| UTHEALTH HOME ABOUT SBMI A-Z WEBMAIL INSIDE THE UNIVERSITY |

|
|||||||
|
Fusion Protein:WNK1-ATP6V1G1 |
Fusion Protein Summary |
 Fusion gene summary Fusion gene summary |
| Fusion partner gene information | Fusion gene name: WNK1-ATP6V1G1 | FusionPDB ID: 99319 | FusionGDB2.0 ID: 99319 | Hgene | Tgene | Gene symbol | WNK1 | ATP6V1G1 | Gene ID | 65125 | 9550 |
| Gene name | WNK lysine deficient protein kinase 1 | ATPase H+ transporting V1 subunit G1 | |
| Synonyms | HSAN2|HSN2|KDP|PPP1R167|PRKWNK1|PSK|p65 | ATP6G|ATP6G1|ATP6GL|ATP6J|Vma10 | |
| Cytomap | 12p13.33 | 9q32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | serine/threonine-protein kinase WNK1WNK lysine deficient protein kinase 1 isoformerythrocyte 65 kDa proteinprostate-derived sterile 20-like kinaseprotein kinase with no lysine 1protein phosphatase 1, regulatory subunit 167serine/threonine-protein ki | V-type proton ATPase subunit G 1ATPase, H+ transporting, lysosomal (vacuolar proton pump), member JATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1V-ATPase 13 kDa subunit 1V-ATPase subunit G 1vacuolar ATP synthase subunit M16vacuolar H(+)-ATP | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | . | O75348 | |
| Ensembl transtripts involved in fusion gene | ENST ids | ENST00000315939, ENST00000340908, ENST00000530271, ENST00000535572, ENST00000537687, ENST00000447667, ENST00000540360, ENST00000574564, | ENST00000473413, ENST00000374050, |
| Fusion gene scores for assessment (based on all fusion genes of FusionGDB 2.0) | * DoF score | 23 X 19 X 15=6555 | 4 X 4 X 2=32 |
| # samples | 37 | 4 | |
| ** MAII score | log2(37/6555*10)=-4.14699860459281 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/32*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context (manual curation of fusion genes in FusionPDB) | PubMed: WNK1 [Title/Abstract] AND ATP6V1G1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint (based on all fusion genes of FusionGDB 2.0) | WNK1(998389)-ATP6V1G1(117359849), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | WNK1-ATP6V1G1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. WNK1-ATP6V1G1 seems lost the major protein functional domain in Hgene partner, which is a CGC by not retaining the major functional domain in the partially deleted in-frame ORF. WNK1-ATP6V1G1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. WNK1-ATP6V1G1 seems lost the major protein functional domain in Hgene partner, which is a essential gene by not retaining the major functional domain in the partially deleted in-frame ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | WNK1 | GO:0006468 | protein phosphorylation | 10660600 |
