SEZ6L2, GTF2F2, GART, ALDOC, PSMD11, EIF6, DFFA, RAP1GDS1, NELFA, CARS, CCT4, P4HB, FSCN1, PSMD13, PSMA5, CCT5, PAPSS1, OSBP, SAE1, ATP6V1A, LMAN2, YWHAE, SRP54, IDE, SNX2, RPSA, BZW1, SND1, UBQLN1, MAPK1, CALR, MARCKSL1, SUGT1, TBC1D4, GLRX3, LETM1, HMGB1, EIF3H, SLC9A3R1, DDX42, LAP3, PRKCSH, EEF2, PSMD6, BCAP31, ANXA11, ACTN1, CCT6A, PSMA4, ANXA5, CCT7, EIF3L, FUBP1, SSB, PGM1, MAPRE1, UBA2, AMPD2, PPAT, CLIC1, CCT2, ECHS1, LTA4H, USP14, CCT8, ANK2, PDIA4, TPR, NRCAM, MAT2A, HTATSF1, THOP1, RANBP1, RSRC2, NMT1, AK2, EEF1G, TPM3, PSMA3, EIF4H, GLUD1, CAP1, TKT, ATP5B, TXNL1, STIP1, HMGB2, GGH, HSPB1, CALU, RPL17, PSMA1, RSRC1, ST13, PRDX6, EIF3J, PNPT1, APEX1, TARS, MARCKS, PSMA6, EEF1A1, COPB2, NUDT5, HSPA1A, ERO1L, ME2, RPH3A, ESD, EIF5, ADSL, RUFY3, DPYSL5, ANXA6, NUDC, NANS, GOT2, GLDC, PSAT1, SET, PAK2, UBA6, UCHL1, L1CAM, PGLS, MAP1B, CCDC124, EIF3M, VAT1, PAFAH1B3, ALDOA, IRGQ, UBQLN2, NACA, OLA1, ACTN4, ARHGDIA, PSMD7, CACYBP, PSMA2, CPNE1, MDH2, PGAM1, DDX46, ACAT2, ALCAM, HARS, CKB, FKBP4, ASNA1, GPI, INA, HYOU1, CHGA, MDH1, EZR, ANP32A, UBA1, ARG1, ANP32B, PAICS, ATIC, TPI1, PPA1, TALDO1, PGK1, DDX39B, TLN1, AKR1B1, ENO1, NGFR, TPTE, GGA1, GGA2, GGA3, USP36, NDFIP1, SOCS2, CDK12, EPHB4, NDUFA4L2, AKT1, SLC5A5, DOT1L, AKT2, SHC4, TRAF4, PTPN6, ERBB2, EGR1, HIPK2, POU4F2, |
| TPR | NUP98, NUP107, NUP205, NUP93, NUP153, MAPK1, GTF2E2, MAP3K4, APC, KAT8, NCOA3, Mad2l1, Tpr, Kifc5b, RAD21, SIRT7, NXF1, U2AF2, RAN, SNRPD1, SRSF5, HRAS, RPL37A, IK, SFPQ, EEF1D, THOC6, RPL18A, EEF1B2, SART1, SF3B6, TAF15, SNRPA1, DDB1, NHP2L1, RRP7A, HNRNPA2B1, RRS1, DDX1, CIRBP, DDX3X, BUD31, SAP18, HNRNPU, EFTUD2, RPL6, SART3, S100A9, RBM4, PRKDC, TOP1, SEPT2, FN1, ITGA4, CBX7, KIAA0101, NPM1, LMNA, HDAC9, MAD1L1, LGR4, STAU1, CUL7, OBSL1, CCDC8, EED, SUMO2, ATL3, GLRX, RRP12, SRSF11, STIP1, MRE11A, NTRK1, gag, MDM2, Katna1, Nup