| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:VMP1-KDM2A |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: VMP1-KDM2A | FusionPDB ID: 98202 | FusionGDB2.0 ID: 98202 | Hgene | Tgene | Gene symbol | VMP1 | KDM2A | Gene ID | 81671 | 22992 |
| Gene name | vacuole membrane protein 1 | lysine demethylase 2A | |
| Synonyms | EPG3|TANGO5|TMEM49 | CXXC8|FBL11|FBL7|FBXL11|JHDM1A|LILINA | |
| Cytomap | 17q23.1 | 11q13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | vacuole membrane protein 1ectopic P-granules autophagy protein 3 homologtransmembrane protein 49transport and golgi organization 5 homolog | lysine-specific demethylase 2ACXXC-type zinc finger protein 8F-box and leucine-rich repeat protein 11F-box/LRR-repeat protein 11[Histone-H3]-lysine-36 demethylase 1AjmjC domain-containing histone demethylation protein 1Ajumonji C domain-containing h | |
| Modification date | 20200327 | 20200320 | |
| UniProtAcc | . | Q9Y2K7 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000262291, ENST00000536180, ENST00000537567, ENST00000539763, ENST00000545362, ENST00000588617, | ENST00000308783, ENST00000526258, ENST00000530342, ENST00000398645, ENST00000529006, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 34 X 17 X 12=6936 | 28 X 27 X 8=6048 |
| # samples | 38 | 32 | |
| ** MAII score | log2(38/6936*10)=-4.19003257489089 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(32/6048*10)=-4.24031432933371 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: VMP1 [Title/Abstract] AND KDM2A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | VMP1(57851246)-KDM2A(66947550), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | VMP1-KDM2A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. VMP1-KDM2A seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. VMP1-KDM2A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. VMP1-KDM2A seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across VMP1 (5'-gene) Fusion gene breakpoints across VMP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
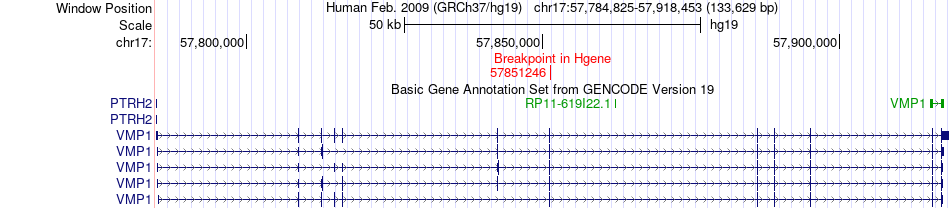 |
 Fusion gene breakpoints across KDM2A (3'-gene) Fusion gene breakpoints across KDM2A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
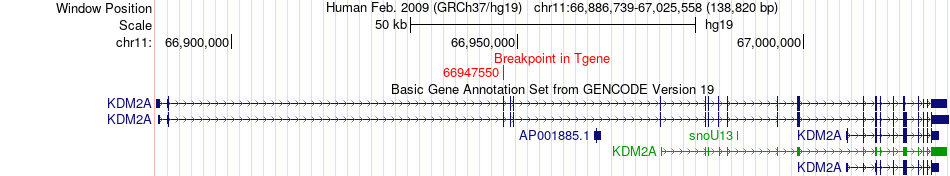 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A1-A0SN-01A | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000262291 | VMP1 | chr17 | 57851246 | + | ENST00000398645 | KDM2A | chr11 | 66947550 | + | 6629 | 1024 | 253 | 3330 | 1025 |
| ENST00000262291 | VMP1 | chr17 | 57851246 | + | ENST00000529006 | KDM2A | chr11 | 66947550 | + | 7503 | 1024 | 253 | 4470 | 1405 |
| ENST00000537567 | VMP1 | chr17 | 57851246 | + | ENST00000398645 | KDM2A | chr11 | 66947550 | + | 6244 | 639 | 327 | 2945 | 872 |
| ENST00000537567 | VMP1 | chr17 | 57851246 | + | ENST00000529006 | KDM2A | chr11 | 66947550 | + | 7118 | 639 | 327 | 4085 | 1252 |
| ENST00000539763 | VMP1 | chr17 | 57851246 | + | ENST00000398645 | KDM2A | chr11 | 66947550 | + | 6308 | 703 | 238 | 3009 | 923 |
| ENST00000539763 | VMP1 | chr17 | 57851246 | + | ENST00000529006 | KDM2A | chr11 | 66947550 | + | 7182 | 703 | 238 | 4149 | 1303 |
| ENST00000536180 | VMP1 | chr17 | 57851246 | + | ENST00000398645 | KDM2A | chr11 | 66947550 | + | 6338 | 733 | 310 | 3039 | 909 |