| Hgene | WNK1 | GO:0010923 | negative regulation of phosphatase activity | 19389623 |
| Hgene | WNK1 | GO:0023016 | signal transduction by trans-phosphorylation | 16669787 |
| Hgene | WNK1 | GO:0035556 | intracellular signal transduction | 10660600 |
 Fusion gene breakpoints across WNK1 (5'-gene) Fusion gene breakpoints across WNK1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
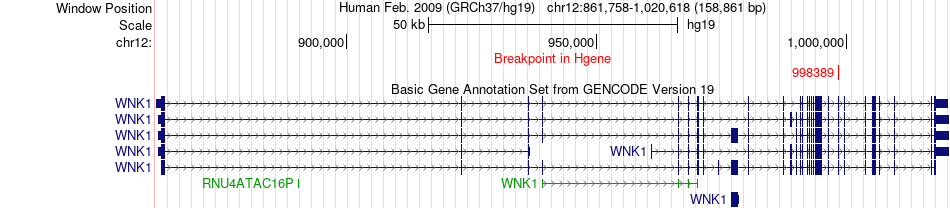 |
 Fusion gene breakpoints across ATP6V1G1 (3'-gene) Fusion gene breakpoints across ATP6V1G1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
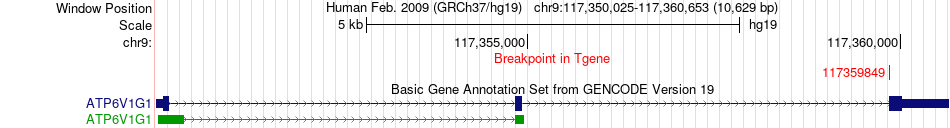 |
Top |
Fusion Gene Sample Information |
 Fusion gene information from FusionGDB2.0. Fusion gene information from FusionGDB2.0. |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A3UA | WNK1 | chr12 | 998389 | + | ATP6V1G1 | chr9 | 117359849 | + |
Top |
Fusion ORF Analysis |
 Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. Fusion information from ORFfinder translation from full-length transcript sequence from FusionPDB. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000535572 | WNK1 | chr12 | 998389 | + | ENST00000374050 | ATP6V1G1 | chr9 | 117359849 | + | 6484 | 5680 | 826 | 5853 | 1675 |
| ENST00000537687 | WNK1 | chr12 | 998389 | + | ENST00000374050 | ATP6V1G1 | chr9 | 117359849 | + | 7675 | 6871 | 496 | 7044 | 2182 |
| ENST00000315939 | WNK1 | chr12 | 998389 | + | ENST00000374050 | ATP6V1G1 | chr9 | 117359849 | + | 6895 | 6091 | 496 | 6264 | 1922 |
| ENST00000530271 | WNK1 | chr12 | 998389 | + | ENST00000374050 | ATP6V1G1 | chr9 | 117359849 | + | 7746 | 6942 | 0 | 7115 | 2371 |
| ENST00000340908 | WNK1 | chr12 | 998389 | + | ENST00000374050 | ATP6V1G1 | chr9 | 117359849 | + | 5031 | 4227 | 0 | 4400 | 1466 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000535572 | ENST00000374050 | WNK1 | chr12 | 998389 | + | ATP6V1G1 | chr9 | 117359849 | + | 0.004286539 | 0.9957135 |
| ENST00000537687 | ENST00000374050 | WNK1 | chr12 | 998389 | + | ATP6V1G1 | chr9 | 117359849 | + | 0.001409968 | 0.99859005 |
| ENST00000315939 | ENST00000374050 | WNK1 | chr12 | 998389 | + | ATP6V1G1 | chr9 | 117359849 | + | 0.001882231 | 0.9981178 |
| ENST00000530271 | ENST00000374050 | WNK1 | chr12 | 998389 | + | ATP6V1G1 | chr9 | 117359849 | + | 0.000868997 | 0.999131 |