98, FOXK2, MCM2, NF2, CDC5L, POU5F1, ZC3HC1, STYX, PDHA1, SDHA, SOD1, UBE2A, BRCA1, TXNIP, CFTR, PTPN4, DUSP19, HDAC4, TNIP2, CHD4, PRKCZ, BPLF1, DCPS, MYC, CDK9, TGOLN2, CANX, CDK7, RAB5A, HIST1H3A, KIAA1429, ACTC1, CHEK2, GBF1, XRCC6, DYRK1A, BIRC3, LRRK2, PLEKHA4, WHSC1, FANCD2, MAP2K1, EMC1, PCCB, NPY2R, ABCC12, NDUFB7, MAP9, NFXL1, ZNF721, PHIP, ADCY10, MNT, ERCC6, CIT, PRC1, DOCK8, SREBF2, SEC62, NDN, Rnf183, NUPR1, RFWD2, BRD4, Apc2, ORF6, CUL4A, DDX58, CD274, WDR5, NAA40, TEN1, MAD2L1, SLFN11, |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| TPR | 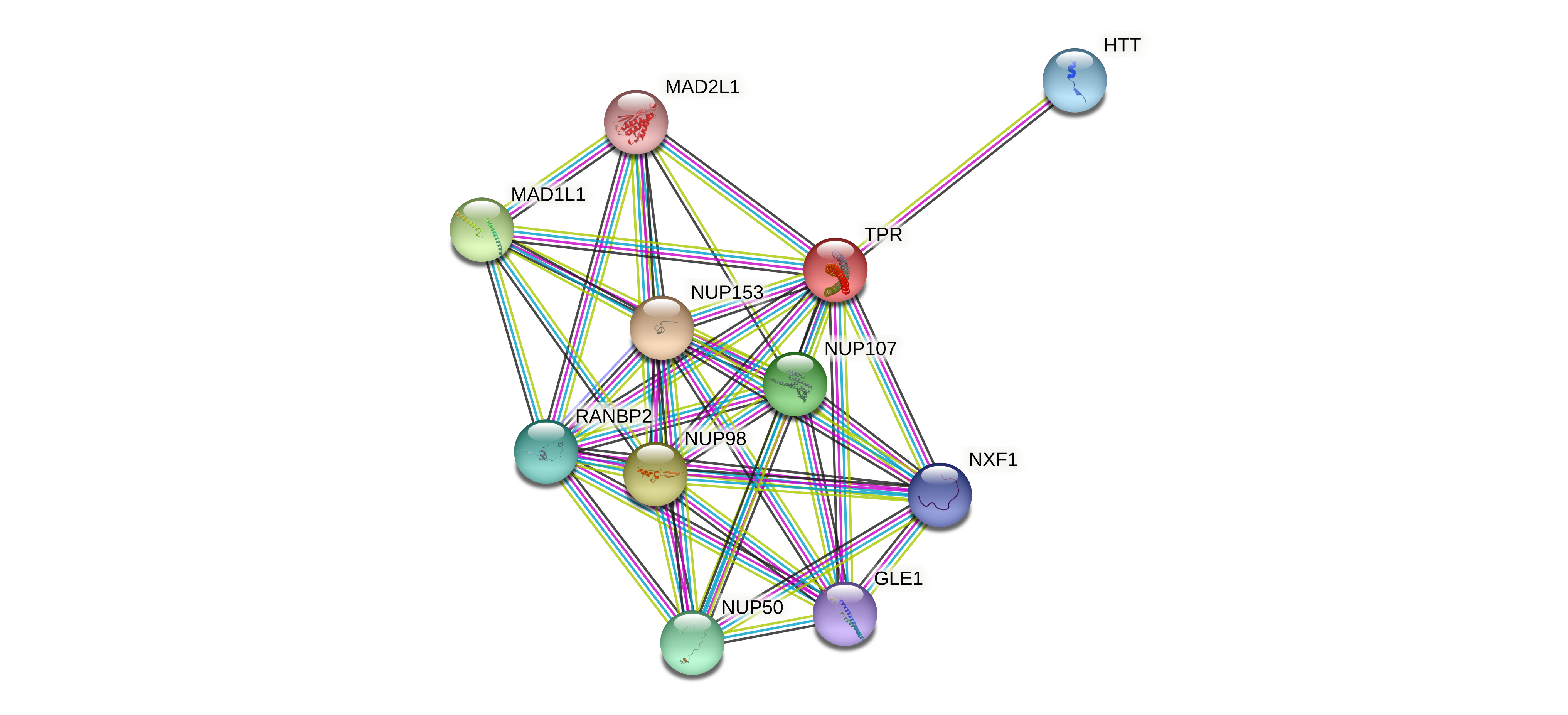 |
| NTRK1 |  |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to TPR-NTRK1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to TPR-NTRK1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
| TPR | NTRK1 | Sarcoma | MyCancerGenome | |
| TPR | NTRK1 | Nos | MyCancerGenome | |
| TPR | NTRK1 | Thyroid Gland Papillary Carcinoma | MyCancerGenome | |
| TPR | NTRK1 | Breast Invasive Ductal Carcinoma | MyCancerGenome | |
| TPR | NTRK1 | Colon Adenocarcinoma | MyCancerGenome | |
| TPR | NTRK1 | Gastric Adenocarcinoma | MyCancerGenome |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
| Hgene | TPR | C0238463 | Papillary thyroid carcinoma | 2 | ORPHANET |
| Hgene | TPR | C0040136 | Thyroid Neoplasm | 1 | CTD_human |
| Hgene | TPR | C0151468 | Thyroid Gland Follicular Adenoma | 1 | CTD_human |
| Hgene | TPR | C0549473 | Thyroid carcinoma | 1 | CTD_human |
| Tgene | NTRK1 | C0020074 | HSAN Type IV | 17 | CTD_human;GENOMICS_ENGLAND;UNIPROT |
| Tgene | NTRK1 | C0238463 | Papillary thyroid carcinoma | 3 | ORPHANET |
| Tgene | NTRK1 | C0002768 | Congenital Pain Insensitivity | 1 | ORPHANET |
| Tgene | NTRK1 | C0005586 | Bipolar Disorder | 1 | CTD_human |
| Tgene | NTRK1 | C0005587 | Depression, Bipolar | 1 | CTD_human |