| ENST00000536180 | VMP1 | chr17 | 57851246 | + | ENST00000529006 | KDM2A | chr11 | 66947550 | + | 7212 | 733 | 310 | 4179 | 1289 |
| ENST00000545362 | VMP1 | chr17 | 57851246 | + | ENST00000398645 | KDM2A | chr11 | 66947550 | + | 6204 | 599 | 41 | 2905 | 954 |
| ENST00000545362 | VMP1 | chr17 | 57851246 | + | ENST00000529006 | KDM2A | chr11 | 66947550 | + | 7078 | 599 | 41 | 4045 | 1334 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000262291 | ENST00000398645 | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + | 0.000649443 | 0.9993506 |
| ENST00000262291 | ENST00000529006 | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + | 0.000423605 | 0.9995764 |
| ENST00000537567 | ENST00000398645 | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + | 0.000914838 | 0.9990852 |
| ENST00000537567 | ENST00000529006 | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + | 0.000432316 | 0.9995677 |
| ENST00000539763 | ENST00000398645 | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + | 0.001262589 | 0.9987374 |
| ENST00000539763 | ENST00000529006 | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + | 0.000561772 | 0.9994382 |
| ENST00000536180 | ENST00000398645 | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + | 0.000777999 | 0.99922204 |
| ENST00000536180 | ENST00000529006 | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + | 0.000440093 | 0.99955994 |
| ENST00000545362 | ENST00000398645 | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + | 0.000812703 | 0.9991873 |
| ENST00000545362 | ENST00000529006 | VMP1 | chr17 | 57851246 | + | KDM2A | chr11 | 66947550 | + | 0.000311457 | 0.9996885 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >98202_98202_1_VMP1-KDM2A_VMP1_chr17_57851246_ENST00000262291_KDM2A_chr11_66947550_ENST00000398645_length(amino acids)=1025AA_BP=257 MRSGAAGVGAAPQELLIYEMAENGKNCDQRRVAMNKEHHNGNFTDPSSVNEKKRREREERQNIVLWRQPLITLQYFSLEILVILKEWTSK LWHRQSIVVSFLLLLAVLIATYYVEGVHQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFPEPP YPDQIICPDEEGTEGTISLWSIISKVRIEACMWGIGTAIGELPPYFMARAARLSGAEPDDEEYQEFEEMLEHAESAQRGTMRRRYEDDGI SDDEIEGKRTFDLEEKLHTNKYNANFVTFMEGKDFNVEYIQRGGLRDPLIFKNSDGLGIKMPDPDFTVNDVKMCVGSRRMVDVMDVNTQK GIEMTMAQWTRYYETPEEEREKLYNVISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKESQTESTNAILEMQYPKVQKYCLMSVRGC YTDFHVDFGGTSVWYHIHQGGKVFWLIPPTAHNLELYENWLLSGKQGDIFLGDRVSDCQRIELKQGYTFVIPSGWIHAVYTPTDTLVFGG NFLHSFNIPMQLKIYNIEDRTRVPNKFRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLSMDLELNGLESGNGDEEAVDREPRRLSS RRSVLTSPVANGVNLDYDGLGKTCRSLPSLKKTLAGDSSSDCSRGSHNGQVWDPQCAPRKDRQVHLTHFELEGLRCLVDKLESLPLHKKC VPTGIEDEDALIADVKILLEELANSDPKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKPHTMKPAPRLTPVRPAAASPIVSGARRR RVRCRKCKACVQGECGVCHYCRDMKKFGGPGRMKQSCVLRQCLAPRLPHSVTCSLCGEVDQNEETQDFEKKLMECCICNEIVHPGCLQMD GEGLLNEELPNCWECPKCYQEDSSEKAQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPTSMLQLIHDPVSPRGMVTRSSPGAGPSD -------------------------------------------------------------- >98202_98202_2_VMP1-KDM2A_VMP1_chr17_57851246_ENST00000262291_KDM2A_chr11_66947550_ENST00000529006_length(amino acids)=1405AA_BP=257 MRSGAAGVGAAPQELLIYEMAENGKNCDQRRVAMNKEHHNGNFTDPSSVNEKKRREREERQNIVLWRQPLITLQYFSLEILVILKEWTSK LWHRQSIVVSFLLLLAVLIATYYVEGVHQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFPEPP YPDQIICPDEEGTEGTISLWSIISKVRIEACMWGIGTAIGELPPYFMARAARLSGAEPDDEEYQEFEEMLEHAESAQRGTMRRRYEDDGI