| ENST00000340908 | ENST00000374050 | WNK1 | chr12 | 998389 | + | ATP6V1G1 | chr9 | 117359849 | + | 0.000883582 | 0.99911636 |
Top |
Fusion Amino Acid Sequences |
 For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. For individual full-length fusion transcript sequence from FusionPDB, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >FusionGDB ID_FusionGDB isoform ID_FGname_Hgene_Hchr_Hbp_Henst_Tgene_Tchr_Tbp_Tenst_length(fusion AA) seq_BP >99319_99319_1_WNK1-ATP6V1G1_WNK1_chr12_998389_ENST00000315939_ATP6V1G1_chr9_117359849_ENST00000374050_length(amino acids)=1922AA_BP=1387 MSVLSEASSGSARTRPAAVPCRPLRGRLGPSRPFRSRIRARSPLSSEPTMSGGAAEKQSSTPGSLFLSPPAPAPKNGSSSDSSVGEKLGA AAADAVTGRTEEYRRRRHTMDKDSRGAAATTTTTEHRFFRRSVICDSNATALELPGLPLSLPQPSIPAAVPQSAPPEPHREETVTATATS QVAQQPPAAAAPGEQAVAGPAPSTVPSSTSKDRPVSQPSLVGSKEEPPPARSGSGGGSAKEPQEERSQQQDDIEELETKAVGMSNDGRFL KFDIEIGRGSFKTVYKGLDTETTVEVAWCELQDRKLTKSERQRFKEEAEMLKGLQHPNIVRFYDSWESTVKGKKCIVLVTELMTSGTLKT YLKRFKVMKIKVLRSWCRQILKGLQFLHTRTPPIIHRDLKCDNIFITGPTGSVKIGDLGLATLKRASFAKSVIGTPEFMAPEMYEEKYDE SVDVYAFGMCMLEMATSEYPYSECQNAAQIYRRVTSGVKPASFDKVAIPEVKEIIEGCIRQNKDERYSIKDLLNHAFFQEETGVRVELAE EDDGEKIAIKLWLRIEDIKKLKGKYKDNEAIEFSFDLERDVPEDVAQEMVESGYVCEGDHKTMAKAIKDRVSLIKRKREQRQLVREEQEK KKQEESSLKQQVEQSSASQTGIKQLPSASTGIPTASTTSASVSTQVEPEEPEADQHQQLQYQQPSISVLSDGTVDSGQGSSVFTESRVSS QQTVSYGSQHEQAHSTGTVPGHIPSTVQAQSQPHGVYPPSSVAQGQSQGQPSSSSLTGVSSSQPIQHPQQQQGIQQTAPPQQTVQYSLSQ TSTSSEATTAQPVSQPQAPQVLPQVSAGKQLPVSQPVPTIQGEPQIPVATQPSVVPVHSGAHFLPVGQPLPTPLLPQYPVSQIPISTPHV STAQTGFSSLPITMAAGITQPLLTLASSATTAAIPGVSTVVPSQLPTLLQPVTQLPSQVHPQLLQPAVQSMGIPANLGQAAEVPLSSGDV LYQGFPPRLPPQYPGDSNIAPSSNVASVCIHSTVLSPPMPTEVLATPGYFPTVVQPYVESNLLVPMGGVGGQVQVSQPGGSLAQAPTTSS QQAVLESTQGVSQVAPAEPVAVAQTQATQPTTLASSVDSAHSDVASGMSDGNENVPSSSGRHEGRTTKRHYRKSVRSRSRHEKTSRPKLR ILNVSNKGDRVVECQLETHNRKMVTFKFDLDGDNPEEIATIMVNNDFILAIERESFVDQVREIIEKADEMLSEDVSVEPEGDQGLESLQG KDDYGFSGSQKLEGEFKQPIPASSMPQQIGIPTSSLTQVVHSAGRRFIVSPVPESRLRESKVFPSEITDTVAASTAQSPGMNLSHSASSL SLQQAFSELRRAQMTEGPNTAPPNFSHTGPTFPVVPPFLSSIAGVPTTAAATAPVPATSSPPNDISTSVIQSEVTVPTEEGIAGVATSTG VVTSGGLPIPPVSESPVLSSVVSSITIPAVVSISTTSPSLQVPTSTSEIVVSSTALYPSVTVSATSASAGGSTATPGPKPPAVVSQQAAG STTVGATLTSVSTTTSFPSTASQLCIQLSSSTSTPTLAETVVVSAHSLDKTSHSSTTGLAFSLSAPSSSSSPGAGVSSYISQPGGLHPLV IPSVIASTPILPQAAGPTSTPLLPQVPSIPPLVQPVANVPAVQQTLIHSQPQPALLPNQPHTHCPEVDSDTQPKAPGIDDIKTLEEKLRS LFSEHSSSGAQHASVSLETSLVIESTVTPGIPTTAVAPSKLLTSTTSTCLPPTNLPLGTVALPVTPVVTPGQVSTPVSTTTSGVKPGTAP SKPPLTKAPVLPVGTELPAGTLPSEQLPPFPGPSLTQSQQPLEDLDAQLRRTLSPEMITVTSAVGALGSRGSCSTEVEKETQEKMTILQT -------------------------------------------------------------- >99319_99319_2_WNK1-ATP6V1G1_WNK1_chr12_998389_ENST00000340908_ATP6V1G1_chr9_117359849_ENST00000374050_length(amino acids)=1466AA_BP=931 MDIKKKDFCSVFVIINSHCCCCPQKDCINEGVKPASFDKVAIPEVKEIIEGCIRQNKDERYSIKDLLNHAFFQEETGVRVELAEEDDGEK IAIKLWLRIEDIKKLKGKYKDNEAIEFSFDLERDVPEDVAQEMVESGYVCEGDHKTMAKAIKDRVSLIKRKREQRQLVREEQEKKKQEES SLKQQVEQSSASQTGIKQLPSASTGIPTASTTSASVSTQVEPEEPEADQHQQLQYQQPSISVLSDGTVDSGQGSSVFTESRVSSQQTVSY GSQHEQAHSTGTVPGHIPSTVQAQSQPHGVYPPSSVAQGQSQGQPSSSSLTGVSSSQPIQHPQQQQGIQQTAPPQQTVQYSLSQTSTSSE ATTAQPVSQPQAPQVLPQVSAGKQLPVSQPVPTIQGEPQIPVATQPSVVPVHSGAHFLPVGQPLPTPLLPQYPVSQIPISTPHVSTAQTG FSSLPITMAAGITQPLLTLASSATTAAIPGVSTVVPSQLPTLLQPVTQLPSQVHPQLLQPAVQSMGIPANLGQAAEVPLSSGDVLYQGFP PRLPPQYPGDSNIAPSSNVASVCIHSTVLSPPMPTEVLATPGYFPTVVQPYVESNLLVPMGGVGGQVQVSQPGGSLAQAPTTSSQQAVLE STQGVSQVAPAEPVAVAQTQATQPTTLASSVDSAHSDVASGMSDGNENVPSSSGRHEGRTTKRHYRKSVRSRSRHEKTSRPKLRILNVSN KGDRVVECQLETHNRKMVTFKFDLDGDNPEEIATIMVNNDFILAIERESFVDQVREIIEKADEMLSEDVSVEPEGDQGLESLQGKDDYGF SGSQKLEGEFKQPIPASSMPQQIGIPTSSLTQVVHSAGRRFIVSPVPESRLRESKVFPSEITDTVAASTAQSPGMNLSHSASSLSLQQAF SELRRAQMTEGPNTAPPNFSHTGPTFPVVPPFLSSIAGVPTTAAATAPVPATSSPPNDISTSVIQSEVTVPTEEGIAGVATSTGVVTSGG LPIPPVSESPVLSSVVSSITIPAVVSISTTSPSLQVPTSTSEIVVSSTALYPSVTVSATSASAGGSTATPGPKPPAVVSQQAAGSTTVGA TLTSVSTTTSFPSTASQLCIQLSSSTSTPTLAETVVVSAHSLDKTSHSSTTGLAFSLSAPSSSSSPGAGVSSYISQPGGLHPLVIPSVIA STPILPQAAGPTSTPLLPQVPSIPPLVQPVANVPAVQQTLIHSQPQPALLPNQPHTHCPEVDSDTQPKAPGIDDIKTLEEKLRSLFSEHS SSGAQHASVSLETSLVIESTVTPGIPTTAVAPSKLLTSTTSTCLPPTNLPLGTVALPVTPVVTPGQVSTPVSTTTSGVKPGTAPSKPPLT KAPVLPVGTELPAGTLPSEQLPPFPGPSLTQSQQPLEDLDAQLRRTLSPEMITVTSAVGALGSRGSCSTEVEKETQEKMTILQTYFRQNR -------------------------------------------------------------- >99319_99319_3_WNK1-ATP6V1G1_WNK1_chr12_998389_ENST00000530271_ATP6V1G1_chr9_117359849_ENST00000374050_length(amino acids)=2371AA_BP=1836 MSGGAAEKQSSTPGSLFLSPPAPAPKNGSSSDSSVGEKLGAAAADAVTGRTEEYRRRRHTMDKDSRGAAATTTTTEHRFFRRSVICDSNA TALELPGLPLSLPQPSIPAAVPQSAPPEPHREETVTATATSQVAQQPPAAAAPGEQAVAGPAPSTVPSSTSKDRPVSQPSLVGSKEEPPP ARSGSGGGSAKEPQEERSQQQDDIEELETKAVGMSNDGRFLKFDIEIGRGSFKTVYKGLDTETTVEVAWCELQDRKLTKSERQRFKEEAE MLKGLQHPNIVRFYDSWESTVKGKKCIVLVTELMTSGTLKTYLKRFKVMKIKVLRSWCRQILKGLQFLHTRTPPIIHRDLKCDNIFITGP TGSVKIGDLGLATLKRASFAKSVIGTPEFMAPEMYEEKYDESVDVYAFGMCMLEMATSEYPYSECQNAAQIYRRVTSGVKPASFDKVAIP EVKEIIEGCIRQNKDERYSIKDLLNHAFFQEETGVRVELAEEDDGEKIAIKLWLRIEDIKKLKGKYKDNEAIEFSFDLERDVPEDVAQEM