| Tgene | NTRK1 | C0017638 | Glioma | 1 | CTD_human |
| Tgene | NTRK1 | C0020075 | Hereditary Sensory Autonomic Neuropathy, Type 5 | 1 | CTD_human;ORPHANET |
| Tgene | NTRK1 | C0024713 | Manic Disorder | 1 | CTD_human |
| Tgene | NTRK1 | C0027796 | Neuralgia | 1 | CTD_human |
| Tgene | NTRK1 | C0027819 | Neuroblastoma | 1 | CTD_human |
| Tgene | NTRK1 | C0033958 | Psychosis, Brief Reactive | 1 | CTD_human |
| Tgene | NTRK1 | C0033975 | Psychotic Disorders | 1 | CTD_human |
| Tgene | NTRK1 | C0036337 | Schizoaffective Disorder | 1 | CTD_human |
| Tgene | NTRK1 | C0036341 | Schizophrenia | 1 | CTD_human |
| Tgene | NTRK1 | C0036358 | Schizophreniform Disorders | 1 | CTD_human |
| Tgene | NTRK1 | C0038870 | Neuralgia, Supraorbital | 1 | CTD_human |
| Tgene | NTRK1 | C0042656 | Neuralgia, Vidian | 1 | CTD_human |
| Tgene | NTRK1 | C0234247 | Neuralgia, Atypical | 1 | CTD_human |
| Tgene | NTRK1 | C0234249 | Neuralgia, Stump | 1 | CTD_human |
| Tgene | NTRK1 | C0259783 | mixed gliomas | 1 | CTD_human |
| Tgene | NTRK1 | C0273115 | Lung Injury | 1 | CTD_human |
| Tgene | NTRK1 | C0338831 | Manic | 1 | CTD_human |
| Tgene | NTRK1 | C0423711 | Neuralgia, Perineal | 1 | CTD_human |
| Tgene | NTRK1 | C0423712 | Neuralgia, Iliohypogastric Nerve | 1 | CTD_human |
| Tgene | NTRK1 | C0555198 | Malignant Glioma | 1 | CTD_human |
| Tgene | NTRK1 | C0598589 | Inherited neuropathies | 1 | GENOMICS_ENGLAND |
| Tgene | NTRK1 | C0751371 | Neuralgia, Ilioinguinal | 1 | CTD_human |
| Tgene | NTRK1 | C0751372 | Nerve Pain | 1 | CTD_human |
| Tgene | NTRK1 | C0751373 | Paroxysmal Nerve Pain | 1 | CTD_human |
| Tgene | NTRK1 | C0752347 | Lewy Body Disease | 1 | CTD_human |
| Tgene | NTRK1 | C1833921 | Familial medullary thyroid carcinoma | 1 | CTD_human;GENOMICS_ENGLAND;ORPHANET |
| Tgene | NTRK1 | C2350344 | Chronic Lung Injury | 1 | CTD_human |