SDDEIEGKRTFDLEEKLHTNKYNANFVTFMEGKDFNVEYIQRGGLRDPLIFKNSDGLGIKMPDPDFTVNDVKMCVGSRRMVDVMDVNTQK GIEMTMAQWTRYYETPEEEREKLYNVISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKESQTESTNAILEMQYPKVQKYCLMSVRGC YTDFHVDFGGTSVWYHIHQGGKVFWLIPPTAHNLELYENWLLSGKQGDIFLGDRVSDCQRIELKQGYTFVIPSGWIHAVYTPTDTLVFGG NFLHSFNIPMQLKIYNIEDRTRVPNKFRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLSMDLELNGLESGNGDEEAVDREPRRLSS RRSVLTSPVANGVNLDYDGLGKTCRSLPSLKKTLAGDSSSDCSRGSHNGQVWDPQCAPRKDRQVHLTHFELEGLRCLVDKLESLPLHKKC VPTGIEDEDALIADVKILLEELANSDPKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKPHTMKPAPRLTPVRPAAASPIVSGARRR RVRCRKCKACVQGECGVCHYCRDMKKFGGPGRMKQSCVLRQCLAPRLPHSVTCSLCGEVDQNEETQDFEKKLMECCICNEIVHPGCLQMD GEGLLNEELPNCWECPKCYQEDSSEKAQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPTSMLQLIHDPVSPRGMVTRSSPGAGPSD HHSASRDERFKRRQLLRLQATERTMVREKENNPSGKKELSEVEKAKIRGSYLTVTLQRPTKELHGTSIVPKLQAITASSANLRHSPRVLV QHCPARTPQRGDEEGLGGEEEEEEEEEEEDDSAEEGGAARLNGRGSWAQDGDESWMQREVWMSVFRYLSRRELCECMRVCKTWYKWCCDK RLWTKIDLSRCKAIVPQALSGIIKRQPVSLDLSWTNISKKQLTWLVNRLPGLKDLLLAGCSWSAVSALSTSSCPLLRTLDLRWAVGIKDP QIRDLLTPPADKPGQDNRSKLRNMTDFRLAGLDITDATLRLIIRHMPLLSRLDLSHCSHLTDQSSNLLTAVGSSTRYSLTELNMAGCNKL -------------------------------------------------------------- >98202_98202_3_VMP1-KDM2A_VMP1_chr17_57851246_ENST00000536180_KDM2A_chr11_66947550_ENST00000398645_length(amino acids)=909AA_BP=141 MDLKYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFPEPPYPDQIICPDEEGTEGTISLWSIISKV RIEACMWGIGTAIGELPPYFMARAARLSGAEPDDEEYQEFEEMLEHAESAQRGTMRRRYEDDGISDDEIEGKRTFDLEEKLHTNKYNANF VTFMEGKDFNVEYIQRGGLRDPLIFKNSDGLGIKMPDPDFTVNDVKMCVGSRRMVDVMDVNTQKGIEMTMAQWTRYYETPEEEREKLYNV ISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKESQTESTNAILEMQYPKVQKYCLMSVRGCYTDFHVDFGGTSVWYHIHQGGKVFWL IPPTAHNLELYENWLLSGKQGDIFLGDRVSDCQRIELKQGYTFVIPSGWIHAVYTPTDTLVFGGNFLHSFNIPMQLKIYNIEDRTRVPNK FRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLSMDLELNGLESGNGDEEAVDREPRRLSSRRSVLTSPVANGVNLDYDGLGKTCRS LPSLKKTLAGDSSSDCSRGSHNGQVWDPQCAPRKDRQVHLTHFELEGLRCLVDKLESLPLHKKCVPTGIEDEDALIADVKILLEELANSD PKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKPHTMKPAPRLTPVRPAAASPIVSGARRRRVRCRKCKACVQGECGVCHYCRDMKK FGGPGRMKQSCVLRQCLAPRLPHSVTCSLCGEVDQNEETQDFEKKLMECCICNEIVHPGCLQMDGEGLLNEELPNCWECPKCYQEDSSEK AQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPTSMLQLIHDPVSPRGMVTRSSPGAGPSDHHSASRDERFKRRQLLRLQATERTMV -------------------------------------------------------------- >98202_98202_4_VMP1-KDM2A_VMP1_chr17_57851246_ENST00000536180_KDM2A_chr11_66947550_ENST00000529006_length(amino acids)=1289AA_BP=141 MDLKYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFPEPPYPDQIICPDEEGTEGTISLWSIISKV RIEACMWGIGTAIGELPPYFMARAARLSGAEPDDEEYQEFEEMLEHAESAQRGTMRRRYEDDGISDDEIEGKRTFDLEEKLHTNKYNANF VTFMEGKDFNVEYIQRGGLRDPLIFKNSDGLGIKMPDPDFTVNDVKMCVGSRRMVDVMDVNTQKGIEMTMAQWTRYYETPEEEREKLYNV ISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKESQTESTNAILEMQYPKVQKYCLMSVRGCYTDFHVDFGGTSVWYHIHQGGKVFWL IPPTAHNLELYENWLLSGKQGDIFLGDRVSDCQRIELKQGYTFVIPSGWIHAVYTPTDTLVFGGNFLHSFNIPMQLKIYNIEDRTRVPNK FRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLSMDLELNGLESGNGDEEAVDREPRRLSSRRSVLTSPVANGVNLDYDGLGKTCRS LPSLKKTLAGDSSSDCSRGSHNGQVWDPQCAPRKDRQVHLTHFELEGLRCLVDKLESLPLHKKCVPTGIEDEDALIADVKILLEELANSD PKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKPHTMKPAPRLTPVRPAAASPIVSGARRRRVRCRKCKACVQGECGVCHYCRDMKK