VESGYVCEGDHKTMAKAIKDRVSLIKRKREQRQLVREEQEKKKQEESSLKQQVEQSSASQTGIKQLPSASTGIPTASTTSASVSTQVEPE EPEADQHQQLQYQQPSISVLSDGTVDSGQGSSVFTESRVSSQQTVSYGSQHEQAHSTGTVPGHIPSTVQAQSQPHGVYPPSSVPRRGRSM SVCVPIFLLLPLCPASLPVLFHPTASTVCTSFSFPPPDCPEETFAEKLSKALESVLPMHSASQRKHRRSSLPSLFVSTPQSMAHPCGGTP TYPESQIFFPTIHERPVSFSPPPTCPPKVAISQRRKSTSFLEAQTHHFQPLLRTVGQSLLPPGGSPTNWTPEAVVMLGTTASRVTGESCE IQVHPMFEPSQVYSDYRPGLVLPEEAHYFIPQEAVYVAGVHYQARVAEQYEGIPYNSSVLSSPMKQIPEQKPVQGGPTSSSVFEFPSGQA FLVGHLQNLRLDSGLGPGSPLSSISAPISTDATRLKFHPVFVPHSAPAVLTHNNESRSNCVFEFHVHTPSSSSGEGGGILPQRVYRNRQV AVDLNQEELPPQSVGLHGYLQPVTEEKHNYHAPELTVSVVEPIGQNWPIGSPEYSSDSSQITSSDPSDFQSPPPTGGAAAPFGSDVSMPF IHLPQTVLQESPLFFCFPQGTTSQQVLTASFSSGGSALHPQAQGQSQGQPSSSSLTGVSSSQPIQHPQQQQGIQQTAPPQQTVQYSLSQT STSSEATTAQPVSQPQAPQVLPQVSAGKQLPVSQPVPTIQGEPQIPVATQPSVVPVHSGAHFLPVGQPLPTPLLPQYPVSQIPISTPHVS TAQTGFSSLPITMAAGITQPLLTLASSATTAAIPGVSTVVPSQLPTLLQPVTQLPSQVHPQLLQPAVQSMGIPANLGQAAEVPLSSGDVL YQGFPPRLPPQYPGDSNIAPSSNVASVCIHSTVLSPPMPTEVLATPGYFPTVVQPYVESNLLVPMGGVGGQVQVSQPGGSLAQAPTTSSQ QAVLESTQGVSQVAPAEPVAVAQTQATQPTTLASSVDSAHSDVASGMSDGNENVPSSSGRHEGRTTKRHYRKSVRSRSRHEKTSRPKLRI LNVSNKGDRVVECQLETHNRKMVTFKFDLDGDNPEEIATIMVNNDFILAIERESFVDQVREIIEKADEMLSEDVSVEPEGDQGLESLQGK DDYGFSGSQKLEGEFKQPIPASSMPQQIGIPTSSLTQVVHSAGRRFIVSPVPESRLRESKVFPSEITDTVAASTAQSPGMNLSHSASSLS LQQAFSELRRAQMTEGPNTAPPNFSHTGPTFPVVPPFLSSIAGVPTTAAATAPVPATSSPPNDISTSVIQSEVTVPTEEGIAGVATSTGV VTSGGLPIPPVSESPVLSSVVSSITIPAVVSISTTSPSLQVPTSTSEIVVSSTALYPSVTVSATSASAGGSTATPGPKPPAVVSQQAAGS TTVGATLTSVSTTTSFPSTASQLCIQLSSSTSTPTLAETVVVSAHSLDKTSHSSTTGLAFSLSAPSSSSSPGAGVSSYISQPGGLHPLVI PSVIASTPILPQAAGPTSTPLLPQVPSIPPLVQPVANVPAVQQTLIHSQPQPALLPNQPHTHCPEVDSDTQPKAPGIDDIKTLEEKLRSL FSEHSSSGAQHASVSLETSLVIESTVTPGIPTTAVAPSKLLTSTTSTCLPPTNLPLGTVALPVTPVVTPGQVSTPVSTTTSGVKPGTAPS KPPLTKAPVLPVGTELPAGTLPSEQLPPFPGPSLTQSQQPLEDLDAQLRRTLSPEMITVTSAVGALGSRGSCSTEVEKETQEKMTILQTY -------------------------------------------------------------- >99319_99319_4_WNK1-ATP6V1G1_WNK1_chr12_998389_ENST00000535572_ATP6V1G1_chr9_117359849_ENST00000374050_length(amino acids)=1675AA_BP=1140 MSVLSEASSGSARTRPAAVPCRPLRGRLGPSRPFRSRIRARSPLSSEPTMSGGAAEKQSSTPGSLFLSPPAPAPKNGSSSDSSVGEKLGA AAADAVTGRTEEYRRRRHTMDKDSRGAAATTTTTEHRFFRRSVICDSNATALELPGLPLSLPQPSIPAAVPQSAPPEPHREETVTATATS QVAQQPPAAAAPGEQAVAGPAPSTVPSSTSKDRPVSQPSLVGSKEEPPPARSGSGGGSAKEPQEERSQQQDDIEELETKAVGMSNDGRFL KFDIEIGRGSFKTVYKGLDTETTVEVAWCELQDRKLTKSERQRFKEEAEMLKGLQHPNIVRFYDSWESTVKGKKCIVLVTELMTSGTLKT YLKRFKVMKIKVLRSWCRQILKGLQFLHTRTPPIIHRDLKCDNIFITGPTGSVKIGDLGLATLKRASFAKSVIGTPEFMAPEMYEEKYDE