FGGPGRMKQSCVLRQCLAPRLPHSVTCSLCGEVDQNEETQDFEKKLMECCICNEIVHPGCLQMDGEGLLNEELPNCWECPKCYQEDSSEK AQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPTSMLQLIHDPVSPRGMVTRSSPGAGPSDHHSASRDERFKRRQLLRLQATERTMV REKENNPSGKKELSEVEKAKIRGSYLTVTLQRPTKELHGTSIVPKLQAITASSANLRHSPRVLVQHCPARTPQRGDEEGLGGEEEEEEEE EEEDDSAEEGGAARLNGRGSWAQDGDESWMQREVWMSVFRYLSRRELCECMRVCKTWYKWCCDKRLWTKIDLSRCKAIVPQALSGIIKRQ PVSLDLSWTNISKKQLTWLVNRLPGLKDLLLAGCSWSAVSALSTSSCPLLRTLDLRWAVGIKDPQIRDLLTPPADKPGQDNRSKLRNMTD FRLAGLDITDATLRLIIRHMPLLSRLDLSHCSHLTDQSSNLLTAVGSSTRYSLTELNMAGCNKLTDQTLIYLRRIANVTLIDLRGCKQIT -------------------------------------------------------------- >98202_98202_5_VMP1-KDM2A_VMP1_chr17_57851246_ENST00000537567_KDM2A_chr11_66947550_ENST00000398645_length(amino acids)=872AA_BP=104 MDLKGPHIASVTLAAYECNSVNFPEPPYPDQIICPDEEGTEGTISLWSIISKVRIEACMWGIGTAIGELPPYFMARAARLSGAEPDDEEY QEFEEMLEHAESAQRGTMRRRYEDDGISDDEIEGKRTFDLEEKLHTNKYNANFVTFMEGKDFNVEYIQRGGLRDPLIFKNSDGLGIKMPD PDFTVNDVKMCVGSRRMVDVMDVNTQKGIEMTMAQWTRYYETPEEEREKLYNVISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKES QTESTNAILEMQYPKVQKYCLMSVRGCYTDFHVDFGGTSVWYHIHQGGKVFWLIPPTAHNLELYENWLLSGKQGDIFLGDRVSDCQRIEL KQGYTFVIPSGWIHAVYTPTDTLVFGGNFLHSFNIPMQLKIYNIEDRTRVPNKFRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLS MDLELNGLESGNGDEEAVDREPRRLSSRRSVLTSPVANGVNLDYDGLGKTCRSLPSLKKTLAGDSSSDCSRGSHNGQVWDPQCAPRKDRQ VHLTHFELEGLRCLVDKLESLPLHKKCVPTGIEDEDALIADVKILLEELANSDPKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKP HTMKPAPRLTPVRPAAASPIVSGARRRRVRCRKCKACVQGECGVCHYCRDMKKFGGPGRMKQSCVLRQCLAPRLPHSVTCSLCGEVDQNE ETQDFEKKLMECCICNEIVHPGCLQMDGEGLLNEELPNCWECPKCYQEDSSEKAQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPT -------------------------------------------------------------- >98202_98202_6_VMP1-KDM2A_VMP1_chr17_57851246_ENST00000537567_KDM2A_chr11_66947550_ENST00000529006_length(amino acids)=1252AA_BP=104 MDLKGPHIASVTLAAYECNSVNFPEPPYPDQIICPDEEGTEGTISLWSIISKVRIEACMWGIGTAIGELPPYFMARAARLSGAEPDDEEY QEFEEMLEHAESAQRGTMRRRYEDDGISDDEIEGKRTFDLEEKLHTNKYNANFVTFMEGKDFNVEYIQRGGLRDPLIFKNSDGLGIKMPD PDFTVNDVKMCVGSRRMVDVMDVNTQKGIEMTMAQWTRYYETPEEEREKLYNVISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKES QTESTNAILEMQYPKVQKYCLMSVRGCYTDFHVDFGGTSVWYHIHQGGKVFWLIPPTAHNLELYENWLLSGKQGDIFLGDRVSDCQRIEL KQGYTFVIPSGWIHAVYTPTDTLVFGGNFLHSFNIPMQLKIYNIEDRTRVPNKFRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLS MDLELNGLESGNGDEEAVDREPRRLSSRRSVLTSPVANGVNLDYDGLGKTCRSLPSLKKTLAGDSSSDCSRGSHNGQVWDPQCAPRKDRQ VHLTHFELEGLRCLVDKLESLPLHKKCVPTGIEDEDALIADVKILLEELANSDPKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKP HTMKPAPRLTPVRPAAASPIVSGARRRRVRCRKCKACVQGECGVCHYCRDMKKFGGPGRMKQSCVLRQCLAPRLPHSVTCSLCGEVDQNE ETQDFEKKLMECCICNEIVHPGCLQMDGEGLLNEELPNCWECPKCYQEDSSEKAQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPT SMLQLIHDPVSPRGMVTRSSPGAGPSDHHSASRDERFKRRQLLRLQATERTMVREKENNPSGKKELSEVEKAKIRGSYLTVTLQRPTKEL HGTSIVPKLQAITASSANLRHSPRVLVQHCPARTPQRGDEEGLGGEEEEEEEEEEEDDSAEEGGAARLNGRGSWAQDGDESWMQREVWMS VFRYLSRRELCECMRVCKTWYKWCCDKRLWTKIDLSRCKAIVPQALSGIIKRQPVSLDLSWTNISKKQLTWLVNRLPGLKDLLLAGCSWS AVSALSTSSCPLLRTLDLRWAVGIKDPQIRDLLTPPADKPGQDNRSKLRNMTDFRLAGLDITDATLRLIIRHMPLLSRLDLSHCSHLTDQ -------------------------------------------------------------- >98202_98202_7_VMP1-KDM2A_VMP1_chr17_57851246_ENST00000539763_KDM2A_chr11_66947550_ENST00000398645_length(amino acids)=923AA_BP=155 MLLAVLIATYYVEGVHQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFPEPPYPDQIICPDEEG TEGTISLWSIISKVRIEACMWGIGTAIGELPPYFMARAARLSGAEPDDEEYQEFEEMLEHAESAQRGTMRRRYEDDGISDDEIEGKRTFD LEEKLHTNKYNANFVTFMEGKDFNVEYIQRGGLRDPLIFKNSDGLGIKMPDPDFTVNDVKMCVGSRRMVDVMDVNTQKGIEMTMAQWTRY YETPEEEREKLYNVISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKESQTESTNAILEMQYPKVQKYCLMSVRGCYTDFHVDFGGTS VWYHIHQGGKVFWLIPPTAHNLELYENWLLSGKQGDIFLGDRVSDCQRIELKQGYTFVIPSGWIHAVYTPTDTLVFGGNFLHSFNIPMQL KIYNIEDRTRVPNKFRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLSMDLELNGLESGNGDEEAVDREPRRLSSRRSVLTSPVANG VNLDYDGLGKTCRSLPSLKKTLAGDSSSDCSRGSHNGQVWDPQCAPRKDRQVHLTHFELEGLRCLVDKLESLPLHKKCVPTGIEDEDALI ADVKILLEELANSDPKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKPHTMKPAPRLTPVRPAAASPIVSGARRRRVRCRKCKACVQ GECGVCHYCRDMKKFGGPGRMKQSCVLRQCLAPRLPHSVTCSLCGEVDQNEETQDFEKKLMECCICNEIVHPGCLQMDGEGLLNEELPNC WECPKCYQEDSSEKAQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPTSMLQLIHDPVSPRGMVTRSSPGAGPSDHHSASRDERFKR -------------------------------------------------------------- >98202_98202_8_VMP1-KDM2A_VMP1_chr17_57851246_ENST00000539763_KDM2A_chr11_66947550_ENST00000529006_length(amino acids)=1303AA_BP=155 MLLAVLIATYYVEGVHQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFPEPPYPDQIICPDEEG TEGTISLWSIISKVRIEACMWGIGTAIGELPPYFMARAARLSGAEPDDEEYQEFEEMLEHAESAQRGTMRRRYEDDGISDDEIEGKRTFD LEEKLHTNKYNANFVTFMEGKDFNVEYIQRGGLRDPLIFKNSDGLGIKMPDPDFTVNDVKMCVGSRRMVDVMDVNTQKGIEMTMAQWTRY YETPEEEREKLYNVISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKESQTESTNAILEMQYPKVQKYCLMSVRGCYTDFHVDFGGTS VWYHIHQGGKVFWLIPPTAHNLELYENWLLSGKQGDIFLGDRVSDCQRIELKQGYTFVIPSGWIHAVYTPTDTLVFGGNFLHSFNIPMQL KIYNIEDRTRVPNKFRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLSMDLELNGLESGNGDEEAVDREPRRLSSRRSVLTSPVANG VNLDYDGLGKTCRSLPSLKKTLAGDSSSDCSRGSHNGQVWDPQCAPRKDRQVHLTHFELEGLRCLVDKLESLPLHKKCVPTGIEDEDALI ADVKILLEELANSDPKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKPHTMKPAPRLTPVRPAAASPIVSGARRRRVRCRKCKACVQ GECGVCHYCRDMKKFGGPGRMKQSCVLRQCLAPRLPHSVTCSLCGEVDQNEETQDFEKKLMECCICNEIVHPGCLQMDGEGLLNEELPNC WECPKCYQEDSSEKAQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPTSMLQLIHDPVSPRGMVTRSSPGAGPSDHHSASRDERFKR RQLLRLQATERTMVREKENNPSGKKELSEVEKAKIRGSYLTVTLQRPTKELHGTSIVPKLQAITASSANLRHSPRVLVQHCPARTPQRGD EEGLGGEEEEEEEEEEEDDSAEEGGAARLNGRGSWAQDGDESWMQREVWMSVFRYLSRRELCECMRVCKTWYKWCCDKRLWTKIDLSRCK AIVPQALSGIIKRQPVSLDLSWTNISKKQLTWLVNRLPGLKDLLLAGCSWSAVSALSTSSCPLLRTLDLRWAVGIKDPQIRDLLTPPADK PGQDNRSKLRNMTDFRLAGLDITDATLRLIIRHMPLLSRLDLSHCSHLTDQSSNLLTAVGSSTRYSLTELNMAGCNKLTDQTLIYLRRIA -------------------------------------------------------------- >98202_98202_9_VMP1-KDM2A_VMP1_chr17_57851246_ENST00000545362_KDM2A_chr11_66947550_ENST00000398645_length(amino acids)=954AA_BP=186 MIYEMAENGKNCDQRRVAMNKEHHNGNFTDPSSVNEKKRREREERQNIVLWRQPLITLQYFSLEILVILKEWTSKLWHRQSIVVSFLLLL AVLIATYYVEGVHQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGIGTAIGELPPYFMARAARLSGAEPDDEEYQEFEEMLE HAESAQRGTMRRRYEDDGISDDEIEGKRTFDLEEKLHTNKYNANFVTFMEGKDFNVEYIQRGGLRDPLIFKNSDGLGIKMPDPDFTVNDV KMCVGSRRMVDVMDVNTQKGIEMTMAQWTRYYETPEEEREKLYNVISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKESQTESTNAI LEMQYPKVQKYCLMSVRGCYTDFHVDFGGTSVWYHIHQGGKVFWLIPPTAHNLELYENWLLSGKQGDIFLGDRVSDCQRIELKQGYTFVI PSGWIHAVYTPTDTLVFGGNFLHSFNIPMQLKIYNIEDRTRVPNKFRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLSMDLELNGL ESGNGDEEAVDREPRRLSSRRSVLTSPVANGVNLDYDGLGKTCRSLPSLKKTLAGDSSSDCSRGSHNGQVWDPQCAPRKDRQVHLTHFEL EGLRCLVDKLESLPLHKKCVPTGIEDEDALIADVKILLEELANSDPKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKPHTMKPAPR LTPVRPAAASPIVSGARRRRVRCRKCKACVQGECGVCHYCRDMKKFGGPGRMKQSCVLRQCLAPRLPHSVTCSLCGEVDQNEETQDFEKK LMECCICNEIVHPGCLQMDGEGLLNEELPNCWECPKCYQEDSSEKAQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPTSMLQLIHD -------------------------------------------------------------- >98202_98202_10_VMP1-KDM2A_VMP1_chr17_57851246_ENST00000545362_KDM2A_chr11_66947550_ENST00000529006_length(amino acids)=1334AA_BP=186 MIYEMAENGKNCDQRRVAMNKEHHNGNFTDPSSVNEKKRREREERQNIVLWRQPLITLQYFSLEILVILKEWTSKLWHRQSIVVSFLLLL AVLIATYYVEGVHQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGIGTAIGELPPYFMARAARLSGAEPDDEEYQEFEEMLE HAESAQRGTMRRRYEDDGISDDEIEGKRTFDLEEKLHTNKYNANFVTFMEGKDFNVEYIQRGGLRDPLIFKNSDGLGIKMPDPDFTVNDV KMCVGSRRMVDVMDVNTQKGIEMTMAQWTRYYETPEEEREKLYNVISLEFSHTRLENMVQRPSTVDFIDWVDNMWPRHLKESQTESTNAI LEMQYPKVQKYCLMSVRGCYTDFHVDFGGTSVWYHIHQGGKVFWLIPPTAHNLELYENWLLSGKQGDIFLGDRVSDCQRIELKQGYTFVI PSGWIHAVYTPTDTLVFGGNFLHSFNIPMQLKIYNIEDRTRVPNKFRYPFYYEMCWYVLERYVYCITNRSHLTKEFQKESLSMDLELNGL ESGNGDEEAVDREPRRLSSRRSVLTSPVANGVNLDYDGLGKTCRSLPSLKKTLAGDSSSDCSRGSHNGQVWDPQCAPRKDRQVHLTHFEL EGLRCLVDKLESLPLHKKCVPTGIEDEDALIADVKILLEELANSDPKLALTGVPIVQWPKRDKLKFPTRPKVRVPTIPITKPHTMKPAPR LTPVRPAAASPIVSGARRRRVRCRKCKACVQGECGVCHYCRDMKKFGGPGRMKQSCVLRQCLAPRLPHSVTCSLCGEVDQNEETQDFEKK LMECCICNEIVHPGCLQMDGEGLLNEELPNCWECPKCYQEDSSEKAQKRKMEESDEEAVQAKVLRPLRSCDEPLTPPPHSPTSMLQLIHD PVSPRGMVTRSSPGAGPSDHHSASRDERFKRRQLLRLQATERTMVREKENNPSGKKELSEVEKAKIRGSYLTVTLQRPTKELHGTSIVPK LQAITASSANLRHSPRVLVQHCPARTPQRGDEEGLGGEEEEEEEEEEEDDSAEEGGAARLNGRGSWAQDGDESWMQREVWMSVFRYLSRR ELCECMRVCKTWYKWCCDKRLWTKIDLSRCKAIVPQALSGIIKRQPVSLDLSWTNISKKQLTWLVNRLPGLKDLLLAGCSWSAVSALSTS SCPLLRTLDLRWAVGIKDPQIRDLLTPPADKPGQDNRSKLRNMTDFRLAGLDITDATLRLIIRHMPLLSRLDLSHCSHLTDQSSNLLTAV -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr17:57851246/chr11:66947550) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | KDM2A |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | FUNCTION: Histone demethylase that specifically demethylates 'Lys-36' of histone H3, thereby playing a central role in histone code. Preferentially demethylates dimethylated H3 'Lys-36' residue while it has weak or no activity for mono- and tri-methylated H3 'Lys-36'. May also recognize and bind to some phosphorylated proteins and promote their ubiquitination and degradation. Required to maintain the heterochromatic state. Associates with centromeres and represses transcription of small non-coding RNAs that are encoded by the clusters of satellite repeats at the centromere. Required to sustain centromeric integrity and genomic stability, particularly during mitosis. Regulates circadian gene expression by repressing the transcriptional activator activity of CLOCK-ARNTL/BMAL1 heterodimer and RORA in a catalytically-independent manner (PubMed:26037310). {ECO:0000269|PubMed:16362057, ECO:0000269|PubMed:19001877, ECO:0000269|PubMed:26037310, ECO:0000269|PubMed:28262558}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 2_43 | 238.0 | 407.0 | Topological domain | Cytoplasmic |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 65_77 | 238.0 | 407.0 | Topological domain | Extracellular |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 