SVDVYAFGMCMLEMATSEYPYSECQNAAQIYRRVTSGVKPASFDKVAIPEVKEIIEGCIRQNKDERYSIKDLLNHAFFQEETGVRVELAE EDDGEKIAIKLWLRIEDIKKLKGKYKDNEAIEFSFDLERDVPEDVAQEMVESGYVCEGDHKTMAKAIKDRVSLIKRKREQRQLVREEQEK KKQEESSLKQQVEQSSASQTGIKQLPSASTGIPTASTTSASVSTQVEPEEPEADQHQQLQYQQPSISVLSDGTVDSGQGSSVFTESRVSS QQTVSYGSQHEQAHSTGTVPGHIPSTVQAQSQPHGVYPPSSVAQGQSQGQPSSSSLTGVSSSQPIQHPQQQGIQQTAPPQQTVQYSLSQT STSSEATTAQPVSQPQAPQVLPQVSAGKQSTQGVSQVAPAEPVAVAQTQATQPTTLASSVDSAHSDVASGMSDGNENVPSSSGRHEGRTT KRHYRKSVRSRSRHEKTSRPKLRILNVSNKGDRVVECQLETHNRKMVTFKFDLDGDNPEEIATIMVNNDFILAIERESFVDQVREIIEKA DEMLSEDVSVEPEGDQGLESLQGKDDYGFSGSQKLEGEFKQPIPASSMPQQIGIPTSSLTQVVHSAGRRFIVSPVPESRLRESKVFPSEI TDTVAASTAQSPGMNLSHSASSLSLQQAFSELRRAQMTEGPNTAPPNFSHTGPTFPVVPPFLSSIAGVPTTAAATAPVPATSSPPNDIST SVIQSEVTVPTEEGIAGVATSTGVVTSGGLPIPPVSESPVLSSVVSSITIPAVVSISTTSPSLQVPTSTSEIVVSSTALYPSVTVSATSA SAGGSTATPGPKPPAVVSQQAAGSTTVGATLTSVSTTTSFPSTASQLCIQLSSSTSTPTLAETVVVSAHSLDKTSHSSTTGLAFSLSAPS SSSSPGAGVSSYISQPGGLHPLVIPSVIASTPILPQAAGPTSTPLLPQVPSIPPLVQPVANVPAVQQTLIHSQPQPALLPNQPHTHCPEV DSDTQPKAPGIDDIKTLEEKLRSLFSEHSSSGAQHASVSLETSLVIESTVTPGIPTTAVAPSKLLTSTTSTCLPPTNLPLGTVALPVTPV VTPGQVSTPVSTTTSGVKPGTAPSKPPLTKAPVLPVGTELPAGTLPSEQLPPFPGPSLTQSQQPLEDLDAQLRRTLSPEMITVTSAVGAL -------------------------------------------------------------- >99319_99319_5_WNK1-ATP6V1G1_WNK1_chr12_998389_ENST00000537687_ATP6V1G1_chr9_117359849_ENST00000374050_length(amino acids)=2182AA_BP=1647 MSVLSEASSGSARTRPAAVPCRPLRGRLGPSRPFRSRIRARSPLSSEPTMSGGAAEKQSSTPGSLFLSPPAPAPKNGSSSDSSVGEKLGA AAADAVTGRTEEYRRRRHTMDKDSRGAAATTTTTEHRFFRRSVICDSNATALELPGLPLSLPQPSIPAAVPQSAPPEPHREETVTATATS QVAQQPPAAAAPGEQAVAGPAPSTVPSSTSKDRPVSQPSLVGSKEEPPPARSGSGGGSAKEPQEERSQQQDDIEELETKAVGMSNDGRFL KFDIEIGRGSFKTVYKGLDTETTVEVAWCELQDRKLTKSERQRFKEEAEMLKGLQHPNIVRFYDSWESTVKGKKCIVLVTELMTSGTLKT YLKRFKVMKIKVLRSWCRQILKGLQFLHTRTPPIIHRDLKCDNIFITGPTGSVKIGDLGLATLKRASFAKSVIGTPEFMAPEMYEEKYDE SVDVYAFGMCMLEMATSEYPYSECQNAAQIYRRVTSGVKPASFDKVAIPEVKEIIEGCIRQNKDERYSIKDLLNHAFFQEETGVRVELAE EDDGEKIAIKLWLRIEDIKKLKGKYKDNEAIEFSFDLERDVPEDVAQEMVESGYVCEGDHKTMAKAIKDRVSLIKRKREQRQLVREEQEK KKQEESSLKQQVEQSSASQTGIKQLPSASTGIPTASTTSASVSTQVEPEEPEADQHQQLQYQQPSISVLSDGTVDSGQGSSVFTESRVSS QQTVSYGSQHEQAHSTGTVPGHIPSTVQAQSQPHGVYPPSSVPQSMAHPCGGTPTYPESQIFFPTIHERPVSFSPPPTCPPKVAISQRRK STSFLEAQTHHFQPLLRTVGQSLLPPGGSPTNWTPEAVVMLGTTASRVTGESCEIQVHPMFEPSQVYSDYRPGLVLPEEAHYFIPQEAVY VAGVHYQARVAEQYEGIPYNSSVLSSPMKQIPEQKPVQGGPTSSSVFEFPSGQAFLVGHLQNLRLDSGLGPGSPLSSISAPISTDATRLK FHPVFVPHSAPAVLTHNNESRSNCVFEFHVHTPSSSSGEGGGILPQRVYRNRQVAVDLNQEELPPQSVGLHGYLQPVTEEKHNYHAPELT VSVVEPIGQNWPIGSPEYSSDSSQITSSDPSDFQSPPPTGGAAAPFGSDVSMPFIHLPQTVLQESPLFFCFPQGTTSQQVLTASFSSGGS ALHPQAQGQSQGQPSSSSLTGVSSSQPIQHPQQQQGIQQTAPPQQTVQYSLSQTSTSSEATTAQPVSQPQAPQVLPQVSAGKQGFPPRLP PQYPGDSNIAPSSNVASVCIHSTVLSPPMPTEVLATPGYFPTVVQPYVESNLLVPMGGVGGQVQVSQPGGSLAQAPTTSSQQAVLESTQG VSQVAPAEPVAVAQTQATQPTTLASSVDSAHSDVASGMSDGNENVPSSSGRHEGRTTKRHYRKSVRSRSRHEKTSRPKLRILNVSNKGDR VVECQLETHNRKMVTFKFDLDGDNPEEIATIMVNNDFILAIERESFVDQVREIIEKADEMLSEDVSVEPEGDQGLESLQGKDDYGFSGSQ KLEGEFKQPIPASSMPQQIGIPTSSLTQVVHSAGRRFIVSPVPESRLRESKVFPSEITDTVAASTAQSPGMNLSHSASSLSLQQAFSELR RAQMTEGPNTAPPNFSHTGPTFPVVPPFLSSIAGVPTTAAATAPVPATSSPPNDISTSVIQSEVTVPTEEGIAGVATSTGVVTSGGLPIP PVSESPVLSSVVSSITIPAVVSISTTSPSLQVPTSTSEIVVSSTALYPSVTVSATSASAGGSTATPGPKPPAVVSQQAAGSTTVGATLTS VSTTTSFPSTASQLCIQLSSSTSTPTLAETVVVSAHSLDKTSHSSTTGLAFSLSAPSSSSSPGAGVSSYISQPGGLHPLVIPSVIASTPI LPQAAGPTSTPLLPQVPSIPPLVQPVANVPAVQQTLIHSQPQPALLPNQPHTHCPEVDSDTQPKAPGIDDIKTLEEKLRSLFSEHSSSGA QHASVSLETSLVIESTVTPGIPTTAVAPSKLLTSTTSTCLPPTNLPLGTVALPVTPVVTPGQVSTPVSTTTSGVKPGTAPSKPPLTKAPV LPVGTELPAGTLPSEQLPPFPGPSLTQSQQPLEDLDAQLRRTLSPEMITVTSAVGALGSRGSCSTEVEKETQEKMTILQTYFRQNRDEVL -------------------------------------------------------------- |
Top |
Fusion Protein Functional Features |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:998389/chr9:117359849) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | ATP6V1G1 |
| FUNCTION: Might normally function as a transcriptional repressor. EWS-fusion-proteins (EFPS) may play a role in the tumorigenic process. They may disturb gene expression by mimicking, or interfering with the normal function of CTD-POLII within the transcription initiation complex. They may also contribute to an aberrant activation of the fusion protein target genes. | FUNCTION: Catalytic subunit of the peripheral V1 complex of vacuolar ATPase (V-ATPase). V-ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells. In aerobic conditions, involved in intracellular iron homeostasis, thus triggering the activity of Fe(2+) prolyl hydroxylase (PHD) enzymes, and leading to HIF1A hydroxylation and subsequent proteasomal degradation (PubMed:28296633). {ECO:0000269|PubMed:28296633}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - Retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000315939 | + | 21 | 28 | 221_479 | 1816.0 | 2383.0 | Domain | Protein kinase |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000340908 | + | 18 | 25 | 221_479 | 1409.0 | 1976.0 | Domain | Protein kinase |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000537687 | + | 