99_109 | 238.0 | 407.0 | Topological domain | Cytoplasmic |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 110_130 | 238.0 | 407.0 | Transmembrane | Helical |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 44_64 | 238.0 | 407.0 | Transmembrane | Helical |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 78_98 | 238.0 | 407.0 | Transmembrane | Helical |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000308783 | 0 | 10 | 148_316 | 0 | 621.0 | Domain | JmjC | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000308783 | 0 | 10 | 889_936 | 0 | 621.0 | Domain | Note=F-box | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000398645 | 1 | 21 | 148_316 | 14.0 | 1490.3333333333333 | Domain | JmjC | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000398645 | 1 | 21 | 889_936 | 14.0 | 1490.3333333333333 | Domain | Note=F-box | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000529006 | 1 | 21 | 148_316 | 14.0 | 1163.0 | Domain | JmjC | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000529006 | 1 | 21 | 889_936 | 14.0 | 1163.0 | Domain | Note=F-box | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000530342 | 0 | 10 | 148_316 | 0 | 724.0 | Domain | JmjC | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000530342 | 0 | 10 | 889_936 | 0 | 724.0 | Domain | Note=F-box | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000308783 | 0 | 10 | 1048_1073 | 0 | 621.0 | Repeat | Note=LRR 3 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000308783 | 0 | 10 | 1074_1103 | 0 | 621.0 | Repeat | Note=LRR 4 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000308783 | 0 | 10 | 1104_1128 | 0 | 621.0 | Repeat | Note=LRR 5 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000308783 | 0 | 10 | 1129_1156 | 0 | 621.0 | Repeat | Note=LRR 6 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000308783 | 0 | 10 | 961_982 | 0 | 621.0 | Repeat | Note=LRR 1 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000308783 | 0 | 10 | 984_1010 | 0 | 621.0 | Repeat | Note=LRR 2 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000398645 | 1 | 21 | 1048_1073 | 14.0 | 1490.3333333333333 | Repeat | Note=LRR 3 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000398645 | 1 | 21 | 1074_1103 | 14.0 | 1490.3333333333333 | Repeat | Note=LRR 4 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000398645 | 1 | 21 | 1104_1128 | 14.0 | 1490.3333333333333 | Repeat | Note=LRR 5 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000398645 | 1 | 21 | 1129_1156 | 14.0 | 1490.3333333333333 | Repeat | Note=LRR 6 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000398645 | 1 | 21 | 961_982 | 14.0 | 1490.3333333333333 | Repeat | Note=LRR 1 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000398645 | 1 | 21 | 984_1010 | 14.0 | 1490.3333333333333 | Repeat | Note=LRR 2 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000529006 | 1 | 21 | 1048_1073 | 14.0 | 1163.0 | Repeat | Note=LRR 3 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000529006 | 1 | 21 | 1074_1103 | 14.0 | 1163.0 | Repeat | Note=LRR 4 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000529006 | 1 | 21 | 1104_1128 | 14.0 | 1163.0 | Repeat | Note=LRR 5 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000529006 | 1 | 21 | 1129_1156 | 14.0 | 1163.0 | Repeat | Note=LRR 6 