21 | 28 | 221_479 | 2076.0 | 2643.0 | Domain | Protein kinase |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000315939 | + | 21 | 28 | 301_304 | 1816.0 | 2383.0 | Nucleotide binding | ATP |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000340908 | + | 18 | 25 | 301_304 | 1409.0 | 1976.0 | Nucleotide binding | ATP |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000537687 | + | 21 | 28 | 301_304 | 2076.0 | 2643.0 | Nucleotide binding | ATP |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000315939 | + | 21 | 28 | 488_555 | 1816.0 | 2383.0 | Region | Autoinhibitory domain |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000340908 | + | 18 | 25 | 488_555 | 1409.0 | 1976.0 | Region | Autoinhibitory domain |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000537687 | + | 21 | 28 | 488_555 | 2076.0 | 2643.0 | Region | Autoinhibitory domain |
| - Not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000574564 | + | 1 | 1 | 221_479 | 0 | 435.0 | Domain | Protein kinase |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000574564 | + | 1 | 1 | 301_304 | 0 | 435.0 | Nucleotide binding | ATP |
| Hgene | WNK1 | chr12:998389 | chr9:117359849 | ENST00000574564 | + | 1 | 1 | 488_555 | 0 | 435.0 | Region | Autoinhibitory domain |
Top |
Fusion Protein-Protein Interaction |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type from validated records (BIOGRID-3.4.160) |
| Gene | PPI interactors |
 Protein-protein interactors based on sequence similarity (STRING) Protein-protein interactors based on sequence similarity (STRING) |
| Gene | STRING network |
| WNK1 | |
| ATP6V1G1 |
 - Retained interactions in fusion protein (protein functional feature from UniProt). - Retained interactions in fusion protein (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost interactions due to fusion (protein functional feature from UniProt). - Lost interactions due to fusion (protein functional feature from UniProt). |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs to WNK1-ATP6V1G1 |
 Drugs used for this fusion-positive patient. Drugs used for this fusion-positive patient. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Drug | Source | PMID |
Top |
Related Diseases to WNK1-ATP6V1G1 |
 Diseases that have this fusion gene. Diseases that have this fusion gene. (Manual curation of PubMed, 04-30-2022 + MyCancerGenome) |
| Hgene | Tgene | Disease | Source | PMID |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |