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000529006 | 1 | 21 | 961_982 | 14.0 | 1163.0 | Repeat | Note=LRR 1 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000529006 | 1 | 21 | 984_1010 | 14.0 | 1163.0 | Repeat | Note=LRR 2 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000530342 | 0 | 10 | 1048_1073 | 0 | 724.0 | Repeat | Note=LRR 3 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000530342 | 0 | 10 | 1074_1103 | 0 | 724.0 | Repeat | Note=LRR 4 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000530342 | 0 | 10 | 1104_1128 | 0 | 724.0 | Repeat | Note=LRR 5 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000530342 | 0 | 10 | 1129_1156 | 0 | 724.0 | Repeat | Note=LRR 6 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000530342 | 0 | 10 | 961_982 | 0 | 724.0 | Repeat | Note=LRR 1 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000530342 | 0 | 10 | 984_1010 | 0 | 724.0 | Repeat | Note=LRR 2 | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000308783 | 0 | 10 | 564_610 | 0 | 621.0 | Zinc finger | CXXC-type | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000308783 | 0 | 10 | 617_678 | 0 | 621.0 | Zinc finger | PHD-type | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000398645 | 1 | 21 | 564_610 | 14.0 | 1490.3333333333333 | Zinc finger | CXXC-type | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000398645 | 1 | 21 | 617_678 | 14.0 | 1490.3333333333333 | Zinc finger | PHD-type | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000529006 | 1 | 21 | 564_610 | 14.0 | 1163.0 | Zinc finger | CXXC-type | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000529006 | 1 | 21 | 617_678 | 14.0 | 1163.0 | Zinc finger | PHD-type | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000530342 | 0 | 10 | 564_610 | 0 | 724.0 | Zinc finger | CXXC-type | |
| Tgene | KDM2A | chr17:57851246 | chr11:66947550 | ENST00000530342 | 0 | 10 | 617_678 | 0 | 724.0 | Zinc finger | PHD-type |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 173_316 | 238.0 | 407.0 | Region | VTT domain |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 131_250 | 238.0 | 407.0 | Topological domain | Extracellular |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 272_273 | 238.0 | 407.0 | Topological domain | Cytoplasmic |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 295_305 | 238.0 | 407.0 | Topological domain | Extracellular |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 327_363 | 238.0 | 407.0 | Topological domain | Cytoplasmic |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 385_406 | 238.0 | 407.0 | Topological domain | Extracellular |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 251_271 | 238.0 | 407.0 | Transmembrane | Helical |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 274_294 | 238.0 | 407.0 | Transmembrane | Helical |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 306_326 | 238.0 | 407.0 | Transmembrane | Helical |
| Hgene | VMP1 | chr17:57851246 | chr11:66947550 | ENST00000262291 | + | 7 | 12 | 364_384 | 238.0 | 407.0 | Transmembrane | Helical |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| VMP1 | |
| KDM2A |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to VMP1-KDM2A |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to VMP1-KDM2A |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

